Abstract
Medulloblastoma (MB) is the most common malignant brain tumor of childhood. Sonic Hedgehog (SHH) signaling drives a minority of MB, correlating with desmoplastic pathology and favorable outcome. The majority, however, arises independently of SHH and displays classic or large cell anaplastic (LCA) pathology and poor prognosis. To identify common signaling abnormalities, we profiled mRNA, demonstrating misexpression of MYCN in the majority of human MB and negligible expression in normal cerebella. We clarified a role in pathogenesis by targeting MYCN (and luciferase) to cerebella of transgenic mice. MYCN-driven MB showed either classic or LCA pathologies, with Shh signaling activated in ∼5% of tumors, demonstrating that MYCN can drive MB independently of Shh. MB arose at high penetrance, consistent with a role for MYCN in initiation. Tumor burden correlated with bioluminescence, with rare metastatic spread to the leptomeninges, suggesting roles for MYCN in both progression and metastasis. Transient pharmacological down-regulation of MYCN led to both clearance and senescence of tumor cells, and improved survival. Targeted expression of MYCN thus contributes to initiation, progression, and maintenance of MB, suggesting a central role for MYCN in pathogenesis.
Keywords: Large cell anaplastic, N-myc, SHH-independent, cerebellum, medulloblastoma, oncogene addiction
Brain tumors are the most common solid tumors of childhood. The embryonal tumor medulloblastoma (MB) is the most frequent childhood brain tumor, with ∼50% overall survival (Davis et al. 1999). Morbidities such as cognitive impairment, psychiatric disorders, endocrine dysfunction, growth retardation, and therapy-induced malignancies are considerable in survivors (Polkinghorn and Tarbell 2007). Risk factors in MB include dissemination (seeding of the cerebrospinal fluid occurs in one-third of patients) and aggressive pathology (Finlay et al. 2007; Polkinghorn and Tarbell 2007). Among pathological subtypes, desmoplastic tumors (∼25%) show round, pale nodules and correlate with favorable outcome. Classic tumors (∼50%) are more uniformly cellular and undifferentiated and suggest intermediate outcome. Large cell anaplastic (LCA) tumors (∼10%) consist of large, angulated cells with marked nuclear atypia and show the poorest outcome (Eberhart and Burger 2003; McManamy et al. 2003).
MB is thought to arise from stem cells or granule progenitor cells of the cerebellum; in the latter case from either sustained proliferation controlled by the Sonic Hedgehog (SHH) pathway, or disruption of signals that cause normal granule neurons to stop dividing (Grimmer and Weiss 2008). The transcription factor Math1 directs granule neuron precursors (GNPs) born in the fourth ventricular zone to migrate and subsequently proliferate in the external granular layer (EGL), a transient, cell-dense structure that lines the surface of the fetal cerebellum (Ben-Arie et al. 1997; Machold and Fishell 2005; Wang et al. 2005). SHH is a morphogen secreted by large Purkinje cells deep to the EGL. SHH and other signals mediate proliferation and migration of GNPs into the cerebellum, where they terminally differentiate and form the internal granular layer (IGL). The initial clues implicating SHH in MB arose from an inherited predisposition syndrome. Patients with Gorlin syndrome were subsequently shown to have germline heterozygosity for PTCH1, a key regulator of SHH signaling (Johnson et al. 1996). Approximately 25% of sporadic MB shows genetic aberrations that drive SHH signaling (Raffel et al. 1997; Eberhart and Burger 2003; Polkinghorn and Tarbell 2007). SHH activation correlates with desmoplastic pathology and favorable clinical outcome.
While signaling in SHH-driven MB has been well characterized, pathways driving SHH-independent tumors remain poorly understood. As some MB express markers normally found in ventricular zone stem cells, these cells could represent cells of origin for some MB tumors (Eberhart 2007). Wnt signaling abnormalities were described recently in ∼15% of MB, corresponding to a minority of tumors with classic pathology and defining a subset of patients with improved outcome (Clifford et al. 2006; Kool et al. 2008; Rogers et al. 2009). Few molecular hallmarks and signaling abnormalities have been identified as drivers for the majority of aggressive tumors, which show classic or LCA pathology. However, several recent reports identified the transcription factor OTX2 to be expressed prominently in classic MB (de Haas et al. 2006; Kool et al. 2008; Adamson et al. 2009).
A number of observations suggest a role for the proto-oncogene MYCN in the pathogenesis of MB. Amplification of MYCN, a relatively rare event in MB, correlates with poor outcome (Pfister et al. 2009). Shh promotes the expression and post-transcriptional stabilization of N-Myc in mice (Kenney et al. 2003; Thomas et al. 2009). Brain-specific deletion of murine N-myc results in cerebellar dysplasia, suggesting that N-myc is critical to proliferation in the developing cerebellum, and that other myc family members cannot compensate for loss of N-myc (Knoepfler et al. 2002). Experiments in genetically engineered mice demonstrate that N-myc is required for MB development (Hatton et al. 2006), suggesting that human tumors could also arise, in part, through misexpression of MYCN (Kenney et al. 2003; Oliver et al. 2003; Hatton et al. 2006; Kessler et al. 2009; Thomas et al. 2009). To date, however, N-myc overexpression (alone or in combination with Gli1, IGF-II, or Bcl-2) has failed to initiate MB in any experimental model (Browd et al. 2006; McCall et al. 2007).
Several genetically engineered models for MB have been described, including models deleted for the Shh pathway antagonist Ptch1 (Goodrich et al. 1997), or activating positive regulators of the Shh pathway (Goodrich et al. 1997; Hallahan et al. 2004; Rao et al. 2004; Uziel et al. 2005; Browd et al. 2006; Mao et al. 2006; Hatton et al. 2008; Schuller et al. 2008; Yang et al. 2008). Two recent studies generated models of LCA MB, either through spontaneous amplification of MYCN in tumors arising through cooperation between MYC and β-catenin (Momota et al. 2008), or by deleting the tumor suppressor genes Rb and p53 (Shakhova et al. 2006). Although loss of p53 accelerates formation of MB in Ptch1-deficient tumors (Wetmore et al. 2001), mutation in p53 is uncommon in human MB (Adesina et al. 1994). Expression of N-myc has not been analyzed in the majority of models, although expression of this gene has been documented in models that have been examined (Lee et al. 2003; Shakhova et al. 2006).
To clarify a role for MYCN in MB, we analyzed mRNA profiles from human cerebella and MB tumors. MYCN mRNA was essentially absent in normal adult cerebella, but was expressed in both fetal cerebella and in the vast majority of MB, suggesting that MYCN was misexpressed in tumors. To determine whether MYCN contributes functionally to the pathogenesis of MB, we targeted its expression to the cerebella of transgenic mice. Transgenic animals developed MB at high penetrance. Tumors showed either classic or LCA pathologies, representative of the majority of human MB. Analysis of the Shh pathway and Math1 levels revealed that a minority of tumors activated Shh and Math1. The majority of murine MB, like their human counterparts, were wild type for p53 and expressed the transcription factor Otx2. Tumors showed a wide range of genomic instability, as revealed by comparative genomic hybridization (CGH) analysis. Both classic and LCA MBs could be transplanted serially, maintaining aggressive growth and histopathology. Down-regulation of Mycn in the model led to both clearance and senescence of tumor cells. These results establish that MYCN contributes to the initiation, progression, and maintenance of MB, and suggest dysregulation of MYCN, observed commonly in human tumors, as a contribution to pathogenesis. Murine MB driven by MYCN models classic and LCA MB tumors, generating a platform to dissect biology and developmental therapeutics in these aggressive subsets of MB.
Results
MYCN is expressed in the majority of human MB
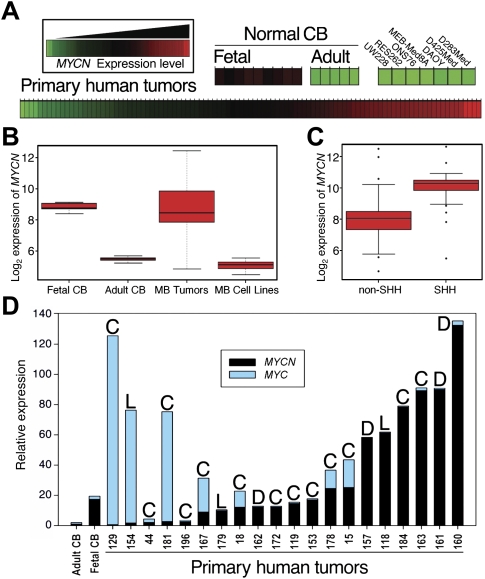
We analyzed mRNA from 14 normal human cerebella (nine fetal and five adult). Expression of MYCN was uniformly high in fetal cerebella and essentially absent in adult cerebella (Fig. 1A,B), consistent with a normal role restricted to cerebellar development (Sawai et al. 1993; Knoepfler et al. 2002; Kenney et al. 2003; Oliver et al. 2003; Hatton et al. 2006). Next, we analyzed 103 primary human MB tumors, demonstrating expression of MYCN in the vast majority. Surprisingly, while all widely used human MB cell lines tested expressed MYC (MYCC), none expressed MYCN (Fig. 1A; Supplemental Fig. 1). The absence of MYCN in D283Med, D425Med, DAOY, MEB-Med8A, ONS76, RES262, and UW228 suggests that these tumor-derived cell lines do not accurately recapitulate an abnormality common to the majority of human MBs.
Figure 1.
