Abstract
Transcriptional enhancers are key determinants of developmentally regulated gene expression. Models of enhancer function must distinguish between analog or digital control of transcription, as well as their requirement to initiate or maintain transcriptional activity of a gene. In light of a recent study by Chong and colleagues (pp. 659–669) providing evidence of a transient requirement of an enhancer associated with the CD4 gene, we discuss possible mechanisms by which transcriptional memory can be propagated in the absence of enhancers.
Keywords: Epigenetics, transcription, chromatin, gene regulation, CD4, T-cell development
Transcriptional enhancers are defined as cis-regulatory sequences that function independently of orientation and distance to the transcription initiation site (Bulger and Groudine 2010). At the time of their discovery, this definition served to distinguish enhancers from gene promoters that are located at a close and often precise position with respect to the transcription initiation site. In contrast, enhancers are typically located at considerable distances both 5′ and 3′ from transcription initiation sites of cellular genes. Enhancers are also important components of locus control regions (LCRs), which include additional cis-regulatory modules that are often defined by their DNase I hypersensitivity (Palstra et al. 2008). In combination with such auxiliary elements, enhancers allow for the correct expression in mouse germline transformation assays. In addition to their varied locations, most early studies on enhancers relied on transfection assays in which an assortment of promoters, enhancers, and genes was combined into plasmids to test the effects on transcription. In such experiments, enhancers activated transcription when located 5′ or 3′ to the reporter gene, thereby cementing the view that they are distance-independent. In reality, enhancers vary considerably in their ability to activate transcription even when tested with recombinant reporter constructs, and the effects of moving enhancers to different positions in the context of cellular genes are not known. Another underappreciated aspect of enhancer function is that most enhancers preferentially activate transcription from some promoters over others. The idea that enhancers act equally on all promoters also stems from the early use of a few well-defined promoter sequences to study enhancer function in transfection assays. Moreover, the presence of multiple promoters in the vicinity of an enhancer can influence the selectivity of promoter activation by enhancers. Thus, any current analysis of enhancer function must incorporate a broader view of such nuances to fully understand the role of an enhancer in a specific gene context.
Enhancer function I: analog versus digital response
At the most basic level, enhancers “work” by recruiting sequence-specific DNA-binding proteins to the gene locus. Typical enhancers contain binding sites for multiple transcription factors, and mutation of these sites affects enhancer function. However, significant reduction of enhancer activity often requires simultaneous mutation of more than one transcription factor-binding site. This observation has been interpreted to indicate functional redundancy (Dang et al. 1998) among the several factors recruited to the enhancer, but may also reflect the nature of assays used to study enhancer function. A subset of enhancer-binding proteins can confer activity in the context of synthetic enhancers that contain multimerized factor-binding sites, whereas others cannot. The logic by which different transcription factor-binding sites are put together to generate functional enhancers remains unclear. Additionally, most enhancer-binding proteins can also function as promoter factors. The lack of obvious functional distinction between enhancer-binding proteins and promoter-associated factors has blurred the distinction between promoters and enhancers.
The close correspondence between enhancer- and promoter-binding proteins is the basis for the most widely accepted model of enhancer function that enhancers increase transcription initiation from promoters. In this model, the enhancer serves as a repository or a targeting site for DNA-binding proteins and associated proteins, including the mediator and RNA polymerase II, which are required to transcribe a gene. The enhancer-associated proteins can be “delivered” to the promoter by direct interactions between the promoter and enhancer with looping out of the intervening DNA. Such loops have indeed been detected with the aid of chromosome conformation capture (3C) assays. For example, the LCR in the murine β-globin locus, which is essential for efficient transcription of all β-like genes, is in physical proximity to the promoter active at the appropriate developmental stage (de Laat et al. 2008). This looping requires the transcription factor CTCF, which binds to DNase I-hypersensitive sites at the 5′ and 3′ ends of the locus (Splinter et al. 2006). Similarly, long-range loops have been detected in antigen receptor loci (Oestreich et al. 2006; Wuerffel et al. 2007) and cytokine gene loci in lymphocytes (Spilianakis and Flavell 2004; Sekimata et al. 2009). Implicit in this model is that promoter factors cannot get recruited or fail to get activated if the enhancer does not deliver the “goods,” including transcriptional coactivators, chromatin remodeling factors, and/or histone-modifying complexes. A stronger enhancer allows more molecules of polymerase to initiate and/or elongate transcription, thereby leading to higher transcriptional levels. Looping also extends the flexibility of different enhancers acting on the same gene promoter, as well as independent regulation of genes in the looped-out portion of the genome. Other models of delivering transcription components have been considered previously, such as tracking of important molecules along the genome from the enhancer to the promoter. In this case, it would be more difficult to independently regulate the intervening DNA between the enhancer and promoter. To date, however, there is no direct evidence for a tracking model of enhancer function.
