Abstract
Background
The high and low alcohol preferring (HAP1 and LAP1) mouse lines were selectively bred for differences in alcohol intake. The HAP1 and LAP1 mice are essentially noninbred lines that originated from the outbred colony of HS/Ibg mice, a heterogeneous stock developed from intercrossing 8 inbred strains of mice.
Methods
A total of 867 informative SNPs were genotyped in 989 HAP1 × LAP1 F2, 68 F1s, 14 parents (6 LAP1, 8 HAP1), as well as the 8 inbred strains of mice crossed to generate the HS/Ibg colony. Multipoint genome wide analyses were performed to simultaneously detect linked QTLs and also fine map these regions using the ancestral haplotypes.
Results
QTL analysis detected significant evidence of association on 4 chromosomes: 1, 3, 5, and 9. The region on chromosome 9 was previously found linked in a subset of these F2 animals using a whole genome microsatellite screen.
Conclusions
We have detected strong evidence of association to multiple chromosomal regions in the mouse. Several of these regions include candidate genes previously associated with alcohol dependence in humans or other animal models.
Keywords: Quantitative Trait Locus, Alcohol Consumption, Association
In the United States, 17.6 million people, about 1 in every 12 adults, abuse alcohol or are alcohol dependent (http://www.niaaa.nih.gov). The use of alcohol occurs in virtually every facet of our society, over many age groups, and is a leading cause of morbidity and premature death in our country (Caces et al., 1995; Campbell et al., 1995; DeBakery et al., 1995).
It has been well documented that genetic factors contribute to the etiology of human alcoholism (Cadoret et al., 1980; Cloninger et al., 1981; Goodwin, 1979; Heath et al., 1997; Kendler et al., 1994; Pickens and Svikis, 1991); however, it is clearly a complex disease comprised of many behavioral phenotypes and influenced by multiple genes (Goldman, 1993). Researchers have attempted to genetically dissect alcoholism into its constituent traits and identify the genes of influence. In human populations, these efforts are complicated by genetic heterogeneity, complex inheritance, diagnostic uncertainties, and environmental factors (Gershenfeld et al., 1997). This has led to the development of animal models to elucidate the genetic basis of alcohol-seeking behavior and to contribute to our knowledge of the etiology of alcoholism.
The use of animal models, with phenotypes similar to human alcoholism, may provide important genetic clues that will improve the efficiency of identifying genes underlying human alcohol-seeking behavior. It may not be possible to develop a perfect model, but traits of the disease (i.e., alcohol preference) may be modeled and studied to yield an understanding of particular aspects of the human disease. One way to study the contributions of individual quantitative behavioral traits, such as alcohol preference, is to locate the genetic information that encodes them (Grisel, 2000). This is accomplished through quantitative trait loci (QTL) mapping. This method has proven to be successful in identifying QTL associated with alcohol phenotypes.
The present study takes advantage of noninbred mouse lines, the high alcohol preferring and low alcohol preferring (HAP and LAP) mice, selectively bred for differences in alcohol preference and consumption (Grahame et al., 1999). The HAP mice consume pharmacologically significant quantities of alcohol; whereas, the LAP mice consume little if any alcohol (Grahame et al., 1999). The HAP and LAP mice originated from the outbred colony of HS/Ibg mice maintained at the Institute of Behavioral Genetics in Boulder, Colorado. This heterogeneous stock, with a wide range of alcohol preference, was developed from intercrossing 8 inbred strains of mice: A, AKR, BALB/c, C3H/2, C57BL/6, DBA/2, Is/Bi, and RIII (McClearn et al., 1970). Therefore, the HS/Ibg mice can, in principle, have 8 alleles at any given locus at which all parent strains differ; although, some fixation of alleles and loss of the original diversity are likely to have occurred in the many generations elapsing as these mice were originally derived (Grahame et al., 1999).
In a recent study, microsatellite markers were used to carry out a genome wide screen in 432 HAP1 × LAP1 F2 mice to detect quantitative trait loci (QTL) that influence alcohol preference and consumption. This study identified a large effect QTL on chromosome 9 (Bice et al., 2006) with a LOD score of 5.04. We have now expanded upon this earlier study and have genotyped the previous 432 F2 mice as well as an additional 557 HAP1 × LAP1 F2 mice as part of a whole genome screen using a denser set of single nucleotide markers. We have also employed a powerful analytic approach allowing us to utilize ancestral recombination as well as recombination in the F2 animals to perform both QTL detection and fine mapping (Mott et al., 2000). This is implemented computationally by calculating the probability that a marker genotype or haplotype is inherited identical by descent from each of the 8 inbred founders crossed to generate the HS/IBG and subsequently the HAP1 × LAP1 F2. This study design and analytic approach were successfully implemented in a recent study of 1904 heterogeneous stock animals to detect 843 QTLs for 97 traits (Valdar et al., 2006).