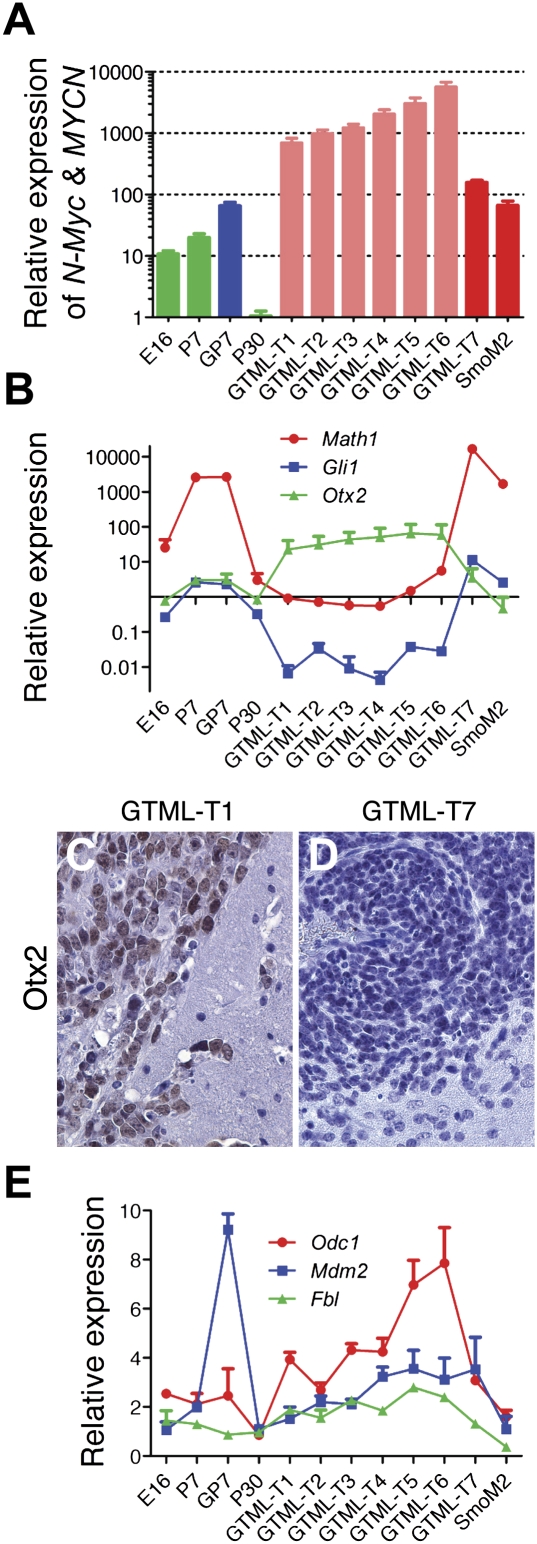
Aberrant expression of MYCN is prominent in human MB. (A) Expression analysis of 103 primary MB tumors (MB Tumors), nine normal fetal cerebella (Fetal CB), five normal adult cerebella (Adult CB), and seven MB cell lines (MB Cell Lines) for MYCN obtained using Affymetrix exon arrays. MYCN was expressed in all normal fetal cerebella and in >90% of primary tumors, but not in normal adult cerebella or in tumor-derived cell lines. (B,C) Results of A in box plot form and quantified for SHH-driven versus SHH-independent tumors. (Center line) Median; (red boxes) upper and lower quartiles; (bars) range of nonoutliers; (circles outside the boxes) outliers. (D) Quantitative real-time PCR analysis in a subset of 20 primary human MB tumors (selected randomly) using normal cerebella as controls. Expression of MYCN was observed in the vast majority of tumors, irrespective of pathology. The highest levels of MYC expression were in tumors with low MYCN expression. (C) Classic; (L) LCA; (D) desmoplastic.
Mean levels of MYCN mRNA in primary tumors were equivalent to levels in fetal cerebella (Fig. 1B). To investigate the correlation of MYCN expression and SHH activation, we also segregated tumors into SHH-driven and SHH-independent groups using criteria described previously (Kool et al. 2008; Northcott et al. 2009). Of 103 primary tumors, 33 showed elevated levels of SHH signaling. On average, MYCN was highest in SHH-driven tumors (Fig. 1C), consistent with MYCN as a target of SHH signaling (Kenney et al. 2003; Oliver et al. 2003; Hatton et al. 2006). Importantly, SHH-independent tumors showed mean levels of MYCN lower than those in SHH-dependent tumors, but higher than levels in adult cerebella.
We next performed quantitative real-time PCR (qRT–PCR) analysis on a subset of tumors (chosen randomly from each tumor subtype, as described in Kool et al. 2008; Northcott et al. 2009), both to verify array results and to correlate levels of MYCN and MYC with histological subtypes (Fig. 1D). This analysis again demonstrated misexpression of MYCN in a majority of tumors. While desmoplastic tumors on average showed the highest level of MYCN, expression of MYCN was not limited to this pathological subset. Three of five tumors with low levels of MYCN expressed high levels of MYC.
MB arise in mice doubly transgenic for Glt1-tTA (glutamate transporter 1-tetracycline transactivator) and tetracycline response element (TRE)-MYCN/luciferase (Luc) (GTML)
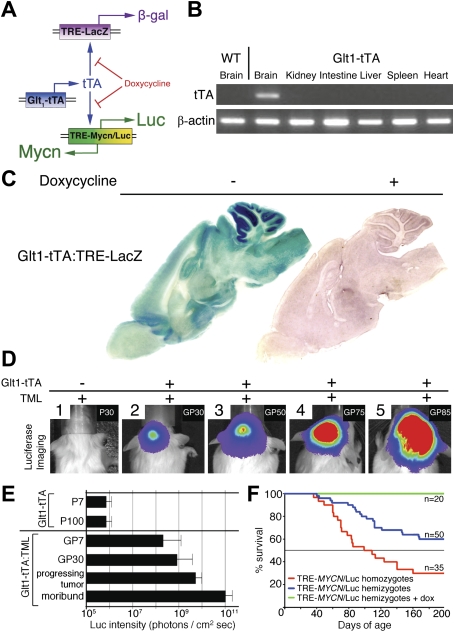
To clarify whether aberrant expression of MYCN could contribute to pathogenesis, we used the Tet system (Gossen and Bujard 1992) to generate transgenic mice expressing both MYCN and firefly Luc from a bidirectional operator (TML) (Fig. 2A). To target expression to cerebellum, we generated mice transgenic for a tTA under control of the mouse Glt1 promoter. Glt1 is expressed in the developing posterior hindbrain from postnatal day 1 (P1) through adulthood in rodents (Furuta et al. 1997). RT–PCR analysis of mouse organs from P14 Glt1-tTA mice (seven founders, two examined with concordant results) showed brain-specific expression of tTA, with undetectable levels in kidney, intestine, liver, spleen, and heart (Fig. 2B). We further characterized expression of tTA in the brain by generating mice transgenic for Glt1-tTA and for LacZ under control of the TRE (TRE-LacZ). β-Galactosidase (β-gal) activity in the resulting Glt1-tTA:TRE-LacZ mice (P60) was evident diffusely, but was higher in cerebellum than in other brain regions (Fig. 2C). Glt1-tTA:TRE-LacZ mice were then treated with doxycycline (dox) in order to monitor inactivation of TRE promoter activity. Mice placed on dox chow for 1 wk (P60) showed substantial reduction of β-gal activity (Fig. 2C).
Figure 2.
Targeting expression of MYCN to the hindbrain gives rise to MB. (A) Expression of tTA protein driven by the murine Glt1 promoter. In the absence of dox, tTA protein drives expression from a promoter element, the TRE. Glt1-tTA mice were mated to mice carrying either TRE-LacZ or TML transgenes. (B) RT–PCR analysis of organs indicated from P14 Glt1-tTA mice showed brain-specific expression of tTA with undetectable levels of tTA mRNA in kidney, intestine, liver, spleen, and heart. (C) Glt1-tTA:TRE-LacZ mice show strong β-gal staining in the brain, especially cerebellum. Mice fed dox chow at P60 showed absence of LacZ activity. (D, panels 1–5) Luc imaging allows robust detection of tumors in Glt1-tTA:TRE-MYCN/Luc (GTML) mice. A single, representative GTML mouse is depicted in panels 2–5. (E) Quantification of luminescence intensities. In GTML P7 mice (GP7), signals from the hindbrain (before tumors arise) were more than two logs greater than the background levels observed in Glt1-tTA mice (P7). At 30 d (GP30), 75 d (progressing tumor), and 100 d (moribund) after birth, progression in GTML mice was evidenced over approximately three logs of dynamic range before sacrifice due to tumor burden. (F) Survival data for GTML mice demonstrate a transgene dose effect. Tumors arising in mice homozygous for TML (doubling the MYCN copy number, red line) showed shortened latency and increased penetrance relative to hemizygous mice (blue line). GTML mice treated with dox from P1 to P30 (green line) survived until sacrifice at P200.
We then generated mice that expressed both MYCN and Luc from the bidirectional TRE (TML). Six founders were generated in an FVB/NJ background. To determine whether MYCN could initiate MB, we analyzed mice doubly transgenic for GTML. All TML lines showed brain-specific expression of Luc when crossed to mice transgenic for Glt1-tTA, verified by in vivo imaging (data not shown), and consistent with the basal low level of Glt1 observed diffusely in the brains of mice transgenic for Glt1-tTA:TRE-LacZ (Fig. 2C). Two separate Glt1-tTA founder lines were crossed with mice transgenic for TML with concordant results. Cerebellar tumors were observed in GTML mice from both Glt1-tTA founder lines.