An alternative view of enhancer function posits that enhancers increase the probability that a promoter will be activated to initiate transcription (Walters et al. 1995; Fiering et al. 2000). The crux of this model, which distinguishes it from the polymerase loading model described above, is that a promoter is sufficient in itself to fully activate transcription. However, in the absence of an enhancer, fewer promoters in a population of cells achieve this state; those that do, however, will transcribe the gene at the same level as when an enhancer is present. Experimentally, the two models can be evaluated only if enhancer activity is assessed at a single-cell level. Indeed, experiments designed to specifically address this issue have yielded results consistent with this idea. For example, both the SV40 enhancer (Weintraub 1988) and the enhancer of the β-globin LCR increase the proportion of stably transfected cells that express the associated reporter. Conversely, deleting the β-globin enhancer from stably transfected plasmids that were transcriptionally active resulted in variegated expression (Walters et al. 1996). That is, some cells expressed the enhancer-deleted reporter at the same level as an enhancer-sufficient reporter, whereas other cells did not express the reporter at all. Similar observations were made by deleting the intronic enhancer (Eμ) of the endogenous immunoglobulin heavy chain (IgH) gene in a B-cell hybridoma (Ronai et al. 1999, 2004). After enhancer deletion, IgH protein was expressed in only a subset of cells. In IgH-expressing cells, however, the level of expression was comparable with that in cells in which the enhancer was intact. Of particular interest, Ronai et al. (1999) determined that the “on” state of enhancer-deleted alleles was stably maintained for ∼100 cell divisions, whereas the “off” state lasted much longer. These observations lead to the idea of epigenetically propagated on and off states, wherein the transition between the states is governed by the Eμ enhancer. Recently, the digital all-or-none transcriptional responses have been studied by directly monitoring nascent transcripts in living cells, which indicated that genes are transcribed in pulses of a characteristic duration, frequency, and intensity (Larson et al. 2009). Moreover, models for stochastic fluctuations in gene expression have been proposed (Pedraza and Paulsson 2008). While the probabilistic model argues against delivery of transcription components, it is quite consistent with spatial juxtaposition of promoters and enhancers. Perhaps the increased probability of a promoter firing in the presence of an enhancer is caused by such promoter/enhancer proximity. Alternatively, enhancers could affect chromatin structure more broadly, thereby leading to unhindered accessibility of the promoter to transcription factors and RNA polymerase. In support of a chromatin model of enhancer function, juxtaposition of the Eμ enhancer with a promoter for bacteriophage T7, instead of a promoter for RNA polymerase II, allowed for efficient access of the prokaryotic promoter in pro-B cells of a transgenic mouse (Jenuwein et al. 1997). Thus, enhancer-dependent promoter accessibility was observed independently of detectable transcription by RNA polymerase II. Most likely, enhancers act by both mechanisms to stimulate promoter function.