METHOD
HAP1 × LAP1 F2 Progenitors
The HAP1/LAP1 progenitors were taken from the 23rd and 24th generations of selection. During this time, selection pressure continued to be applied. Mean alcohol consumption scores for the 8 HAP1 and 6 LAP1 progenitors and 989 F2 animals are shown in Table 1.
Table 1.
Alcohol Consumption
| Sample | n | Mean | SEM |
|---|---|---|---|
| HAP1 progenitors | 8 | 23.47 | 1.26 |
| LAP1 progenitors | 6 | 0.87 | 0.05 |
| All F2 | 989 | 5.43 | 0.18 |
| F2 Family 1: male | 113 | 2.29 | 0.19 |
| F2 Family 1: female | 120 | 5.26 | 0.44 |
| F2 Family 2: male | 126 | 4.31 | 0.34 |
| F2 Family 2: female | 112 | 12.64 | 0.68 |
| F2 Family 3: male | 118 | 1.28 | 0.11 |
| F2 Family 3: female | 120 | 4.34 | 0.41 |
| F2 Family 4: male | 152 | 4.02 | 0.25 |
| F2 Family 4: female | 128 | 9.65 | 0.51 |
SEM, standard error of measurement.
All values are in g/kg/d.
Development of HAP1 × LAP1 F2 Mice
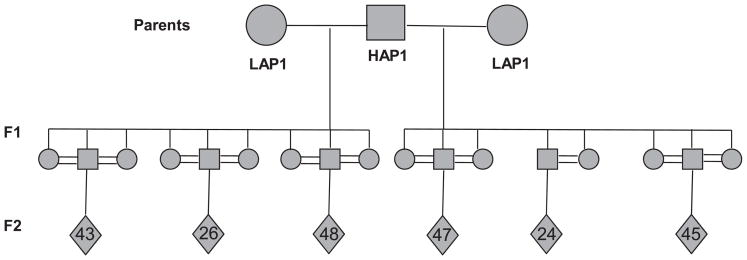
Four independent, 3 generational families were established for the purpose of creating the F2 animals used in this study. For family 1, 1 male HAP1 mouse was paired with 2 female LAP1 mice to produce 17 F1 progeny. Family 2 consisted of 2 male LAP1 mice paired with 2 femaleHAP1 mice to produce 16 F1 progeny. For family 3, 1 male HAP1mouse was paired with 2 female mice to produce 18 F1 progeny. Family 4 consisted of 1 male LAP1 mouse paired with 4 female HAP1 mice to produce 18 F1 progeny; 2 additional parental females were added to family 4 because fecundancy was low. Within each family, the F1 mice were intercrossed to produce a total of 989 F2 mice: Family 1, 2, 3, and 4 each produced 233, 238, 238, and 280 F2 mice, respectively. Figure 1 provides an example of the breeding pedigree used to develop the F2 mice for this study.
Fig. 1.
Breeding scheme for the HAP1 × LAP1 F2 mice. Shown is the pedigree for the breeding of family 1. Circles represent female mice; squares represent male mice; diamonds represent the total number of F2 offspring produced by each F1 pair. Cross-joined lines indicate mating pairs. Those with a single line are related animals that are mated; those with a double line are related animals (consanguineous mating).
Sample Preparation for CIDR
The study sample consisted of DNA samples from all 989 F2 animals as well as 68 F1 animals and the 8 HAP1 and 6 LAP1 progenitors. In addition, the dataset also included 1 DNA sample from a male animal from each of the 8 inbred strains (A/J, AKR/J, BALB/cJ, C3H/2J, C57BL/6J, DBA/2J, Is/CamRKJ, and RIII/ImrNhsJ). DNA was isolated in our laboratory using the Puregene kit (Gentra Systems, Minneapolis, MN) according to the manufacturer’s protocol.
This study applied for and was granted access to the genotyping facility at the Center for Inherited Disease Research (CIDR) operated by National Human Genome Research Institute (NHGRI). CIDR genotyped the Illumina Medium Density Mouse Linkage Panel which consists of 1,449 SNPs chosen from the Wellcome-CTC Mouse Strain SNP Genotype Set (http://www.well.ox.ac.uk/mouse/ INBREDS, Valdar et al., 2006). The SNPs included in this mouse linkage panel were selected based on available genotypic data from 10 inbred strains (129S1/SvlmJ, AKR/J, BALB/cJ, C3H/H3J, C57BL/6J, CBA/J, DBA/2J, FVB/NJ, NOD/LtJ, and SJL/J). The panel contains approximately 3 SNPs in each 5 Mb interval across the genome and was selected such that at least 1 SNP in each interval is informative for 85% of the 10 strain combinations.