A cohort of 30 Glt1-tTA mice was imaged at multiple time points between P7 and P100 to establish baseline values for Luc imaging. As expected, in the absence of a Luc transgene, bioluminescent signals remained at background levels (106 photons/cm2sec) (Fig. 2D,E). To establish an upper threshold for Luc signal strength, a cohort of 30 GTML mice with robust, increasing Luc signals (>108 photons/cm2sec) was followed from P30 until sacrifice due to tumor burden. A representative mouse imaged at multiple time points is shown in Figure 2D, with tabulated results for the cohort displayed in Figure 2E. Although variability existed in tumor progression times and age at sacrifice, Luc signal magnitudes from tumors of moribund mice were tightly grouped at approximately five orders of magnitude over background-level luminescence (1011 photons/cm2sec), having progressed through approximately three orders of magnitude (from 108 photons/cm2sec). All 30 mice in this cohort progressed to a moribund state requiring sacrifice before P200, demonstrating the utility of Luc imaging in detecting progressing lesions in this model.
In order to assess latency and penetrance, we followed 50 doubly hemizygous GTML mice to P200. In this cohort, overall survival was 80% at 100 d and 60% at 200 d (Fig. 2F). To assess a role for dosage of the TML transgene, we next generated a second cohort of 35 mice homozygous for the TML transgene and again followed tumor incidence to P200. In these mice, overall survival fell to 50% at 100 d and 25% after 200 d (Fig. 2F). The acceleration of tumor progression by transgene dosage suggests that MYCN contributes to malignant progression, and is reflective of data from human cancer correlating high levels of MYCN (in MYCN-amplified tumors) with aggressive disease (Brodeur et al. 1984; Seeger et al. 1985; Eberhart et al. 2004; Stearns et al. 2006; Pfister et al. 2009).
MYCN drives two distinct pathologies and promotes metastases in murine MB
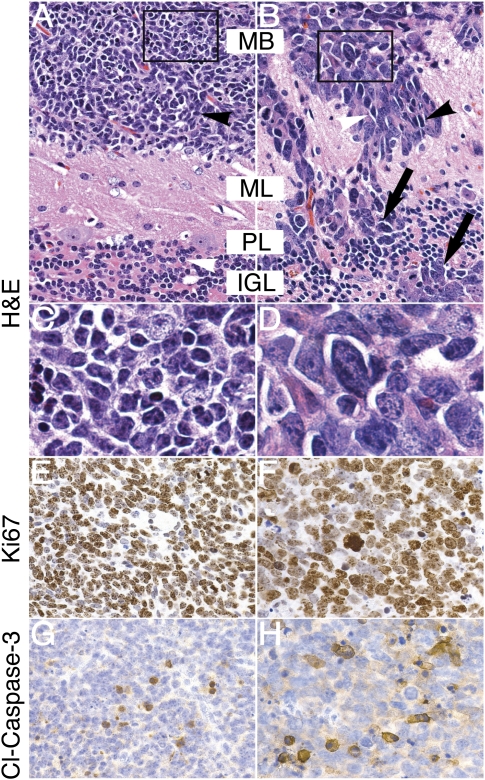
We analyzed 50 GTML animals sacrificed based on high Luc signal (1011 photons/cm2sec) and on clinical signs (poor grooming, weight loss, inactivity, circling, and ataxia). Full necropsies revealed no tumors outside of the craniospinal axis. CNS tumors in this cohort showed histology evenly split between classic and LCA (Fig. 3A,B). Classic tumors consisted of cells with moderate stromal content, low to intermediate vascularity, and variable anaplasia (Fig. 3C). While the classic tumor cells were similar in size to mature granule cells in the IGL, LCA tumor cells were larger and exhibited high nuclear-to-cytoplasmic ratios, polygonal cell morphology, and high levels of anaplasia and invasion (Fig. 3D). To characterize neuronal and glial differentiation, we stained tumors of both histologies for the glial marker glial fibrillary acidic protein (GFAP) and for the neuronal markers synaptophysin and NeuN. A population of cells in both LCA and classic tumors were highly immunoreactive for the astrocytic marker GFAP. Both LCA and classic MB were negative for NeuN and showed moderate expression of synaptophysin, suggesting a low level of differentiation (Supplemental Fig. 2).
Figure 3.
GTML tumors represent two distinct, aggressive MB histologies. (A) Tumor cells of classic histology (black arrowhead) were similar in size to mature granule cells in the IGL (white arrowhead). (B) When compared with classic tumors, LCA tumors exhibited increased nuclear-to-cytoplasmic ratios (black arrowhead), polygonal cell morphology (white arrowhead), and high levels of anaplasia. Invasion was prominent in LCA tumors, with disruption of molecular layer, Purkinje layer, and IGL (ML, PL, and IGL respectively; black arrows). (C) Inset from A shows detail of small, round, blue cells of the classic histology. (D) Inset from B shows detail of large, angular, pleiomorphic LCA cells. (E,F) Immunohistochemical staining for the proliferative marker Ki67 shows strong staining in both classic and LCA GTML tumors, suggesting aggressive tumor growth. (G,H) Immunohistochemical staining for the marker of apoptosis, cleaved Caspase-3 (Cl-Caspase-3), shows positive staining in both classic and LCA GTML tumors, respectively. All pictures except inset (1000×) are shown at 400× magnification.
Leptomeningeal spread of MB cells through the cerebrospinal fluid and down the spine (so-called drop metastasis) occurs in patients with advanced disease and correlates with poor prognosis (Hsieh et al. 2008). To determine whether this biology was recapitulated in the GTML model, we screened 100 mice with established primary tumors (>5 × 109 photons/cm2sec by Luc imaging) and found 15 mice with elevated spinal Luc signals (>108 photons/cm2sec). Background luminescence from the proximal cranial tumors tended to obscure spinal Luc signals. Our inability to fully shield this cranial signal therefore limited precise measurement of the much weaker spinal signal. To compensate for this, we decalcified spinal bone and examined spinal and paraspinal tissues pathologically. Two of 15 animals showed disseminated tumor in the lumbrosacral region, partially encasing nerve roots in the subarachnoid spaces adjacent to the spinal cord (Supplemental Fig. 3). Spinal metastases were discontinuous with primary tumors, suggesting leptomeningeal spread through cerebrospinal fluid (analogous to that seen in children with drop metastases). Histology among this limited sample of metastatic tumors corresponded to that of the primary tumors (Supplemental Fig. 3). Thus, targeted expression of MYCN in GTML mice can recapitulate leptomeningeal spread, a characteristic feature of malignant progression observed in human MB.
Down-regulation of MYCN leads to cell cycle arrest and senescence
To address a role for MYCN in initiation of MB, we blocked expression of MYCN in newborn mice, placing a cohort of 20 GTML mice on dox treatment from P1. Dox was then withdrawn at P30, and mice were serially imaged through P200. All mice survived through P200. Baseline Luc signals did not change over time, and no tumors were observed upon subsequent necropsy (data not shown). These data suggest that MYCN contributes to the initiation of MB in GTML mice, and that a window of vulnerability for development of MB exists from birth through P30.
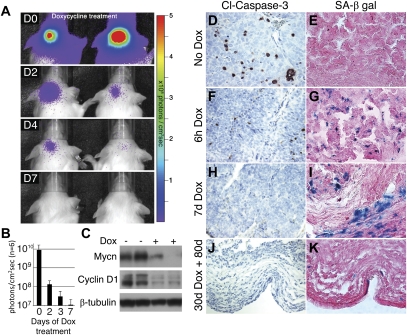
To characterize a role for MYCN in maintenance, a second cohort of 20 adult GTML mice with large, established tumors (>5 × 109 photons/cm2sec) was placed on dox treatment. All mice again survived through 200 d of treatment, suggesting that maintenance of tumors requires ongoing expression of MYCN. Dox treatment resulted in a 1000-fold drop in Luc signal intensity by 7 d (Fig. 4A,B). To verify that TRE-driven expression of MYCN was also abrogated, we analyzed cerebellar lysates from six mice treated with dox for 7 d. Mycn protein levels were reduced substantially on immunoblot, with decreased levels of cyclin D1 (Fig. 4C).
Figure 4.
GTML tumors require sustained expression of MYCN. (A) Administration of dox chow for a week (D0–D7) to GTML mice with established tumors decreased Luc signal intensity to near background levels in two representative mice. (B) Quantified luminescence demonstrated a three-log decrease in cerebellar Luc signal intensity after 7 d on dox. (C) Western blot analysis of GTML tumors treated with dox for 7 d shows reduced levels of Mycn protein and cyclin D1. A representative untreated GTML tumor shows moderate apoptosis by cleaved Caspase-3 staining (D) and no senescent cells, as shown by senescence-associated β-galactosidase (SA-β-gal) staining (E). (F,G) Acute dox treatment after i.p. injections of dox (6h Dox) leads to a reduction of apoptotic cells but an increase of senescent cells, as indicated. (H,I) A representative GTML mouse after 7 d of dox (7d Dox) shows no apoptosis but several senescent cells, respectively. (J,K) A mouse with a large tumor treated for 30 d with dox and for 80 d without dox (30d Dox + 80d) presents no apoptotic cells and a few scattered senescent cells in tumor remains, respectively. Shown in 400× magnification.
GTML tumors have rather high levels of intrinsic apoptosis, as shown when staining for cleaved Caspase-3 (Fig. 4D). After acute dox treatment (6 h after i.p. injections of dox), fewer apoptotic cells were found (Fig. 4F), and several cells close to the blood vessels entered senescence (as compared with mock-treated GTML control tumors) (Fig. 4E,G; Supplemental Fig. 4A). Regions around the senescent cells also appeared necrotic (Supplemental Fig. 4B). At P7, neither markers of proliferation (PCNA and Ki67) nor apoptosis (cleaved Caspase-3) were evident in the remaining tumor mass (Fig. 4H; Supplemental Fig. 4C,D). However, levels of senescence were still highly elevated (Fig. 4I). These data suggest that Mycn drives both proliferation and apoptosis in murine MB. Subsequent withdrawal of Mycn leads to clearance of tumor regions in which apoptosis was initiated (positive for cleaved Caspase-3), with senescence of the remaining nonapoptotic regions.