Enhancer function II: initiation or maintenance
An aspect of enhancer function that has direct relevance to their mechanisms of action is whether the function of an enhancer is required once to initiate transcription from a promoter, or is required continuously to maintain transcription of a gene. The question arises from biochemical studies of transcription wherein once a preinitiation complex, composed of several general transcription factors, has been generated on a TATA box, it is sufficient to maintain several transcription initiation events. Thus, it is possible that, once an enhancer has activated a promoter, and therefore resulted in the formation of a functional promoter complex, the enhancer may no longer be required to maintain transcription from that promoter. However, the biochemical studies on which these ideas were based were carried out on naked DNA templates, and the stability of preinitiation complexes in more natural contexts has yet to be evaluated systematically. Moreover, these in vitro studies did not address what would happen to a promoter-based transcription preinitiation complex after DNA replication. Thus, whether or not an enhancer is required to maintain transcriptional activity of a promoter must be examined in the presence of cell division.
It is not a coincidence that the earliest studies designed to directly test the initiation versus maintenance question involved the first characterized viral and tissue-specific enhancers. Wang and Calame (1986) used a temperature-sensitive cell line that permitted SV40T antigen-dependent replication of an extrachromosomal plasmid to inducibly amplify the copy number of SV40 enhancers present in a cell. Concurrently, they recorded expression from a reporter plasmid that was kept at a fixed copy number, and was controlled by the SV40 enhancer. In this experimental design, amplification of SV40 enhancer sequences would titrate off SV40 enhancer-binding proteins from the SV40 enhancer that controlled reporter gene expression. Wang and Calame (1986) found that competition from SV40 enhancer sequences indeed reduced reporter activity if competition occurred concurrently with establishment of transcriptional competence of the reporter. However, if reporter transcription was established first, then a subsequent increase of SV40 enhancer sequences did not affect reporter expression. Wang and Calame (1986) interpreted the results to indicate that the SV40 enhancer was not required once the reporter was transcriptionally active.
Almost simultaneously Grosschedl and Marx (1988) developed an elegant strategy to test whether the tissue-specific IgH enhancer was required to maintain transcription of a functionally rearranged IgH gene. They generated constructs in which the IgH enhancer, together with a gpt gene as positive and negative selection marker, was flanked by recombination signal sequences that could be used to delete the enhancer after establishment of stable transfectants in an Abelson virus-transformed cell line. An enhancerless IgH gene located close to this cassette was shown to be dependent on the gpt-associated enhancer for transcription. Grosschedl and Marx (1988) found that IgH-expressing clones lost transcription upon recombination-induced deletion of the accompanying enhancer. Ergo, the IgH enhancer was continuously required to maintain IgH transcription. The significant differences in the experiment design of the two studies preclude direct comparison. However, the different results obtained underscore the importance of this question in terms of basic understanding of the mechanism of enhancer function.
The first analogous study to examine the role of enhancers in the context of an endogenous gene was carried out by Groudine and colleagues (Reik et al. 1998). They introduced cre and flp recombinase target sites into the human β-globin gene locus by targeted recombination. The human chromosome carrying the modified locus was transferred to mouse erythroleukemia cells by gene fusion. Having ascertained that the targeted human β-globin locus was transcriptionally active in these cells, Reik et al. (1998) transiently introduced cre, or flp, recombinase to delete different combinations of DNase I-hypersensitive sites that comprise the β-globin LCR. They found that transcription was significantly reduced on LCR-deleted alleles, leading to the conclusion that the LCR was continuously required to maintain high-level transcription of β-globin genes. Interestingly, LCR-deleted alleles retained several aspects of “open” chromatin, such as generalized DNase I sensitivity, indicating that these features were insufficient to load RNA polymerase II at the promoter. More recent studies have confirmed these chromatin structural conclusions in primary erythrocytes from LCR-deleted mice (Epner et al. 1998; Bender et al. 2000). However, these new studies are not directly pertinent to the initiation versus maintenance question, because LCR-deleted alleles were probably never transcribed at high levels.