A total of 25 duplicate samples were included in the genotyped dataset. The genotypic concordance rate for the duplicate samples was 100%(0 event/23,882 paired genotypes), as reported by CIDR. All genotypic data were evaluated for non-Mendelian inheritance in the 3 generations of marker data using the program PEDCHECK (O’Connell and Weeks, 1998). All genotypic inconsistencies (0.1% error rate) were reviewed and resolved or removed prior to the QTL analyses.
Phenotypic Measure
To determine ethanol consumption scores, mice at 45 days of age were individually caged and allowed to drink from two 25 ml-graduated cylinders. One cylinder contained 10% ethanol (v/v) and the other contained distilled water. Food was available ad libitum. The volumes consumed were measured 3 times per week, and the cylinders containing fluids were switched to control for position bias. Mice were weighed on the first Monday of each week during testing. Testing continued for a period of 4 weeks. Consumption scores were defined as grams per kilogram per day averaged over a 4-week access period. The drinking procedure was identical to that described in Grahame et al., 1999 for the selection of the HAP and LAP mice.
Among the 4 families, there was extensive variation in the distribution of the alcohol consumption values as well as substantial gender effects within each family (see Table 1). Such heterogeneity can result in spurious inflation of the resulting QTL analysis results. Therefore, both family and gender effects were included in the model, yielding more robust analytic results.
Genome Scan
We employed a multipoint dynamic-programming algorithm and regression models to perform genomewide QTL analyses (HAPPY; Mott et al., 2000; and custom code written in R). Given genotypes of the heterogeneous stock animals and the founders, HAPPY uses a hidden Markov model to calculate the expected frequency FLi(s, t) of a haplotype pair from strains s and t in the marker interval L of animal i, and also calculates the expected marginal frequency ALi (s) of haplotype s. We modeled the effect of each locus genotype on the phenotype by fitting the linear model
where for animal i of sex w[i] and family f[i], yi is the phenotype (log-transformed to approximate symmetry and normality), βw is the effect of sex w, βf is the effect of family f, and gi is either an 8-vector of elements ALi(s) for all s, representing an additive genetic model, or a 36-vector of elements FLi (s, t) for all {s, t}, representing an additive plus dominance (i.e., “full”) genetic model. For either genetic model we tested for the presence of a QTL using a partial F-test that evaluates the probability that βg ≠ 0.
To evaluate the genomewide significance of the QTL analyses, we performed genome scans on simulated phenotypes that were affected by only covariate and sibship effects. Specifically, we fit the linear mixed effects model E(yi) = μ + βw[i] + βf [i] + uk[i] where uk[i] is the effect of sibship k of animal i and , and then simulated new values of the phenotype for each animal from this fitted model. A total of 200 replicates were generated and then analyzed in the same way as was performed in the original data. The most significant result (i.e., highest logp, where logp = logarithm to the base 10 of the F-test p-value) from each genome scan was recorded. A generalized extreme value distribution was fitted to the 200 simulated maximum-logp scores, and estimated quantiles from this fitted distribution were used to estimate genome-wide significance thresholds (Valdar et al., 2006). This highly conservative approach gave 5% thresholds of logp = 6.78 and 6.90 for the additive and full genetic models, respectively.
Multiple QTL Analysis
The model averaging approach of Valdar and colleagues (2006) was used for the multiple QTL analysis. First, a selection of marker intervals were chosen to represent candidate QTL peaks. An interval was chosen as a peak if it had the highest logp above the genomewide threshold of all markers within a radius of 5 cm. The animals were then re-sampled with replacement to create bootstrap samples of the F2s. At each resample, a forward-selection algorithm was used to select uncorrelated candidate QTL peaks. At each step in the forward selection, the effect of adding each candidate QTL was tested conditional upon the presence of the covariates and the previously selected QTLs using a partial F-test. The QTL with the most significant partial-logp was added to the QTL set provided its partial logp exceeded the genomewide significance level. The procedure terminated when no more significant QTLs could be added. From 1000 bootstraps, we counted the proportion of times each QTL was selected to give an empirical model inclusion probability that the QTL association is robust. Like the analysis of Valdar and colleagues (2006), we report multiple QTL data for the additive model only; unlike that analysis, we included the X-chromosome in our multiple QTL models, statistically treating hemizygous male genotypes as if they were homozygote.