To analyze the fate of these senescent regions over time, we treated three animals with dox for 30 d, and then followed them for 80 additional days off dox. All showed evidence of prior regressed tumor (Fig. 4J,K) and stained negatively for NeuN and other markers of differentiation (Supplemental Fig. 4E,F). The cells remaining in these long-term dox-treated tumors showed no signs of mitosis or apoptosis, whereas a few scattered senescent cells were evident (Fig. 4J,K; Supplemental Fig. 4G,H). Residual tumor was not evident elsewhere in the brain.
GTML tumors are Shh-independent and show a classic MB profile
We used qRT–PCR analysis to compare levels of MYCN mRNA in 20 GTML tumors, with levels in both Math1-driven SmoM2 mutant murine MB (Schuller et al. 2008) and in normal cerebellum. The human MYCN transgene was expressed in the developing cerebellum (P7) of GTML mice (Fig. 5A), as predicted from Luc signals (Fig. 2E). Nineteen out of 20 tumors had levels of MYCN increased beyond that observed in GTML cerebella at P7 (six of which, GTML-T1–6, are shown in Fig. 5A). The remaining tumor had levels comparable with P7 GTML cerebella (GTML-T7) (Fig. 5A). As expected, the Math1-driven SmoM2 tumor showed elevated levels of endogenous N-myc (Fig. 5A). These data are consistent with observations that Smo/Shh signaling leads to increased levels of N-myc (Kenney et al. 2003; Oliver et al. 2003). However, levels of endogenous N-Myc in GTML-T7 were even higher than that of SmoM2 (Supplemental Fig. 5A). To characterize this tumor, we demonstrated decreased expression of Ptch1, a negative regulator of Shh signaling, in the Shh-dependent GTML-T7 tumor (Supplemental Fig. 5B). This result potentially explains the increase in endogenous expression of N-Myc found in this tumor, as Ptch1 loss is correlated with increased levels of N-myc (Kenney et al. 2003; Oliver et al. 2003; Hatton et al. 2006).
Figure 5.
Expression analysis of GTML tumors generally describing a classic MB profile. (A) Expression analysis by qRT–PCR detecting both N-Myc (endogenous murine promoter) and transgene level (MYCN) in seven GTML tumors (GTML-T1–7), one Math1–SmoM2 tumor (SmoM2), and controls of cerebella from normal (E16, P7, and P30) and GTML (GP7) mice. Normal controls are presented in green bars, GP7 is shown in blue, Shh− tumors (GTML-T1-6) are indicated in light red, and Shh+ tumors (GTML-T7 and SmoM2) are presented in dark red. (B) Expression analysis by qRT–PCR for markers of Shh activation (Gli1), granule cell precursor lineage (Math1), and nondesmoplastic MB (Otx2). (C,D). IHC showing elevated Otx2 protein expression in the GTML-T1 tumor, but no (or low) expression in the GTML–T7 tumor, respectively (400× magnification). (E) Up-regulated expression of N-Myc-target genes Odc1, Mdm2, and Fbl (>1) in almost all tumors generally correlating with levels of MYCN. The expression levels are all relative to expression of total RNA prepared from normal adult (P60) cerebellum, and data are representative of two experiments performed in triplicate and normalized to Gapdh.
To clarify the human MB subtype represented by GTML tumors, we analyzed expression of the EGL marker Math1 and the Shh-target Gli1, both of which are typically elevated in SHH-driven desmoplastic human tumors. Levels of Math1 and Gli1 were comparable in the majority of GTML tumors (19 of 20, as shown by GTML-T1–6) (Fig. 5B) and were similar in GTML and wild-type cerebella at P7. As expected, elevated expression of Math1 and Gli1 was observed in P7 cerebella from both control and GTML mice, with comparable levels found in the Shh-driven SmoM2 tumor and in the GTML-T7 tumor (Fig. 5B). Collectively, these findings demonstrate that Shh activation is an infrequent event in GTML-driven MB tumors, with targeted expression of MYCN in GTML leading to Shh-driven tumors in only 5% of the cohort analyzed.
To further characterize this set of tumors, we next studied expression of Otx2, which is commonly and specifically expressed in nondesmoplastic MB (Adamson et al. 2009). As expected, Otx2 levels were elevated in all tumors except for the Shh-driven SmoM2 and GTML-T7 tumors. We next analyzed Otx2 protein levels using immunohistochemistry (IHC). Shh-negative tumors GTML-T1–6 stained positively for Otx2, whereas the Shh-driven GTML7 tumors showed low Otx2 protein levels (Fig. 5C,D). To verify that MYCN expression was functional and could appropriately up-regulate expression of known downstream targets, we analyzed three known Mycn-targets:Odc1, Mdm2, and Fbl (Coller et al. 2000; Slack et al. 2005; Hogarty et al. 2008). All three were up-regulated in GTML tumors compared with controls (Fig. 5E), with Mdm2 in particular expressed to high levels in P7 GTML cerebella. The mRNA levels of all three genes generally correlated with levels of MYCN in the GTML tumors. Since synaptophysin levels are typically higher in Shh-independent than in Shh-driven tumors (Buhren et al. 2000), we next verified higher levels of synaptophysin in Shh-independent GTML-T1–6 tumors compared with Shh-driven GTML-T7 and SmoM2 tumors (Supplemental Fig. 5C). Finally, and as compared with GTML-T1–6, we saw a higher protein expression of NeuN in the GTML-T7 case (Supplemental Fig. 5D,E), which also correlates with a desmoplastic subtype reported in human MB (Preusser et al. 2006).
EGL cells in GTML mice are normal, and GTML tumors can be transplanted
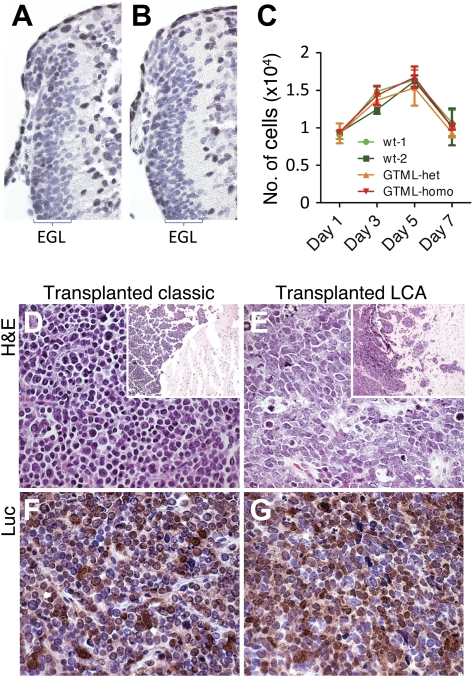
To understand the cellular response to misexpression of MYCN during cerebellar development, we sectioned the cerebella of three GTML mice and wild-type controls at P7. Levels of apoptosis were comparable in GTML and control cerebella (Supplemental Fig. 5F,G). Neither dysplastic nor early neoplastic lesions were observed in the EGL at this time point (Fig. 6A,B). We next isolated EGL cells from the cerebella of mice hemizygous or homozygous for GTML (n = 3 for each group) using percoll gradient centrifugation, followed by culture on poly-D-lysine-coated plates (serum-free conditions with 10 ng/mL Shh-N). Proliferation and survival of GTML cells, monitored for 7–10 d, were identical to P7 wild-type EGL culture controls (Fig. 6C). Lastly, we demonstrated that GTML tumors were transplantable and therefore contained true tumor-initiating/propagating cells. We isolated classic and LCA tumor cells and transfected 104 cells separately into the cerebella of adult nude mice. Recipient mice became symptomatic at 60–70 d post-transplantation (Fig. 6B) with engrafted tumors retaining both the appropriate classic or LCA pathology (Fig. 6D,E) and expression of Luc (Fig. 6F,G). These data also suggest that transplanted tumor cells sustained expression of the TML transgene (Fig. 2A), enabling monitoring of bioluminescence when grafted in vivo (data not shown).
Figure 6.
EGL cell proliferation is normal in GTML mice, and GTML tumors are transplantable. The width of the EGL in the cerebellum of a P7 wild-type (wt) mouse (A) is not different from the EGL in a P7 GTML mouse (B). EGL cells are visualized here with hematoxylin (blue), and non-EGL cells are stained with Sox9 (brown) that is positive in ependymal and glial cells. Choroid plexus is cut away to the right of the pictures. (C) Fluorescence from an EGL cell proliferation assay (CyQUANT) showing the calculated cell number (per 96 wells) at days 1, 3, 5, and 7 after representative outgrowth from two wild-type control mice, one GTML hemizygous mouse, and one GTML doubly homozygous mouse (from quadruplicate measures). (D,E) Classic and LCA GTML tumors retain their classic and LCA histopathology (stained with H&E) after transplantation, respectively. Inset shows tumors (darker) on the right in brain of hosts. (F,G) Tumors are positive for transgenic Luc, as shown using an antibody directed against the Luc protein, in respective samples. All pictures except inset (40×) are shown at 400× magnification.
Genomic instability in MYCN-driven murine MB
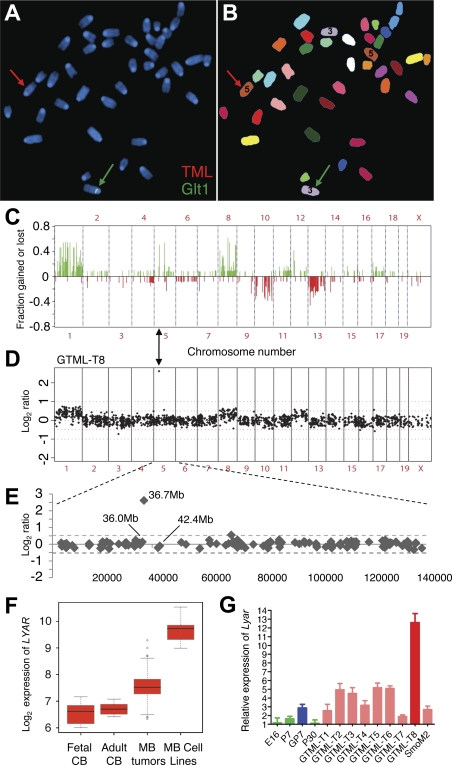
We performed metaphase fluorescence in situ hybridization (FISH) using labeled probes for the two transgenes. The Glt1-tTA transgene mapped to chromosome 3, and the TML transgene mapped to chromosome 5 (Fig. 7A,B; Supplemental Fig. 6).