A new definitive study
A recent study in Genes & Development from the Littman laboratory (Chong et al. 2010) addresses the initiation versus maintenance question in the most definitive way currently available. They analyzed the role of the T-lymphocyte-specific enhancer (E4p) that is located 13 kb 5′ of the murine Cd4 gene. During T-lymphocyte differentiation in the thymus, Cd4 is activated first in so-called double-positive (DP) thymocytes that express CD4 and CD8 coreceptors on the cell surface. DP thymocytes also express the heterodimeric T-cell receptor (TCR), and comprise the largest proportion of cells in the thymus. While most DP cells die in the thymus, a small proportion of DP cells are “positively selected” to further differentiate into single-positive (SP) thymocytes that express either a CD4 or CD8 coreceptor. SP thymocytes are the most mature T cells in the thymus and, upon export out of the thymus, generate the peripheral pool of functional CD4+ or CD8+ T cells. Earlier transgenic studies showed that E4p was by itself active in DP cells and both CD4+ and CD8+ SP cells. Lack of CD4 expression in CD8+ cells is presently understood to be determined by the Cd4 silencer (S), also characterized by the Littman laboratory (Taniuchi and Littman 2004). Germline deletion of E4p in the present study abolished CD4 expression on the majority of DP thymocytes. Despite a lack of CD4 expression in DP thymocytes, CD4 expression was evident on 40% of cells that had been positively selected and virtually 100% of cells that reached the most mature CD4+ SP stage in the thymus. However, the average level of CD4 surface expression was lower and more broadly distributed in E4p-deleted CD4+ SP cells in both the thymus and the spleen.
Chong et al. (2010) also found that this E4p-independent CD4 expression was lost upon proliferation of E4p-deleted CD4+ T cells. They inferred that a presently undefined regulatory sequence partially compensated for the absence of E4p after positive selection to activate Cd4 expression. However, this element did not fully recapitulate the pattern of expression of the normal locus. To rigorously determine whether loss of Cd4 expression in proliferating E4p-deleted cells indicated the continuous requirement for E4p, Chong et al. (2010) “floxed” E4p, allowing CD4 cells to develop normally. When E4p was deleted in mature CD4 T cells by retroviral expression of cre recombinase, CD4 expression was stably maintained over several rounds of cell division despite the absence of E4p. The striking conclusion is that CD4 T cells that developed with an intact E4p do not need it to maintain expression through cell division. Chong et al. (2010) conclude that the presence of E4p during development creates an epigenetic state that is propagated through proliferation without E4p.
Mechanisms of enhancer-induced transcriptional memory
These observations raise several important questions for future analyses. First, what is the mechanism by which E4p “marks” the Cd4 locus so that this information can be propagated through cell division even in the absence of E4p? Chong et al. (2010) addressed this issue by examining histone modifications associated positively or negatively with transcriptional activity, at various locations in the Cd4 locus. When E4p was deleted in the germline, they found that the Cd4 locus did not contain acetylated histone H3 (H3ac) or H3 trimethylated at Lys 4 (H3K4me3) in DP thymocytes (which do not express Cd4), whereas CD4+ thymocytes contained H3ac and low levels of H3K4me3, consistent with lower transcriptional activity of the E4p-deleted gene compared with E4p-sufficient alleles. During proliferation-induced variegation of CD4 expression in E4p-deleted spleen cells, the H3ac levels fell precipitously, whereas they were unchanged in wild-type CD4 cells. While these are important steps toward an answer, these observations do not really provide insight as to how early presence of E4p marks the locus to preserve H3ac during subsequent proliferation. Furthermore, a distinct mechanism may be responsible for lower and more variegated transcriptional activity of CD4 in E4p-deleted cells.