QTL Confidence Intervals
To assess the uncertainty in location of detected QTLs, we determined confidence intervals using positional bootstrapping (Visscher et al., 1996). For a candidate QTL peak, we rescaned all marker intervals within a 20 cm radius using a bootstrap resample of the data and then recorded the maximally associated interval. We repeated this procedure 500 times to generate a distribution of the maxima and used the region covered by the central 95% of this distribution as our estimate of the confidence interval.
RESULTS
Although 1,449 SNPs were genotyped by CIDR, data for only 917 SNPs were released by CIDR at the conclusion of genotyping. For these 917 SNPs both alleles were observed within our genotyped samples. The genotypic data for the remaining 532 SNPs included in the Illumina Medium Density Mouse Linkage Panel were not released by CIDR for various reasons. For 34 SNPs, the marker was informative, but the SNP genotypes were not released due to poor performance. In the case of a further 16 SNPs, the marker was informative, but more than 3 genotypes were observed. For the remaining 482 SNPs, only 1 allele was observed and the SNP was not informative in our sample.
Further quality assessment resulted in the removal of 50 additional SNPs. For 48 SNPs, genotypic data was missing for at least one of the inbred strains and therefore the SNP could not be used in the analysis. Two SNPs were removed from further analysis because an allele was present in the F2 but was not observed in any of the 8 inbred strains. Ten additional SNPs were removed because genotypic data were missing for the entire sample. The final dataset consisted of 989 F2 animals with genotypic data for 822 autosomal SNPs and 45X chromosome SNPs. The average physical distance between informative SNPs was 2.7 Mb.
We also evaluated whether the SNPs to be included in the analysis were sufficiently informative to provide the needed power to detect QTL across the genome. To address this issue, we computed the proportion of genotyped SNPs with minor allele frequency (MAF) less than 10%. When utilizing the rather limited number of samples available in the HAP1/LAP1 generation, 15.1% of the SNPs included in the analysis had a MAF less than 10%. This estimate was quite similar to that estimated in the F1 generation (15.0%) and from the nonindependent F2 generation (15.6%). Thus, the SNPs selected for genotyping in this sample are still overall relatively informative in this particular population of mice.
Review of the genotypic data provided further insight regarding the effect of the breeding strategy used to maintain the HAP1 and LAP1 animals. Among the 8 HAP1 and 6 LAP1 mice genotyped, the average number of homozygous markers per animal was 616 and 596, respectively. When the genotypes were reviewed at each SNP, 231 markers were homozygous for the same allele in the HAP1 mice and 228 markers were homozygous for the same allele in the LAP1 mice.
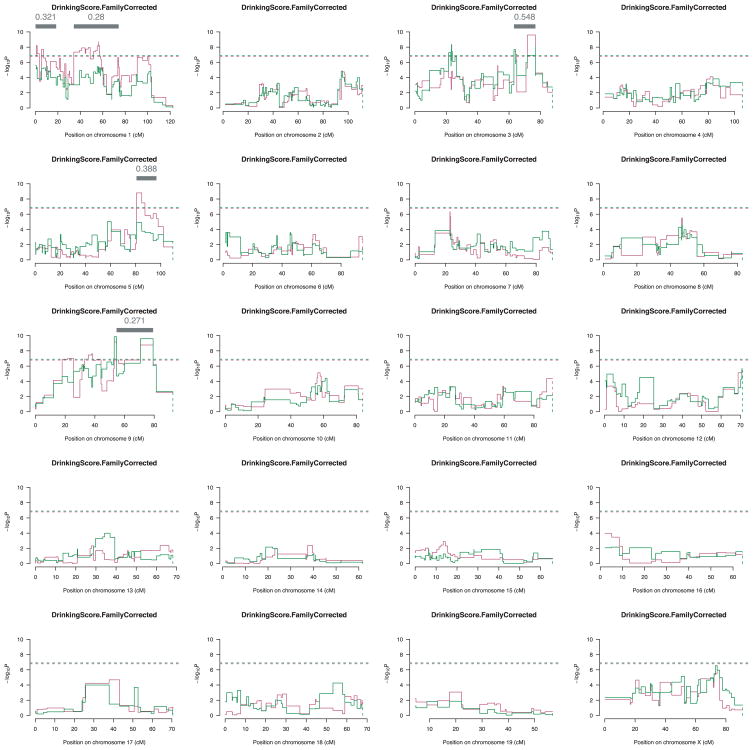
QTL analysis detected significant evidence of association to 4 chromosomes: 1, 3, 5, and 9 (Fig. 2). On chromosome 1, 2 regions exceeded the genomewide screening threshold. In each region, multiple SNPs support the evidence of association and the data appear more compatible with an additive model of genetic effect. In the more centromic QTL, the primary QTL is centered on rs13475706 located at 1.0 cm; therefore, 95% confidence interval can only extend distally and includes nearly 19 cm. The multiple QTL model inclusion probability of this QTL is 0.32. The second QTL on chromosome 1 is also broad with the most associated SNP being rs13476012 at 56 cm. The 95% confidence interval includes nearly 40 cm, the maximum that can be estimated (34 to 74 cm). The inclusion probability for this QTL is 0.28.