Figure 7.
Genomic instability and Shh independence in GTML tumors. (A) Spectral karyotyping (SKY) and FISH in metaphase of a doubly hemizygous GTML mouse. The red arrow indicates hybridization of the TML probe (red) on chromosome 5. The green arrow indicates hybridization of the Glt1-tTA probe (Glt1; green) on chromosome 3. (B) Chromosomes shown in SKY display colors with chromosomes 3 and 5 indicated (the color scheme for all chromosomes is presented in Supplemental Fig. 6A). (C) Frequency plot of genomic changes in GTML tumors by array CGH. Tumors showed gains at chromosomes 1 and 11q and losses at chromosome 13 (syntenic with human chromosomes 6, 17q, and 1q, respectively), three sites frequently altered in human poor-risk MB. (D,E) CGH analysis for one tumor, GTML-T8, showed a subchromosomal amplification (36.7 Mb) on mouse chromosome 5. (F) Box plots showing human LYAR expression levels in MB samples and controls (as described in Fig. 1). (G) Expression analysis by qRT–PCR detecting the candidate gene Lyar in mice using previous set of controls in green and blue (see Fig. 5A). All GTML tumors (except GTML-T8 in dark red) are now presented in light-red bars.
To examine chromosomal copy number changes, we analyzed 13 GTML tumors by bacterial artificial chromosome (BAC) array-based CGH. Both whole- and subchromosomal copy number abnormalities (CNAs) were observed, suggesting (in concert with focal tumors found on pathology) that tumors resulted from progressive accumulation of genetic lesions, rather than from broad transformation of cerebellar tissue, as typically observed in Shh-driven murine models. Representative CGH plots from five tumors, as well as frequency analyses of 13 tumors, are shown (Fig. 7C; Supplemental Fig. 7A–E). Several recurrent CNAs were observed across multiple tumors, suggesting clonal selection and expansion of tumor cells harboring these CNAs. Tumors commonly showed gain of chromosomes 1 and 8, as well as loss of chromosomes 10 and 13. Cluster analysis suggested that gain of chromosome 1 was frequently associated with other CNAs, most notably loss of chromosome 10 (Supplemental Fig. 7F).
To identify additional genetic events in these tumors, we sequenced all exons of Trp53 from the Shh-driven tumor (GTML7) as well as Shh-independent classic tumors (n = 2) and LCA tumors (n = 3). No mutations were identified in Trp53 (data not shown). To verify this result, and since elevated levels of p53 typically result from p53 mutation, we also analyzed six tumors and matched somatic control tissues for expression of p53 and Arf proteins, with no differences noted on IHC staining (data not shown). The findings suggest that p53 is functional in the GTML model, and that mutation in p53 is unlikely to contribute to tumor development.
Rare tumors showed subchromosomal gains or losses by array CGH. Two clones on chromosome 5, mapping 35 Mb apart, showed increased copy number in >50% of tumor samples. In both cases, BAC clones within this 35-Mb interval did not show consistent gain, suggesting that the gains observed in these regions represent distinct events. The frequent gain of one but not both of these regions could potentially be explained by amplification of TML. The proximal clone on chromosome 5 at 36.7 Mb (RP23-118A15) showed high-level amplification in a subset of tumors (example shown in Fig. 7D,E). To identify endogenous genes in this small amplicon that may contribute to selection for localized amplification, we analyzed expression of the 25 RefSeq genes mapping between the flanking clones in a database of human MB (Northcott et al. 2009). Among these, LYAR (Ly-1 antibody-reactive clone) showed elevated expression in human primary tumors and cell lines, compared with normal cerebella (Fig. 7F).
To determine whether Lyar might also play a role in GTML tumors, we used real-time PCR to analyze expression of Lyar and other candidate genes in the tumor with the highest amplification of chromosome 5 (GTML-T8) (Fig. 7D,E) and in seven GTML tumors profiled previously (Fig. 5). Lyar was up-regulated prominently in seven of nine GTML tumors (Fig. 7G). Lyar was also up-regulated to a lesser extent in preneoplastic cerebella from GTML mice (P7) as compared with controls (Fig. 7G), whereas two other gene candidates in the amplicon, Drd5 and Slc2a9, were not differentially expressed (Supplemental Fig. 7G,H). The relative expression of LYAR was highest in GTML-T8, the tumor harboring the subchromosomal gain on chromosome 5. Expression levels of Drd5 and Slc2a9 were also elevated in GTML-T8 (Supplemental Fig. 7G,H), although expression of these genes was not increased in other GTML tumors (data not shown). These data suggest (1) a possible role for LYAR in Mycn signaling and MB biology, and (2) utility of the GTML model to identify candidate genes in MB.
Discussion
To address a role for MYCN in MB, we analyzed expression in human cerebella and in MB tumors. In support of an embryonal origin for MB, and consistent with data generated by others (Eberhart et al. 2004), MYCN was expressed in human fetal but not adult cerebella. Since expression of MYCN was essentially absent after cerebellar development, we conclude that the elevated levels of expression observed in the vast majority of human MB tumors (Fig. 1) represent misexpression of MYCN (Pomeroy et al. 2002; Eberhart et al. 2004; Gajjar et al. 2004; Gilbertson 2004).
To test whether misexpression of MYCN could initiate MB, we targeted expression of MYCN to the cerebellum under the control of the Tet system. Tumors arising in this model showed distinct classic or LCA pathologies, both of which are correlated with unfavorable outcome clinically, and are poorly represented in current genetically engineered murine models of MB. LCA and classic tumors together account for the majority of human tumors, including the most aggressive subset (Eberhart and Burger 2003), and arose in GTML mice at equal incidence. A minority of mice with advanced GTML tumors displayed leptomeningeal spread and metastases to spine, hallmarks of advanced human disease. Taken together, these results suggest that MYCN-driven MB tumors in mice recapitulate both histological and biological features observed in human MB.
Low-level copy gain of MYCN and MYC (twofold to fivefold using FISH) occurs commonly in both desmoplastic and classic MB (Takei et al. 2009). Genomic amplifications of MYCN and MYC are relatively infrequent in these subtypes; however, both amplicons have been linked to anaplasia in human MB (Brown et al. 2000; Leonard et al. 2001; Aldosari et al. 2002; Eberhart et al. 2002), with overexpression of MYC driving anaplasia in xenograft models (Stearns et al. 2006), and through cooperation with β-catenin in a genetically engineered model (Momota et al. 2008). Could misexpression of MYCN be linked to all pathologies of human MB? Our data are consistent with a model in which classic and LCA tumors form a continuum, with MYCN contributing to both pathologies. Since a majority of MB tumors misexpress MYCN, it is possible that low-level expression of MYCN serves as an initiating event in this embryonal tumor, with subsequent amplification or high-level expression of MYCN (or MYC) and/or the acquisition of mutations in additional genes driving LCA pathology.
The Glt1 promoter used to drive MYCN is not normally expressed in EGL progenitor cells (Furuta et al. 1997), the likely cells of origin for desmoplastic MB. Glt1 is a sodium-dependent transporter that regulates the levels of the excitatory neurotransmitter glutamate in CNS (Tanaka et al. 1997). Interestingly, Glt1 has also been identified as one of many genes up-regulated in classic MB (Pomeroy et al. 2002). Consistent with a non-EGL origin, most GTML tumors were negative for the EGL marker Math1 and positive for the classic pathology marker Otx2. In addition, EGL cells derived from GTML animals failed to show a growth advantage in culture. Collectively, these observations suggest a non-EGL origin for the majority of GTML tumors.
Several chromosomal changes occur in regions syntenic to those altered in human tumors, suggesting that GTML tumors mirror some genetic features of human MB. Tumors in GTML mice showed a range of copy number aberrations by CGH. Genomic gain of chromosome arm 17q is among the most frequent chromosomal abnormalities in MB, and correlates with poor prognosis (Pfister et al. 2009). Regions of human 17q are syntenic with mouse chromosome 11 and showed frequent gains in GTML mice (Fig. 7C). Loss of 1q occurs in 20%–40% of human MB (Kraus et al. 1996), and shows synteny with corresponding losses on chromosome 13 also observed commonly in GTML mice. Gain of human chromosome 6q is associated with poor prognosis, and is syntenic with a frequent gain in GTML mice, on chromosome 1.
Shh-driven tumors from Ptch1+/− mice have demonstrated relatively little genomic instability beyond the loss of heterozygosity at Ptch1 (Zindy et al. 2007). However, mice doubly mutant at Ptch1 and p18Ink4c (a Cdk inhibitor) developed MB with considerable genomic instability, and with frequent trisomy of chromosomes 6 and 14 (Zindy et al. 2007). Gains of chromosomes 6 and 14 are relatively rare in GTML tumors (Fig. 7C), suggesting that GTML tumors select for secondary genomic events distinct from those observed in Ptch1/p18Ink4c knockout mice.