Consideration of the published literature provides some possibilities to understand these observations. One example is the liver-specific tyrosine-amino transferase (Tat) gene that is regulated by glucocorticoids. The inducible responsiveness to glucocorticoids has been mapped to a position ∼2.5 kb 5′ of the transcription initiation site that contains several binding sites for the glucocorticoid receptor and other transcription factors. This region of the gene undergoes rapid glucocorticoid-induced chromatin remodeling and DNA demethylation coincident with inducible gene transcription. Interestingly, although chromatin remodeling and transcription are reversible upon withdrawal of corticosteroids, the 2.5-kb region remains demethylated (Thomassin et al. 2001). Subsequent induction of the Tat gene occurs more rapidly and to higher levels. Thomassin et al. (2001) of this study found evidence for a methylation-sensitive transcription factor that binds to this region that may propagate the memory of the initial corticosteroid treatment. In a more physiological context, demethylation of this DNA region occurs during rat embryogenesis between fetal day 15 and birth, presumably in response to an increase in corticosteroid concentrations that occurs around this time. Notably, the Tat gene remains transcriptionally silent during this period of DNA demethylation. The working model is that demethylation of the −2.5-kb region “prepares” the gene for robust induction subsequently. The beauty of the model is that it is easy to visualize how the memory of a DNA demethylated region can be propagated even through many rounds of cell division.
A similar mechanism has been proposed for effective induction of interleukin 2 (IL-2) gene after a secondary stimulus (Bruniquel and Schwartz 2003; Murayama et al. 2006). Using human T lymphocytes, Murayama et al. (2006) showed that a specific CpG site in the IL-2 gene promoter was specifically demethylated after activation of the cells via the TCR. The demethylated region bound the constitutive transcription factor Oct-1, which remained bound to the IL-2 promoter after the activation stimulus was removed. Murayama et al. (2006) suggest that an Oct-1-bound demethylated promoter propagates the memory of prior IL-2 gene activation for effective responses upon secondary stimulation. The concept that emerges from both these examples is very similar. Namely, inducible DNA demethylation, and consequent binding of a transcription factor, marks a locus for efficient transcription during secondary stimulation (Fig. 1). We note that both these examples are of genes that are programmed to be inducibly activated during physiological responses. Whether these principles apply to other noninducible tissue-specific genes, such as the Cd4 gene studied by Littman and colleagues (Chong et al. 2010), or even to all tissue-specific genes, remains to be established. Chong et al. (2010) note that their earlier studies indicate that the CD4 gene is not controlled by DNA demethylation. However, that observation was in the context of silencer function, and it remains an open question whether DNA methylation events contribute to E4p-dependent memory in mature CD4 T cells.
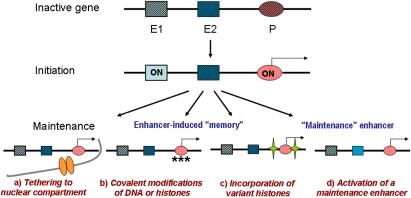
Figure 1.
Models of enhancer-induced transcriptional memory. The top line shows a hypothetical gene with two enhancers (E1 and E2) and a promoter (P) in the off state (hatched). (Middle line) Gene transcription is initiated by activation of a developmentally regulated enhancer (E1). Thereafter, gene transcription may be maintained by several mutually nonexclusive mechanisms discussed in the text, such as tethering to a specialized nuclear subcompartment (a); covalent epigenetic modifications, such as histone methylation and DNA demethylation of the promoter or other cis-regulatory sequences (stars) (b); incorporation of variant histones, such as H3.3 and H2A.Z (green stars) (c), and activation of a “maintenance” enhancer (E2) (d).