Fig. 2.
Genomewide analysis results from 989 F2 progeny. Results are shown for the additive (red) and full (green) genetic models. The Y-axis gives the negative log10 of the p value (−logp) of the test of association. The X-axis gives the chromosomal position of the analyzed SNPs. The additive model is shown with a red line, while the full model is shown with the green line. The gray bar denotes the 95% confidence interval for QTLs that exceed the genomewide significance threshold and which also have a greater than 95% posterior probability.
On chromosome 3, while there are several small regions that exceed the genomewide threshold of significance; however, the primary QTL is in the more distal region and best fits an additive model. This QTL is centered on rs13477439 located at 74 cm with a 95% confidence interval encompassing 14 cm (63 to 77 cm). The inclusion probability for this QTL is 0.55. On chromosome 5, a single QTL was identified in the more distal region of the chromosome, with the most associated SNP (rs8239888) located at 83 cm. This QTL spans a 95% confidence interval of 16 cm (81 to 97 cm). The inclusion probability of this QTL is 0.39.
On chromosome 9, the genomewide significance threshold is exceeded in several regions of the chromosome, but the primary evidence for a QTL is contained in the more distal region of the chromosome. The SNP on which the confidence interval is based is rs13480399 located at 75 cm. The 95% confidence interval spans 25 cm (55 to 79 cm). The inclusion probability of this QTL is 0.27.
DISCUSSION
We have employed a relatively novel approach to detect QTLs contributing to the variation in alcohol consumption in an F2 cross of phenotypically selected, noninbred mice strains. Using genotypic data from the 8 inbred lines crossed to generate the progenitors of the parental animals, we were able to demonstrate significant evidence of association to multiple chromosomal regions.
In previous analyses of a subset of the HAP1 × LAP1 F2 (Bice et al., 2006) that employed microsatellite markers with an average intermarker distance of 11.2 cm across the mouse genome, we detected significant evidence of linkage to chromosome 9 with a LOD score of 5.04. This QTL was estimated to account for 11% of the phenotypic variance associated with the phenotype and best fit an additive model of inheritance. Gender-specific analyses supported a greater effect of the QTL among female mice (LOD score = 5.19; p = 0.0008) than the male mice (LOD score = 1.19; p = 0.715). In the results provided herein of an expanded set of HAP1 × LAP1 F2, we were able to again detect strong evidence of association to chromosome 9 in the same region previously reported; however, we did not observe significant evidence of a sex-specific QTL effect. The chromosome 9 QTL (30 to 40 cm) has also been linked to consumption/preference in multiple other mouse QTL studies (Belknap and Atkins, 2001; Phillips et al., 1998; Tarantino et al., 1998). The fact that we detected a larger number of QTLs in the present study, compared to the initial F2 study, is not surprising because we doubled our sample size, increased marker coverage, and used a more powerful analytical method.
There are interesting candidate genes in each of the QTL regions identified in this study that have been implicated in the alcohol drinking phenotype in other studies. The chromosome 9 QTL region is syntenic to chromosome 8 (q24) in the rat and chromosome 11 (q22-23) and 15 (q21-q24) in the human. In the region between 55 and 79 cm, an interesting candidate gene is Cck, cholecystokinin, located at 71 cm. Cck is a peptide hormone of the gastrointestinal system responsible for stimulating the digestion of fat and protein. Recent evidence has suggested that it also plays a major role in inducing drug tolerance to opioids like morphine and heroin, and is partly implicated in experiences of pain hypersensitivity during opioid withdrawal (Fukazawa et al., 2007; Kissin et al., 2000). Consistent with this hypothesis, investigators have identified QTLs for cocaine-related behavior (70 cm) and behavioral responses to methamphetamines (71 cm) within this region (Grisel et al.,1997; Jones et al., 1999; Palmer et al., 2005).