Lyar was identified as up-regulated among 30 genes in a small amplicon present in 50% of tumors examined by CGH. Lyar is a nucleolar protein with zinc finger motifs that was initially isolated from a mouse T-cell leukemia line (Su et al. 1993). LYAR was oncogenic in NIH3T3 fibroblasts (Su et al. 1993), and was shown recently to regulate self-renewal and differentiation of embryonal stem cells (Li et al. 2009). The LYAR gene was overexpressed in both human MB and in murine GTML tumors, and was elevated specifically in MYCN-driven GTML P7 cerebella compared with MYCN-independent wild-type P7 cerebellar controls. FISH analysis showed that TRE-MYCN transgene integration was localized to chromosome 5. Transgene amplification has been reported as a selection advantage during tumor progression in other models, including a neuroblastoma model driven by a MYCN transgene (Hackett et al. 2003). Although expression of LYAR was also increased significantly in human MB, we cannot exclude the TML transgene as the target of this CNA.
In line with human data showing that mutation in p53 is rare in MB (Adesina et al. 1994), sequencing of p53 in the GTML tumors revealed no mutations. In contrast, distinct genomic CNAs were common in GTML tumors, which suggests that secondary genetic events contribute to tumor formation. These observations imply that MYCN is necessary but not sufficient to drive MB in the mouse, a result aligned with that of Fults and coworkers (Browd et al. 2006), who modeled MB by injecting retroviruses into a small field of cerebellar progenitor cells in vivo and established that N-myc could cooperate with Shh in driving MB. However, N-myc could not by itself drive tumor formation (Browd et al. 2006); and no tumors were generated even when ectopically expressing a mutationally stabilized form of N-myc, alone or in combination with the anti-apoptotic protein Bcl-2 (Browd et al. 2006; McCall et al. 2007). Similarly, in our experiments, misexpression of a MYCN transgene in a larger field of cells led to MB tumors only through acquisition of secondary genetic abnormalities. Importantly, and consistent with previous studies (Browd et al. 2006; Kessler et al. 2009; Thomas et al. 2009), in the single Shh-dependent tumor identified, Ptch1 (a negative regulator of Shh) was expressed at very low levels, suggesting that loss of Ptch1 cooperated with MYCN to initiate rare GTML tumors driven by Shh.
Our results suggest a pleiotropic role for MYCN in the initiation, progression, and maintenance of MB. MYCN is misexpressed in the majority of human MB. Targeted expression of MYCN can induce both classic and LCA pathology in genetically engineered mice. Interestingly, we observed apoptosis at baseline in GTML tumors, an event commonly associated with transformation by Myc family genes (Evan et al. 1992; Nesbit et al. 1998). Withdrawal of Mycn led to senescence of tumor cells, a result aligned with clinical observations demonstrating both quiescence and differentiation in human MB after therapy (Kane and Aronson 1967; Kudo et al. 1990; Cai et al. 2000). The GTML mouse provides an ideal system in which to investigate high-risk subclasses of human MB. Furthermore, the use of Luc as a surrogate of tumor burden and the ability to modulate MYCN expression using dox makes this model amenable to studies of developmental therapeutics and oncogene dependence.
Materials and methods
Generation of transgenic mice
To generate Glt1-tTA mice, we used a 13-kb genomic NotI fragment of murine Glt1 containing ∼10 kb of promoter, the first exon, and part of the first intron. We introduced a unique Fse1 site upstream of the mapped transcription initiation site for Glt1, and subcloned the tTA gene with polyA/transcription terminator sequence into this site. We used a linearized NotI fragment for transgene production, generating five founders. Two of these showed identical expression of LacZ (Fig. 2) when crossed to TRE-LacZ animals. The TML construct was generated using a pBI expression vector (Clontech, Inc.) containing a bidirectional TRE (Gossen and Bujard 1992), by cloning 2 kb of the pGL3 Luc coding sequence (Promega, Inc.) into MCS-II. Then, 1.6 kb of sequence encompassing the human MYCN cDNA (NM_005378) was inserted into MCS-I. Correct tTA-driven expression of the TML construct was verified by transient cotransfection into cultured cell lines using CMV-driven constructs expressing either rtTA or tTA transactivators, immunoblotting for Mycn protein expression, and assessing Luc expression by Luc bioimaging of the culture plates. The primers used for PCR were GLT1-tTA (sense, GGACAACAGAAAAGGGACAACG; antisense, AACATCTCAATGGCTAAGGCGTC) and MYCN (sense, TCATCCTCCAGGACTGCATGTG; antisense, CTTCATCATCTGAATCGCTCAGG). In addition, a TaqMan assay was performed using a TaqMan Gene Expression Master Mix (according to Applied Biosystem's guidelines) in order to monitor transgene copy number levels from genomic DNA generated from mouse tails. The primers and probes used were directed to MYCN (Hs00232074_m1; Applied Biosystems) and to tTA (as designed by Applied Biosystems). Obtained expression data were normalized to β-Actin using primers Actb (Mm00607939_s1; Applied Biosystems). All mouse experiments were approved by and performed according to the guidelines of the Institutional Animal Care and Use Committee of the University of California at San Francisco.
Dox treatment
Dox was administered in mouse chow at 200 mg/kg (Bio-Serve) without other food sources. For studies of transgene inactivation, mice were fed dox chow for 7 d prior to sacrifice or injected i.p. with 15 mg/kg dox (Sigma) for acute (6 h) dox treatments. For tumor initiation studies, mice were fed dox chow from P1 to P30. For intervention studies, mice with Luc-positive tumors were fed dox chow for >200 d.
Luc imaging
Mice were injected with 75 mg/kg sodium luciferin (LUCNA; Gold Biotechnology) in saline prior to imaging in the IVIS Lumina (Caliper Life Sciences) using Living Image 2.5 software (Caliper Life Sciences). Circular regions of interest (ROI) were drawn around cranial signals, and mean values of photons/cm2sec were recorded. Error bars on Luc signal magnitudes represent ±1 standard deviation.
Expression analysis of MYC and MYCN in human MB
Trizol-extracted total RNA isolated from 103 primary human MB was converted to cDNA prior to qRT–PCR (Northcott et al. 2009). Quantitative PCR for MYCC (sense, AGAGTCTGGATCACCTTCTGCT; antisense, ACACTGTCCAACTTGACCCTCT) and MYCN (sense, ATCCACCAGCAGCACAACTAT; antisense, TGTCCTCCGAGTCAGAGTTTC) was performed using Platinum SYBR Green qPCR SuperMix (Invitrogen) on an ABI StepOnePlus Real-Time PCR system. Expression of MYC and MYCN was determined using the Ct method by normalizing to ACTB and calibrating expression relative to pooled (n = 5) normal adult human cerebellum (Biochain). Statistical significance of differential MYC and MYCN expression in the unique MB subgroups was determined using one-way ANOVA. Affymetrix exon arrays were hybridized and processed, and were divided into SHH-dependent and SHH-independent subgroups as described (Northcott et al. 2009).
Real-time PCR gene expression analysis in GTML mice
GTML tumors were collected, flash-frozen in 2-methyl butane (Sigma), and homogenized in Trizol solution (Invitrogen), and total RNA was prepared according to the manufacturer's protocol (Invitrogen). Total RNA was similarly prepared from samples P60, P30, P7, and E16.5 (abbreviated to E16 everywhere in the text and figures) isolated from total cerebellum from mice aged as indicated. Sample GP7 was total cerebellum isolated from a 7-d-old GTML hemizygote mouse. Cerebellum (with choroid plexus) from all ages was removed carefully and did not include any spinal cord or midbrain tissue. Real-time PCR was carried out on a 7900HT Fast Real-Time PCR system (Applied Biosystems) or a Stratagene Mx3005P (Stratagene) using SYBR Green and analyzed as described in Johansson et al. (2005). The primer sequences are presented in the Supplemental Material.
IHC
Samples were prepared as described previously (Johansson et al. 2005; Chesler et al. 2006) using the following antibodies: Ki67 (1:200; M7249; Dako), Ki67 (1:100; NB600-1252; Novus Biologicals), cleaved Caspase-3 (1:200; 9661; Cell Signaling Technology), Synaptophysin (1:200; N1566; Dako), GFAP (1:200; Z0334; Dako), Otx2 (1:500; AB9566; Chemicon International), Sox9 (1:500; AB5535; Millipore), NeuN (1:500; MAB377; Millipore), p53 (1:500; CM5; Vector Laboratories, Inc.), and Luc (1:1000; L2164; Sigma-Aldrich).
LacZ staining and SA-β-gal staining
Mice at P60 were perfused with 2% paraformaldehyde and 0.2% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4), and brains were post-fixed in the same solution (for 4 h at 4°C) with shaking. Fifty-micromolar sections were prepared on a vibratome, washed three times for 30 min in 100 mM sodium phosphate buffer (pH 7.4), 17 mM MgCl2, 0.01% NaDOC, and 0.02% Nonidet-P-40; and stained in 100 mM sodium phosphate buffer (pH 7.4), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, and 1 mg/mL 5-bromo-4-chloro-3-indolyl-d-galactoside (X-gal). Senescence was detected on 16-μm free-floating or 6-μm frozen sections fixed and stained using a Senescence β-Galactosidase Staining Kit according to the manufacturer's instructions (Cell Signaling Technology).
Western blot analysis
Tumors were homogenized, samples were prepared, and Western blots were developed as described previously (Chesler et al. 2006). The antisera used included Mycn (1:500; NCM-II-100; AbCam), polyclonal rabbit antisera to Cyclin D1 (1:200; sc-718; Santa Cruz Biotechnologies), and β-tubulin (1:3000; 05-661; Upstate Biotechnologies).