How else might a gene remain marked for proliferation-independent expression in the absence of an enhancer? Following up quantitative studies of gene transcription that involve the analysis of individual transcripts in real time (Larson et al. 2009), Chubb and colleagues (Muramoto et al. 2010) showed recently that the average transcription frequency of a modified act5 gene is maintained during cell divisions of Dictyostelium. By introducing multiple copies of the RNA recognition sequence for a bacteriophage MS2-GFP fusion protein into the 5′ untranslated region (UTR) of the act5 gene, they could determine that the modified act5 gene is transcribed in a characteristic pulse of 5.5 min per hour. Interestingly, mutations in the gene encoding the histone H3K4 methyltransferase set1 resulted in a loss of the maintenance of pulse frequency of act5 transcription between generations, without altering the steady-state level of transcription (Muramoto et al. 2010). In contrast, no effect on the maintenance of transcription frequency was observed in mutants for the H3K36 methyltransferase Set2 and for the DNA methyltransferase dnmA (Muramoto et al. 2010). These data suggest that H3K4 methylation may be required to propagate an existing transcription state, in addition to its widely recognized association with active transcription. Thus, H3K4 may present a mark that endows the daughter cell with a memory of the transcription program of the mother (Fig. 1). We note that H3K4 monomethylation has also been linked to a specific chromatin signature of enhancers (Cui et al. 2009), and future experiments will have to determine whether the H3K4 mark is also a component of enhancer-dependent transcriptional memory in higher eukaryotes.
Additionally, the histone variant H3.3 may be involved in propagating epigenetic memory of a transcriptionally active state. Ng and Gurdon (2008) studied the expression of MyoD gene in nuclear transplanted embryos that were generated by transferring nuclei from MyoD-expressing somite cells into enucleated fertilized eggs. They found that more than half of the resulting embryos expressed the MyoD gene in the wrong cell type. Ng and Gurdon (2008) inferred that this “inappropriate” expression resulted from a “memory” of the transcriptional state of the gene in nuclei used for transplantation, and provided evidence that it was mediated by H3.3. Although the mechanism by which H3.3 is targeted to transcriptionally active genes is not clear, the role of enhancers in establishing gene transcription suggests that they may be involved directly or indirectly. Once the variant histone is incorporated into an active gene, propagation of this state to daughter cells may provide the means to maintain transcriptional activity of the gene without the continued need for an enhancer. Implicit in this model is the notion that functional promoter complexes can be assembled in H3.3-marked domains in the absence of an enhancer. This view is reminiscent of the probabilistic model of enhancer activation and may happen, for example, by exclusion of the linker histone H1 from promoters (Braunschweig et al. 2009). H3.3 has also been shown to associate with another histone variant, H2A.Z (Jin et al. 2009), to form unstable double-variant nucleosomes that may increase transcription factor access to promoters to activate transcription. One reason why histone variants may be more attractive candidates to propagate an epigenetic state is because modifications like histone acetylation are present in a state of dynamic equilibrium, as evidenced, for example, by the huge increase in acetylated histone levels upon treatment of cells with histone deacetylase inhibitors such as trichostatin A.
Another mechanism to propagate the memory of gene activation could involve changes in the nuclear positioning of a gene (Fig. 1). Several recent studies highlight the relocation of tissue-specific genes from the nuclear periphery to more central parts of the nucleus (Ruault et al. 2008; Takizawa et al. 2008), the association of transcriptionally active genes in the RNA polymerase-enriched regions referred to as transcription factories (Sutherland and Bickmore 2009; Cook 2010), and the spatial association of functionally related genes (Spilianakis et al. 2005; Schoenfelder et al. 2010). Such observations indicate the presence of specialized nuclear subcompartments and regulated movements of genes in and out of them. In yeast, the GAL locus moves to the nuclear periphery concomitant with induction; this is mediated by subunits of the SAGA histone acetyltransferase complex (Cabal et al. 2006). Interestingly, peripherally located GAL1 and INO1 genes are associated with the histone variant H2A.Z, which is essential for their rapid reactivation (Brickner et al. 2007). It is likely that gene regulation due to nuclear location is also mediated by cis-regulatory sequences in higher eukaryotes. Indeed, LCR-deleted β-globin loci fail to move from the nuclear periphery during erythrocyte differentiation, and IgH Eμ enhancer-deleted alleles are preferentially located near the nuclear periphery compared with wild-type alleles (R Sen, unpubl.). It needs to be determined whether other classical enhancers also mediate nuclear movement of gene loci.