On chromosome 1, the QTL region around 0 to 19 cm includes Oprk1, the opioid receptor, kappa 1 gene (at 5.5 cm). Opioid receptors and their endogenous peptide ligands play important roles in the reward and reinforcement of drugs such as heroin, cocaine, and alcohol (Gerra et al., 2007; Xuei et al., 2006). Xuei and colleagues (2006) found that variations in the genes encoding the kappa-opioid receptor (OPRK1) and its ligand (PDYN) are associated with the risk for alcohol dependence. Other mouse and rat studies have also identified QTLs for alcohol preference within this narrowed QTL region (Peirce et al., 1998; Whatley et al., 1999), including a QTL on rat chromosome 5 in the high and low alcohol drinking (HAD and LAD) rats (Carr et al., 2003). In the QTL region between 34 and 74 cm, QTLs for cocaine-induced activation, morphine-withdrawal severity, behavioral response to methamphetamines, seizure susceptibility, and ethanol induced loss of righting reflex have been identified (Bennett and Carosone-Link, 2006; Ferraro et al., 1999; Gill and Boyle, 2003; Grisel et al., 1997; Kest et al., 2004).
On chromosome 3, the region between 63 and 76 cm contains the alcohol dehydrogenase polypeptide genes, Adh1, 4, 5, 6a, 6b, and 7, which are the key enzymes responsible for the oxidation of alcohol to acetaldehyde (Edenberg, 2007; Luo et al., 2006). SNPs in ADH4 have been associated with an increased risk for alcohol dependence (Edenberg and Foroud, 2006; Luo et al., 2006) while variation in ADH1 and ADH7 has been shown to reduce the risk of alcohol dependence (Edenberg, 2007; Osier et al., 2004).
We employed stringent significance thresholds. It is known that thresholds based on permutation tests are anticonservative for populations with family structure (Churchill and Doerge, 2008). Because the F1 parents in our design are in fact progeny from outbred stock, our F2 population is likely to have family structure beyond that of a traditional F2. Therefore, we estimated a set of thresholds that takes into account sibship effects to establish a strict upper threshold. Furthermore, there were significant differences in alcohol consumption in the 4 “families” used in the analysis. This may suggest that there are unique genes contributing to alcohol preference in certain families. By employing sibship effects, which inherently also correct for family effects, we have increased our ability to detect QTL that may only be segregating in some of the families.
Using multiple QTL modeling, we sought to eliminate false-positive associations due to long-range linkage disequilibrium effects of complex pedigree structure. Determining model inclusion probabilities for each QTL allowed us to prioritize those QTL regions most likely to harbor a gene(s) contributing to the variation in alcohol preference. Of note, the 5 QTL regions on the 4 chromosomes all have a greater than 25% inclusion probability, a threshold employed in previous studies (Valdar et al., 2006). Several of the QTLs, particularly those on chromosomes 1 and 9, have relatively large 95% confidence intervals. In estimating the confidence intervals for this study, we have employed an approach that limits the search radius of confidence intervals to 20 cm in either direction from the most associated SNP. We chose this cut off as a compromise between 2 competing factors. First, because our cross offers higher resolution than an F2, choosing a wider search radius causes the positional bootstrap procedure to often include other nearby QTLs and therefore result in poorly identified confidence intervals. Second, a tighter search radius creates intervals that are artificially small. On chromosomes 1 and 9, we thus speculate that the confidence intervals are broad because there are likely to be multiple small effect QTLs within the interval. Review of the data from Fig. 2 would seem to support this hypothesis, as each broad interval typically encompasses what could be considered a distinct QTL interval.
A potential limitation of this study was the genotyping of a set of SNPs that were not selected for their informativeness in this particular cross but were selected to maximize the informativeness across many mouse lines. As a result, there are likely to be some regions of the genome in which an optimal set of SNPs were not genotyped and therefore, the ability to detect a QTL may be suboptimal. To evaluate the likely scope of this limitation, we first examined not only the average spacing between informative SNPs, but also identified the largest gaps in genomic coverage. While our average intermarker distance was 2.7 Mb, we did have 23 intervals in which the distance between markers was greater than 10 Mb. These gaps were not clustered in any specific genomic region but rather were scattered on different chromosomes. The likely effect of the incomplete coverage of the entire genome and the more limited informativeness of a subset of markers due to low MAF is reduced power to detect all relevant QTL. These limitations are unlikely to result in the identification of erroneous QTLs.
The strength of the present study was our powerful analytic approach, utilizing both ancestral recombination and F2 recombination. This methodology allowed us to detect and at the same time fine map QTL for alcohol consumption to small chromosomal intervals in the noninbred HAP1 and LAP1 mice, thus making this strategy relatively cost effective as compared to a more traditional inbred F2 design that typically results in large QTL intervals. The QTL regions identified in the current study can now be narrowed further by genotyping additional SNPs within our large F2 sample and inbred ancestral strains. Using this approach to identify and narrow QTL regions is a powerful method to reduce the number of candidate genes that after characterization will provide targets for prevention intervention in the treatment of alcoholism in humans.