EGL cell proliferation assay
EGL cells were isolated from wild-type, doubly hemizygous, and homozygous GTML mice at P7. Briefly, the cerebellum (with choroid plexus) was removed, dissociated, and incubated (37°C) for 20 min in activated papain (10 U/mL; Worthington) in Hanks' Balanced Salt Solution without Ca/Mg salts (D-HBSS; obtained from the UCSF Cell Culture Facility) containing 200 μg/mL L-cysteine and 250 U/mL DNase (Sigma). Larger cerebellar cells were removed using a percoll gradient 35%–60% as described previously (Uziel et al. 2005). Purified EGL cells were cultured in 96-well plates (104 cells per well) coated with poly-D-lysine (100 μg/mL; Millipore) and growth factor-reduced BD Matrigel Matrix (diluted 1:60; BD Biosciences), and were cultured in serum-free Neurobasal medium with N2 and B27 supplement, sodium pyruvate, L-glutamine, and penicillin/streptomycin (all from GIBCO). IHC of GFAP showed that glial cells accounted for <10% of cells in the cultures. After addition of 10 ng/mL Shh-N (R&D Systems), cells were counted (in quadruplicates) after 1, 3, 5, and 7 d using a CyQUANT NF Cell Proliferation Assay Kit (according to the manufacturer's instructions; Invitrogen) in a Synergy2 microplate reader (BioTek).
Tumor isolation and transplantation
GTML tumor cells were removed from tumor lesions and digested in papain as described above. The remaining tumors were put in 10% PFA and analyzed for pathology after H&E staining (performed by the UCSF Pathology Core). Cells were resuspended in Neurobasal media (with supplements as above and 20 ng/mL EGF and 20 ng/mL FGF) and cultured overnight. After cells were recovered, spun down, and digested for 10 min (37°C) in Accutase (Worthington), cells were counted using a hemocytometer and diluted in cold PBS. Six-week-old to 8-wk-old athymic nude mutant mice were anesthetized using 100 mg/kg ketamine (Ketaset; Fort Dodge Animal Health) and 10 mg/kg xylazine (AnaSed; LLOYD Laboratories), and were positioned in a rodent stereotactic headframe (David Kopf Instruments). An incision was made in the skin over the cerebellum, and a hole was made in the skull 1 mm lateral of midline. Cells (104) in a 5-μL suspension were stereotactically injected 2 mm deep at a 30° angle to the surface of the cerebellum over 2 min using a microsyringe (Hamilton Medical). The needle was left in place for another minute to avoid reflux. Injected mice received 2.5 mg/kg antipamezole (Antisedan; Orion Pharma) after being removed from the frame.
Array CGH and FISH
DNA from frozen tumor and spleen tissue was isolated using two rounds of phenol-chloroform extraction and labeled essentially as described previously (Hackett et al. 2003). Briefly, tumor DNA was labeled with Cy3-dUTP, and sample-matched spleen DNA was labeled with Cy5-dUTP (GE Biosciences) using the Bioprime labeling kit (Invitrogen). Labeled tumor DNA was cohybridized with labeled spleen DNA from the same mouse to 2896-clone mouse BAC arrays printed at the UCSF Comprehensive Cancer Center following protocols described on the Array Core Web site (http://cancer.ucsf.edu/array/index.php). Fluorescent images were collected using a custom-configured CCD camera. Automated image analysis and spot normalization were performed using custom software (Jain et al. 2002). Genomic profiles from all tumors were further analyzed using the aCGH package in the R statistical language. The FISH analysis was performed as described previously (Hackett et al. 2003).
Sequencing of Trp53
Tumor DNA obtained as described in the “Array CGH and FISH” section above was sequenced after PCR amplification using platinum Taq polymerase (Invitrogen) at conditions recommended by the manufacturer for 30 cycles (30 sec at 94°C; 30 sec at 58°C; 45 sec at 72°C). All exons of Trp53 were sequenced using both forward and reverse primers, and sequencing was performed by Elim Biopharmaceuticals, Inc. Alignment analysis was performed using Sequencher 4.8 (Gene Codes Corporation). The primers used are presented in the Supplemental Material.
Additional methods with primer sequences are presented in the Supplemental Material.
Acknowledgments
We thank Nigel Kileen and the UCSF Transgenic Core for generating Glt1-tTA and TML mice, Bruce Conklin for providing TRE-LacZ mice, Vivi Heine and David Rowitch for the Math1-SmoM2 MB samples, Tamar Uziel and Martine Roussel for the EGL isolation protocol and for TaqMan primers, and the UCSF Pathology Core for sample processing. We acknowledge support from Accelerate Brain Cancer Cure, Children's Brain Tumor Foundation, NIH R01 CA133091, NIH R01 CA148699, the Pediatric Brain Tumor Foundation, the Swedish Research Council, the Samuel Waxman Cancer Research Foundation, and the Stewart Trust.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1907510.
Supplemental material is available at http://www.genesdev.org.
References
- Adamson DC, Shi Q, Wortham M, Northcott PA, Di C, Duncan CG, Li J, McLendon RE, Bigner DD, Taylor MD, et al. 2009. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res 70: 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesina AM, Nalbantoglu J, Cavenee WK 1994. p53 gene mutation and mdm2 gene amplification are uncommon in medulloblastoma. Cancer Res 54: 5649–5651 [PubMed] [Google Scholar]
- Aldosari N, Bigner SH, Burger PC, Becker L, Kepner JL, Friedman HS, McLendon RE 2002. MYCC and MYCN oncogene amplification in medulloblastoma. A fluorescence in situ hybridization study on paraffin sections from the Children's Oncology Group. Arch Pathol Lab Med 126: 540–544 [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY 1997. Math1 is essential for genesis of cerebellar granule neurons. Nature 390: 169–172 [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM 1984. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Browd SR, Kenney AM, Gottfried ON, Yoon JW, Walterhouse D, Pedone CA, Fults DW 2006. N-myc can substitute for insulin-like growth factor signaling in a mouse model of Sonic hedgehog-induced medulloblastoma. Cancer Res 66: 2666–2672 [DOI] [PubMed] [Google Scholar]
- Brown HG, Kepner JL, Perlman EJ, Friedman HS, Strother DR, Duffner PK, Kun LE, Goldthwaite PT, Burger PC 2000. ‘Large cell/anaplastic’ medulloblastomas: A Pediatric Oncology Group Study. J Neuropathol Exp Neurol 59: 857–865 [DOI] [PubMed] [Google Scholar]
- Buhren J, Christoph AH, Buslei R, Albrecht S, Wiestler OD, Pietsch T 2000. Expression of the neurotrophin receptor p75NTR in medulloblastomas is correlated with distinct histological and clinical features: Evidence for a medulloblastoma subtype derived from the external granule cell layer. J Neuropathol Exp Neurol 59: 229–240 [DOI] [PubMed] [Google Scholar]
- Cai DX, Mafra M, Schmidt RE, Scheithauer BW, Park TS, Perry A 2000. Medulloblastomas with extensive posttherapy neuronal maturation. Report of two cases. J Neurosurg 93: 330–334 [DOI] [PubMed] [Google Scholar]
- Chesler L, Schlieve C, Goldenberg DD, Kenney A, Kim G, McMillan A, Matthay KK, Rowitch D, Weiss WA 2006. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res 66: 8139–8146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford SC, Lusher ME, Lindsey JC, Langdon JA, Gilbertson RJ, Straughton D, Ellison DW 2006. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle 5: 2666–2670 [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci 97: 3260–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FG, McCarthy BJ, Freels S, Kupelian V, Bondy ML 1999. The conditional probability of survival of patients with primary malignant brain tumors: Surveillance, epidemiology, and end results (SEER) data. Cancer 85: 485–491 [PubMed] [Google Scholar]
- de Haas T, Oussoren E, Grajkowska W, Perek-Polnik M, Popovic M, Zadravec-Zaletel L, Perera M, Corte G, Wirths O, van Sluis P, et al. 2006. OTX1 and OTX2 expression correlates with the clinicopathologic classification of medulloblastomas. J Neuropathol Exp Neurol 65: 176–186 [DOI] [PubMed] [Google Scholar]
- Eberhart CG 2007. In search of the medulloblast: Neural stem cells and embryonal brain tumors. Neurosurg Clin N Am 18: 59–69 [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Burger PC 2003. Anaplasia and grading in medulloblastomas. Brain Pathol 13: 376–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart CG, Kratz JE, Schuster A, Goldthwaite P, Cohen KJ, Perlman EJ, Burger PC 2002. Comparative genomic hybridization detects an increased number of chromosomal alterations in large cell/anaplastic medulloblastomas. Brain Pathol 12: 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart CG, Kratz J, Wang Y, Summers K, Stearns D, Cohen K, Dang CV, Burger PC 2004. Histopathological and molecular prognostic markers in medulloblastoma: c-myc, N-myc, TrkC, and anaplasia. J Neuropathol Exp Neurol 63: 441–449 [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69: 119–128 [DOI] [PubMed] [Google Scholar]
- Finlay JL, Erdreich-Epstein A, Packer RJ 2007. Progress in the treatment of childhood brain tumors: No room for complacency. Pediatr Hematol Oncol 24: 79–84 [DOI] [PubMed] [Google Scholar]
- Furuta A, Rothstein JD, Martin LJ 1997. Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J Neurosci 17: 8363–8375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajjar A, Hernan R, Kocak M, Fuller C, Lee Y, McKinnon PJ, Wallace D, Lau C, Chintagumpala M, Ashley DM, et al. 2004. Clinical, histopathologic, and molecular markers of prognosis: Toward a new disease risk stratification system for medulloblastoma. J Clin Oncol 22: 984–993 [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ 2004. Medulloblastoma: Signalling a change in treatment. Lancet Oncol 5: 209–218 [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277: 1109–1113 [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci 89: 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer MR, Weiss WA 2008. BMPs oppose Math1 in cerebellar development and in medulloblastoma. Genes & Dev 22: 693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett CS, Hodgson JG, Law ME, Fridlyand J, Osoegawa K, de Jong PJ, Nowak NJ, Pinkel D, Albertson DG, Jain A, et al. 2003. Genome-wide array CGH analysis of murine neuroblastoma reveals distinct genomic aberrations which parallel those in human tumors. Cancer Res 63: 5266–5273 [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, et al. 2004. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res 64: 7794–7800 [DOI] [PubMed] [Google Scholar]