Concluding remarks
Accumulating evidence supports a broader role for enhancers beyond delivering transcriptional coactivators and/or RNA polymerase to responsive promoters. This broader role is most likely manifested at the level of chromatin structure, and may involve histone modifications, incorporation of variant histones, DNA demethylation, and/or changes in nuclear location. However, the effect of an enhancer on one or more of these chromatin structural features may vary depending on the gene. For example, deletion of the IgH enhancer Eμ results in significant reduction of histone acetylation up to several hundred kilobases (Chakraborty et al. 2009), whereas deletion of the β-globin LCR does not affect histone acetylation at all. Yet, both deletions lead to drastically reduced transcription and maintenance of deleted alleles at the nuclear periphery. Reduced histone acetylation characterizes enhancer-deleted alleles of all antigen receptor genes (Mathieu et al. 2000; McMurry and Krangel 2000), and it remains to be determined whether this is a special feature of these genes or, perhaps, a distinction between enhancers that work by themselves or in the context of LCRs. The chromatin structural model of enhancer function does not preclude a requirement for looping of enhancers to other parts of the locus. Indeed, one possibility is that structural chromatin changes created by enhancers may even be necessary to permit looping. The altered chromatin structure may facilitate recruitment of chromatin remodeling complexes or looping-associated DNA-binding proteins, such as CTCF, YY1, and SATB family members. The complexity of enhancer function is exemplified by the observation that the binding of YY1 to the Eμ enhancer impairs looping without affecting IgH locus transcription (Liu et al. 2007). Perhaps this kind of multitasking by enhancers is one reason for their complex organization and the presence of multiple protein-binding sites.
Finally, what about initiation versus maintenance models of enhancer function? Whereas the data presented by Chong et al. (2010) definitively demonstrate that E4p is required to initiate, but not maintain, Cd4 gene expression, it remains an open question whether this ability is a common feature of enhancers. It is likely that the dispensability of an enhancer after initiation of gene expression will depend on the presence of other regulatory sequences within the locus, the composition of promoters that are activated by an enhancer, and the regulatory complexities associated with the gene. For example, the expression of the Cd4 gene does not seem to require regulation after maturation of cells to the CD4 SP stage. Therefore, the “responsibility” of maintaining gene expression can be transferred to another less-complex enhancer or the Cd4 promoter itself. In contrast, genes that respond to signals by up- or down-regulation may continuously require an enhancer that ensures appropriate modulation of transcriptional activity. Irrespective of the properties of individual enhancers, they initiate a cascade of locus-specific events. Analysis of the sequence of these events and understanding of the function of the molecular players involved remain important goals for the future.
Acknowledgments
Work in our laboratories is supported by the Intramural Research Program of the National Institute on Aging (Baltimore, MD) (R.S.) and the Max Planck Society and German Research Foundation (R.G.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1930610.
References
- Bender MA, Bulger M, Close J, Groudine M 2000. β-Globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol Cell 5: 387–393 [DOI] [PubMed] [Google Scholar]
- Braunschweig U, Hogan GJ, Pagie L, van Steensel B 2009. Histone H1 binding is inhibited by histone variant H3.3. EMBO J 28: 3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5: e81 doi: 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH 2003. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol 4: 235–240 [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M 2010. Enhancers: The abundance and function of regulatory sequences beyond promoters. Dev Biol 339: 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773 [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Perlot T, Subrahmanyam R, Jani A, Goff PH, Zhang Y, Ivanova I, Alt FW, Sen R 2009. A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation. J Exp Med 206: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MMW, Simpson N, Ciofani M, Chen G, Collins A, Littman DR 2010. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes & Dev 24: 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR 2010. A model for all genomes: The role of transcription factories. J Mol Biol 395: 1–10 [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Nikolajczyk BS, Sen R 1998. Exploring functional redundancy in the immunoglobulin mu heavy-chain gene enhancer. Mol Cell Biol 18: 6870–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, Splinter E, Grosveld F 2008. Three-dimensional organization of gene expression in erythroid cells. Curr Top Dev Biol 82: 117–139 [DOI] [PubMed] [Google Scholar]