Acknowledgments
This work was supported by a grant from the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health (Grant R01 AA015933) and APC of P60 AA007611-21. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract Number N01-HG-65403. William Valdar is funded by a grant from the European Union Framework 6 Programme, contract no: LHSG-CT-2003-503265. We also wish to give special thanks to Christina Best for developing and phenotyping the F2 mice that were used in the genome screen. This study was supported by NIH/NIAAA Grant R01 AA015933, N01-HG-65403, APC of P60 AA007611-21, and LHSG-CT-2003-503265.
References
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P. Replication of small effect quantitative trait loci for behavioral traits facilitated by estimation of effect size from independent cohorts. Genes Brain Behav. 2006;5:404–412. doi: 10.1111/j.1601-183X.2005.00174.x. [DOI] [PubMed] [Google Scholar]
- Bice PJ, Foroud T, Carr LG, Zhang L, Liu L, Grahame NJ, Lumeng L, Li TK, Belknap JK. Identification of QTLs influencing alcohol preference in the high alcohol preferring (HAP) and low alcohol preferring (LAP) mouse lines. Behav Genet. 2006;36:248–260. doi: 10.1007/s10519-005-9019-6. [DOI] [PubMed] [Google Scholar]
- Caces M, Stinson FS, Dufour MC. Trends in alcohol-related morbidity among short-stay community hospital discharges, United States. NIAAA Surveillance Report #3 1995 [Google Scholar]
- Cadoret RJ, Cain CA, Grove WM. Development of alcoholism in adoptees raised apart from alcoholic biologic relatives. Arch Gen Psychiatry. 1980;37:561–563. doi: 10.1001/archpsyc.1980.01780180075008. [DOI] [PubMed] [Google Scholar]
- Campbell KE, Zobeck TS, Bertolucci D. Trends in alcohol-related fatal traffic crashes, United States, 1977–1993. NIAAA Surveillance Report #3 1995 [Google Scholar]
- Carr LG, Habegger K, Spence J, Ritchotte A, Liu L, Lumeng L, Li TK, Foroud T. Analyses of quantitative trait loci contributing to alcohol preference in HAD1/LAD1 and HAD2/LAD2 rats. Alcoho Clin Exp Res. 2003;27:1710–1717. doi: 10.1097/01.ALC.0000097161.51093.71. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Naïve application of permutation testing leads to inflated type I error rates. Genetics. 2008;178:609–610. doi: 10.1534/genetics.107.074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR, Bohman M, Sigvardsson S. Inheritance of alcohol abuse. Cross-fostering analysis of adopted men. Arch Gen Psychiatry. 1981;38:861–868. doi: 10.1001/archpsyc.1981.01780330019001. [DOI] [PubMed] [Google Scholar]
- DeBakery SF, Stinson FS, Grant BF, Dufour MC. Liver cirrhosis mortality in the United States, 1970–92. NIAAA Surveillance Report #37 1995 [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, St Jean P, Schork NJ, Mulholland N, Ballas C, Schill J, Buono RJ, Berrettini WH. Mapping loci for pentylenetetrazol-induced seizure susceptibility in mice. J Neurosci. 1999;19:6733–6739. doi: 10.1523/JNEUROSCI.19-16-06733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Maeda T, Kiguchi N, Tohya K, Kimura M, Kishioka S. Activation of spinal cholecystokinin and neurokinin-1 receptors is associated with the attenuation of intrathecal morphine analgesia following electroacupuncture stimulation in rats. J Pharmacol Sci. 2007;104:159–166. doi: 10.1254/jphs.fp0070475. [DOI] [PubMed] [Google Scholar]
- Gerra G, Leonardi C, Cortese E, D’Amore A, Lucchini A, Strepparola G, Serio G, Farina G, Magnelli F, Zaimovic A, Mancini A, Turci M, Manfredini M, Donnini C. Human kappa opioid receptor gene (OPRK1) polymorphism is associated with opiate addiction. Am J Med Genet B Neuropsychiatr Genet. 2007;144:771–775. doi: 10.1002/ajmg.b.30510. [DOI] [PubMed] [Google Scholar]
- Gershenfeld HK, Neumann PE, Mathis C, Crawley JN, Li X, Paul SM. Mapping quantitative trait loci for open-field behavior in mice. Behav Genet. 1997;27:201–210. doi: 10.1023/a:1025653812535. [DOI] [PubMed] [Google Scholar]
- Gill KJ, Boyle AE. Confirmation of quantitative trait loci for cocaine-induced activation in the AcB/BcA series of recombinant congenic strains. Pharmacogenetics. 2003;13:329–338. doi: 10.1097/00008571-200306000-00004. [DOI] [PubMed] [Google Scholar]