- Hatton BA, Knoepfler PS, Kenney AM, Rowitch DH, de Alboran IM, Olson JM, Eisenman RN 2006. N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res 66: 8655–8661 [DOI] [PubMed] [Google Scholar]
- Hatton BA, Villavicencio EH, Tsuchiya KD, Pritchard JI, Ditzler S, Pullar B, Hansen S, Knoblaugh SE, Lee D, Eberhart CG, et al. 2008. The Smo/Smo model: Hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer Res 68: 1768–1776 [DOI] [PubMed] [Google Scholar]
- Hogarty MD, Norris MD, Davis K, Liu X, Evageliou NF, Hayes CS, Pawel B, Guo R, Zhao H, Sekyere E, et al. 2008. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res 68: 9735–9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh PC, Wu CT, Lin KL, Jaing TH, Tseng CK, Lui TN, Jung SM 2008. The clinical experience of medulloblastoma treatment and the significance of time sequence for development of leptomeningeal metastasis. Childs Nerv Syst 24: 1463–1467 [DOI] [PubMed] [Google Scholar]
- Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D 2002. Fully automatic quantification of microarray image data. Genome Res 12: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson FK, Goransson H, Westermark B 2005. Expression analysis of genes involved in brain tumor progression driven by retroviral insertional mutagenesis in mice. Oncogene 24: 3896–3905 [DOI] [PubMed] [Google Scholar]
- Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH Jr, et al. 1996. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272: 1668–1671 [DOI] [PubMed] [Google Scholar]
- Kane W, Aronson SM 1967. Gangliogliomatous maturation in cerebellar medulloblastoma. Acta Neuropathol 9: 273–279 [DOI] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH 2003. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130: 15–28 [DOI] [PubMed] [Google Scholar]
- Kessler JD, Hasegawa H, Brun SN, Yang ZJ, Dutton JW, Wang F, Wechsler-Reya RJ 2009. N-myc alters the fate of preneoplastic cells in a mouse model of medulloblastoma. Genes & Dev 23: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN 2002. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes & Dev 16: 2699–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, et al. 2008. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 3: e3088 doi: 10.1371/journal.pone.0003088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus JA, Koch A, Albrecht S, Von Deimling A, Wiestler OD, Pietsch T 1996. Loss of heterozygosity at locus F13B on chromosome 1q in human medulloblastoma. Int J Cancer 67: 11–15 [DOI] [PubMed] [Google Scholar]
- Kudo M, Shimizu M, Akutsu Y, Imaya H, Chen MN, Miura M 1990. Ganglioglial differentiation in medulloblastoma. Acta Pathol Jpn 40: 50–56 [DOI] [PubMed] [Google Scholar]
- Lee Y, Miller HL, Jensen P, Hernan R, Connelly M, Wetmore C, Zindy F, Roussel MF, Curran T, Gilbertson RJ, et al. 2003. A molecular fingerprint for medulloblastoma. Cancer Res 63: 5428–5437 [PubMed] [Google Scholar]
- Leonard JR, Cai DX, Rivet DJ, Kaufman BA, Park TS, Levy BK, Perry A 2001. Large cell/anaplastic medulloblastomas and medullomyoblastomas: Clinicopathological and genetic features. J Neurosurg 95: 82–88 [DOI] [PubMed] [Google Scholar]
- Li H, Wang B, Yang A, Lu R, Wang W, Zhou Y, Shi G, Kwon SW, Zhao Y, Jin Y 2009. Ly-1 antibody reactive clone is an important nucleolar protein for control of self-renewal and differentiation in embryonic stem cells. Stem Cells 27: 1244–1254 [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G 2005. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48: 17–24 [DOI] [PubMed] [Google Scholar]
- Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, McMahon AP 2006. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res 66: 10171–10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall TD, Pedone CA, Fults DW 2007. Apoptosis suppression by somatic cell transfer of Bcl-2 promotes Sonic hedgehog-dependent medulloblastoma formation in mice. Cancer Res 67: 5179–5185 [DOI] [PubMed] [Google Scholar]
- McManamy CS, Lamont JM, Taylor RE, Cole M, Pearson AD, Clifford SC, Ellison DW 2003. Morphophenotypic variation predicts clinical behavior in childhood non-desmoplastic medulloblastomas. J Neuropathol Exp Neurol 62: 627–632 [DOI] [PubMed] [Google Scholar]
- Momota H, Shih AH, Edgar MA, Holland EC 2008. c-Myc and β-catenin cooperate with loss of p53 to generate multiple members of the primitive neuroectodermal tumor family in mice. Oncogene 27: 4392–4401 [DOI] [PubMed] [Google Scholar]
- Nesbit CE, Grove LE, Yin X, Prochownik EV 1998. Differential apoptotic behaviors of c-myc, N-myc, and L-myc oncoproteins. Cell Growth Differ 9: 731–741 [PubMed] [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, et al. 2009. The miR-17/92 polycistron is up-regulated in Sonic hedgehog-driven medulloblastomas and induced by N-myc in Sonic hedgehog-treated cerebellar neural precursors. Cancer Res 69: 3249–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, Wickramasinghe R, Scott MP, Wechsler-Reya RJ 2003. Transcriptional profiling of the Sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci 100: 7331–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S, Remke M, Benner A, Mendrzyk F, Toedt G, Felsberg J, Wittmann A, Devens F, Gerber NU, Joos S, et al. 2009. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol 27: 1627–1636 [DOI] [PubMed] [Google Scholar]
- Polkinghorn WR, Tarbell NJ 2007. Medulloblastoma: Tumorigenesis, current clinical paradigm, and efforts to improve risk stratification. Nat Clin Pract Oncol 4: 295–304 [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, et al. 2002. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415: 436–442 [DOI] [PubMed] [Google Scholar]
- Preusser M, Laggner U, Haberler C, Heinzl H, Budka H, Hainfellner JA 2006. Comparative analysis of NeuN immunoreactivity in primary brain tumours: Conclusions for rational use in diagnostic histopathology. Histopathology 48: 438–444 [DOI] [PubMed] [Google Scholar]
- Raffel C, Jenkins RB, Frederick L, Hebrink D, Alderete B, Fults DW, James CD 1997. Sporadic medulloblastomas contain PTCH mutations. Cancer Res 57: 842–845 [PubMed] [Google Scholar]
- Rao G, Pedone CA, Del Valle L, Reiss K, Holland EC, Fults DW 2004. Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene 23: 6156–6162 [DOI] [PubMed] [Google Scholar]
- Rogers HA, Miller S, Lowe J, Brundler MA, Coyle B, Grundy RG 2009. An investigation of WNT pathway activation and association with survival in central nervous system primitive neuroectodermal tumours (CNS PNET). Br J Cancer 100: 1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai S, Shimono A, Wakamatsu Y, Palmes C, Hanaoka K, Kondoh H 1993. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development 117: 1445–1455 [DOI] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, et al. 2008. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D 1985. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med 313: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Shakhova O, Leung C, van Montfort E, Berns A, Marino S 2006. Lack of Rb and p53 delays cerebellar development and predisposes to large cell anaplastic medulloblastoma through amplification of N-Myc and Ptch2. Cancer Res 66: 5190–5200 [DOI] [PubMed] [Google Scholar]
- Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, Shohet JM 2005. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci 102: 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns D, Chaudhry A, Abel TW, Burger PC, Dang CV, Eberhart CG 2006. c-myc overexpression causes anaplasia in medulloblastoma. Cancer Res 66: 673–681 [DOI] [PubMed] [Google Scholar]
- Su L, Hershberger RJ, Weissman IL 1993. LYAR, a novel nucleolar protein with zinc finger DNA-binding motifs, is involved in cell growth regulation. Genes & Dev 7: 735–748 [DOI] [PubMed] [Google Scholar]
- Takei H, Nguyen Y, Mehta V, Chintagumpala M, Dauser RC, Adesina AM 2009. Low-level copy gain versus amplification of myc oncogenes in medulloblastoma: Utility in predicting prognosis and survival. Laboratory investigation. J Neurosurg Pediatr 3: 61–65 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, Iwama H, Nishikawa T, Ichihara N, Kikuchi T, et al. 1997. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science 276: 1699–1702 [DOI] [PubMed] [Google Scholar]
- Thomas WD, Chen J, Gao YR, Cheung B, Koach J, Sekyere E, Norris MD, Haber M, Ellis T, Wainwright B, et al. 2009. Patched1 deletion increases N-Myc protein stability as a mechanism of medulloblastoma initiation and progression. Oncogene 28: 1605–1615 [DOI] [PubMed] [Google Scholar]
- Uziel T, Zindy F, Xie S, Lee Y, Forget A, Magdaleno S, Rehg JE, Calabrese C, Solecki D, Eberhart CG, et al. 2005. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes & Dev 19: 2656–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY 2005. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron 48: 31–43 [DOI] [PubMed] [Google Scholar]
- Wetmore C, Eberhart DE, Curran T 2001. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res 61: 513–516 [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schuller U, Machold R, Fishell G, Rowitch DH, et al. 2008. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 14: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Uziel T, Ayrault O, Calabrese C, Valentine M, Rehg JE, Gilbertson RJ, Sherr CJ, Roussel MF 2007. Genetic alterations in mouse medulloblastomas and generation of tumors de novo from primary cerebellar granule neuron precursors. Cancer Res 67: 2676–2684 [DOI] [PubMed] [Google Scholar]