- Epner E, Reik A, Cimbora D, Telling A, Bender MA, Fiering S, Enver T, Martin DI, Kennedy M, Keller G, et al. 1998. The β-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse β-globin locus. Mol Cell 2: 447–455 [DOI] [PubMed] [Google Scholar]
- Fiering S, Whitelaw E, Martin DI 2000. To be or not to be active: The stochastic nature of enhancer action. Bioessays 22: 381–387 [DOI] [PubMed] [Google Scholar]
- Grosschedl R, Marx M 1988. Stable propagation of the active transcriptional state of an immunoglobulin μ gene requires continuous enhancer function. Cell 55: 645–654 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Forrester WC, Fernandez-Herrero LA, Laible G, Dull M, Grosschedl R 1997. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385: 269–272 [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Singer RH, Zenklusen D 2009. A single molecule view of gene expression. Trends Cell Biol 19: 630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, et al. 2007. Yin Yang 1 is a critical regulator of B-cell development. Genes & Dev 21: 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu N, Hempel WM, Spicuglia S, Verthuy C, Ferrier P 2000. Chromatin remodeling by the T cell receptor (TCR)-β gene enhancer during early T cell development: Implications for the control of TCR-β locus recombination. J Exp Med 192: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry MT, Krangel MS 2000. A role for histone acetylation in the developmental regulation of VDJ recombination. Science 287: 495–498 [DOI] [PubMed] [Google Scholar]
- Muramoto T, Muller I, Thomas G, Melvin A, Chubb JR 2010. Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr Biol 20: 397–406 [DOI] [PubMed] [Google Scholar]
- Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, et al. 2006. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J 25: 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB 2008. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 10: 102–109 [DOI] [PubMed] [Google Scholar]
- Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM 2006. Regulation of TCRβ gene assembly by a promoter/enhancer holocomplex. Immunity 24: 381–391 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, de Laat W, Grosveld F 2008. β-Globin regulation and long-range interactions. Adv Genet 61: 107–142 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, Paulsson J 2008. Effects of molecular memory and bursting on fluctuations in gene expression. Science 319: 339–343 [DOI] [PubMed] [Google Scholar]
- Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M 1998. The locus control region is necessary for gene expression in the human β-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol 18: 5992–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronai D, Berru M, Shulman MJ 1999. Variegated expression of the endogenous immunoglobulin heavy-chain gene in the absence of the intronic locus control region. Mol Cell Biol 19: 7031–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronai D, Berru M, Shulman MJ 2004. The epigenetic stability of the locus control region-deficient IgH locus in mouse hybridoma cells is a clonally varying, heritable feature. Genetics 167: 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruault M, Dubarry M, Taddei A 2008. Re-positioning genes to the nuclear envelope in mammalian cells: Impact on transcription. Trends Genet 24: 574–581 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB 2009. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-γ locus. Immunity 31: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645 [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes & Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA 2009. Transcription factories: Gene expression in unions? Nat Rev Genet 10: 457–466 [DOI] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T 2008. The meaning of gene positioning. Cell 135: 9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi I, Littman DR 2004. Epigenetic gene silencing by Runx proteins. Oncogene 23: 4341–4345 [DOI] [PubMed] [Google Scholar]
- Thomassin H, Flavin M, Espinas ML, Grange T 2001. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J 20: 1974–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DI 1995. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci 92: 7125–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin DI 1996. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes & Dev 10: 185–195 [DOI] [PubMed] [Google Scholar]
- Wang XF, Calame K 1986. SV40 enhancer-binding factors are required at the establishment but not the maintenance step of enhancer-dependent transcriptional activation. Cell 47: 241–247 [DOI] [PubMed] [Google Scholar]
- Weintraub H 1988. Formation of stable transcription complexes as assayed by analysis of individual templates. Proc Natl Acad Sci 85: 5819–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL 2007. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity 27: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]