- Goldman D. Recent developments in alcoholism:genetic transmission. Recent Dev Alcohol. 1993;11:231–248. [PubMed] [Google Scholar]
- Goodwin DW. The cause of alcoholism and why it runs in families. Br J Addict Alcohol Other Drugs. 1979;74:161–164. doi: 10.1111/j.1360-0443.1979.tb02424.x. [DOI] [PubMed] [Google Scholar]
- Grahame NJ, Li TK, Lumeng L. Limited access alcohol drinking in high- and low-alcohol preferring selected lines of mice. Alcohol Clin Exp Res. 1999;23:1015–1022. [PubMed] [Google Scholar]
- Grisel JE. Quantitative trait locus analysis. Alcohol Res Health. 2000;24:169–174. [PMC free article] [PubMed] [Google Scholar]
- Grisel JE, Belknap JK, O’Toole LA, Helms ML, Wenger CD, Crabbe JC. Quantitative trait loci affecting methamphetamine responses in BXD recombinant inbred mouse strains. J Neurosci. 1997;17:745–754. doi: 10.1523/JNEUROSCI.17-02-00745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Jones BC, Tarantino LM, Rodriguez LA, Reed CL, McClearn GE, Plomin R, Erwin VG. Quantitative-trait loci analysis of cocaine-related behaviours and neurochemistry. Pharmacogenetics. 1999;9:607–617. [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am J Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kest B, Palmese CA, Juni A, Chesler EJ, Mogil JS. Mapping of a quantitative trait locus for morphine withdrawal severity. Mamm Genome. 2004;15:610–617. doi: 10.1007/s00335-004-2367-3. [DOI] [PubMed] [Google Scholar]
- Kissin I, Bright CA, Bradley EL., Jr Acute tolerance to continuously infused alfentanil: the role of cholecystokinin and N-methyl-D-aspartatenitric oxide systems. Anesth Analg. 2000;91:110–116. doi: 10.1097/00000539-200007000-00021. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology. 2006;31:1085–1095. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Wilson JR, Meridith W. In: Contributions to Behavior- Genetic Analysis: The Mouse as a Prototype. Lindzey G, Thiessen DD, editors. Appleton-Century-Crofts; New York: 1970. pp. 3–22. [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci U S A. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier MV, Lu RB, Pakstis AJ, Kidd JR, Huang SY, Kidd KK. Possible epistatic role of ADH7 in the protection against alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2004;126(1):19–22. doi: 10.1002/ajmg.b.20136. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Verbitsky M, Suresh R, Kamens HM, Reed CL, Li N, Burkhart-Kasch S, McKinnon CS, Belknap JK, Gilliam TC, Phillips TJ. Gene expression differences in mice divergently selected for methamphetamine sensitivity. Mamm Genome. 2005;16:291–305. doi: 10.1007/s00335-004-2451-8. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Derr R, Shendure J, Kolata T, Silver LM. A major influence of sex-specific loci on alcohol preference in C57Bl/6 and DBA/2 inbred mice. Mamm Genome. 1998;9:942–948. doi: 10.1007/s003359900904. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Brown KJ, Burkhart-Kasch S, Wenger CD, Kelly MA, Rubinstein M, Grandy DK, Low MJ. Alcohol preference and sensitivity are markedly reduced in mice lacking dopamine D2 receptors. Nat Neurosci. 1998;1:610–615. doi: 10.1038/2843. [DOI] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS. Genetic influences in human substance abuse. J Addict Dis. 1991;10:205–213. doi: 10.1300/J069v10n01_14. [DOI] [PubMed] [Google Scholar]
- Tarantino LM, McClearn GE, Rodriguez LA, Plomin R. Confirmation of quantitative trait loci for alcohol preference in mice. Alcohol Clin Exp Res. 1998;22:1099–1105. [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Thompson R, Haley C. Confidence intervals in QTL mapping by bootstrapping. Genetics. 1996;143:1013–1020. doi: 10.1093/genetics/143.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatley VJ, Johnson TE, Erwin VG. Identification and confirmation of quantitative trait loci regulating alcohol consumption in congenic strains of mice. Alcohol Clin Exp Res. 1999;23:1262–1271. doi: 10.1111/j.1530-0277.1999.tb04287.x. [DOI] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J, Jr, Tischfield J, Kuperman S, Porjesz B, Begleiter H, Foroud T, Edenberg HJ. Association of the kappaopioid system with alcohol dependence. Mol Psychiatry. 2006;11:1016–1024. doi: 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]