Summary
Many promoters in eukaryotes have nucleosome depleted regions (NDR) containing transcription factor binding sites (TFBS). However, the functional significance of NDR is not well understood. Here, we examine NDR function in two cell-cycle-regulated promoters, CLN2pr and HOpr, by varying nucleosomal coverage of the binding sites of their activator SBF (SCBs) and probing the corresponding transcriptional activity in individual cells using time-lapse microscopy. Nucleosome-embedded SCBs do not significantly alter peak expression levels. Instead, they induce bimodal, “on/off” activation in individual cell cycles, which displays short-term memory, or epigenetic inheritance, from the mother cycle. In striking contrast, the same SCBs localized in NDR lead to highly reliable activation, once in every cell cycle. We further demonstrate that the high variability in Cln2p expression induced by the nucleosomal SCBs reduces cell fitness. Therefore, we propose that the NDR function in limiting stochasticity in gene expression promotes the ubiquity and conservation of promoter NDR.
Introduction
Transcription activation pathways usually start with binding of activators to their cognate sites in promoters. TFBS localization in NDR is generally believed to be important in enhancing transcription factor binding and facilitating subsequent transcription based on several lines of evidence. Studies on many different factors suggested that nucleosomes limit their TFBS accessibility both in vitro (for review, see Owen-Hughes and Workman, 1994) and in vivo (Liu et al., 2006; Morohashi et al., 2007; Sekinger et al., 2005). Well-studied promoters, such as GAL1-10pr and PHO5pr, have their major TFBS positioned within constitutive NDR, regardless of the transcriptional status of the gene (Lohr, 1997; Svaren and Horz, 1997). Genome-wide studies on nucleosome positioning in Saccharomyces cerevisiae revealed NDR upstream of transcription start sites (TSSs) in most promoters (Field et al., 2008; Lee et al., 2007; Mavrich et al., 2008a; Shivaswamy et al., 2008; Whitehouse et al., 2007; Yuan et al., 2005), and promoters in higher organisms are also frequently nucleosome-deficient (Mavrich et al., 2008b; Ozsolak et al., 2007; Schones et al., 2008). Such configuration is hardly perturbed by global transcriptional profile change (Zawadzki et al., 2009). The ubiquity and evolutionary conservation of NDR indicates its important function.
However, NDR localization of TFBS is not essential for gene activation. Many factors can bind to nucleosomal TFBS and form ternary complexes (DNA, nucleosome and factor) in vitro (Owen-Hughes and Workman, 1994), which might be promoted by the spontaneous “wrapping/unwrapping” of nucleosomal DNA (Li et al., 2005). Gal4p is able to disrupt nucleosomes positioned over its TFBS in vivo, accompanied with transcriptional activation (Balasubramanian and Morse, 1999; Morse, 1993; Xu et al., 1998). Pho4p is shown to occupy its nucleosomal binding site in the PHO5pr in vivo without nucleosome disassembly (Adkins et al., 2004), and a mutant PHO5pr containing solely two nucleosome-embedded high-affinity Pho4p binding sites still allows reasonable PHO5 induction (Lam et al., 2008). In addition, despite the enrichment of NDR in promoters, many functional promoters lack NDR (Field et al., 2008; Lee et al., 2007; Mavrich et al., 2008a; Shivaswamy et al., 2008; Whitehouse et al., 2007; Yuan et al., 2005).
Thus, the functional significance of NDR on transcription remains to be tested directly and quantitatively (Morse, 2007). Previous studies mostly presented bulk experiments, which were limited to measure average transcription level. Cell-to-cell variability is now recognized as an important feature of transcriptional regulation, which could directly influence cell phenotype (Acar et al., 2005; Avery, 2006; Blake et al., 2006; Colman-Lerner et al., 2005). The correlation between gene expression variability and nucleosome localization on promoters has not yet been subjected to direct experimental tests.
Here, we studied NDR function on two G1/S cell-cycle regulated promoters, CLN2pr and HOpr, which are both activated by the transcription factor complex SBF with binding site SCB. These promoters are highly dynamic and only expressed within a brief time window early in a cell cycle, and thus present a stringent test system for NDR in quantitative gene expression. Also, the genes driven by these promoters have important physiological functions: the endogenous gene driven by CLN2pr, G1 cyclin CLN2, is a key regulator mediating G1/S transition (Start). In the absence of a close analog, CLN1, lack of CLN2 leads to death (unbudded arrest) in ~30% of the cell population (Skotheim et al., 2008). The HO gene encodes an endonuclease required for mating-type switching, a critical event in the life-cycle of homothallic yeasts. Therefore, the regulatory properties of CLN2pr and HOpr have direct physiological implications.
We constructed a series of comparable promoters with SCBs either nucleosome-embedded or exposed in NDR. Across this series, promoters with nucleosome-embedded SCBs led to highly variable, “on/off” expression in individual cell cycle, while comparable promoters with NDR-localized SCBs reliably activated transcription every cell cycle with low expression noise (as was observed with the wt CLN2pr). The “on/off” expression displayed partial inheritance from the previous mother cycle with a half life ~1 cell cycle. We further explored the molecular mechanism of this bimodal transcription, and showed that it was related to the lowered accessibility of SBF to nucleosomal SCBs. Finally, we directly demonstrated that high variability in CLN2 gene expression induced by nucleosome-embedded SCBs led to reduced cell fitness, implying an evolutionary driving force for the conservation of promoter NDR.
Results
CLN2pr SCBs are located in NDR
CLN2 expression depends mainly on the transcription factor complex SBF (Swi6p-Swi4p), and secondarily on a closely related complex, MBF (Swi6p-Mbp1p) (Cross et al., 1994; Stuart and Wittenberg, 1994). The upstream activating sequences (UAS) in CLN2pr contain three consensus SCBs (Figure 1A), which are required for efficient CLN2 transcription, and two potential MBF binding sites (MCBs) which partially overlap the SCBs (Cross et al., 1994; Stuart and Wittenberg, 1994). SBF is loaded onto CLN2pr starting in late mitosis (Koch et al., 1996) without immediately inducing promoter activation, primarily because of Whi5 inhibition (Costanzo et al., 2004; de Bruin et al., 2004). At cell cycle Start, Whi5 is inactivated, and SBF-dependent transcription of CLN2pr is initiated. Later, CLN2pr is turned off due to SBF unloading from the promoter (Koch et al., 1996). The effect of chromatin structure on this cyclical regulatory pathway is unknown.
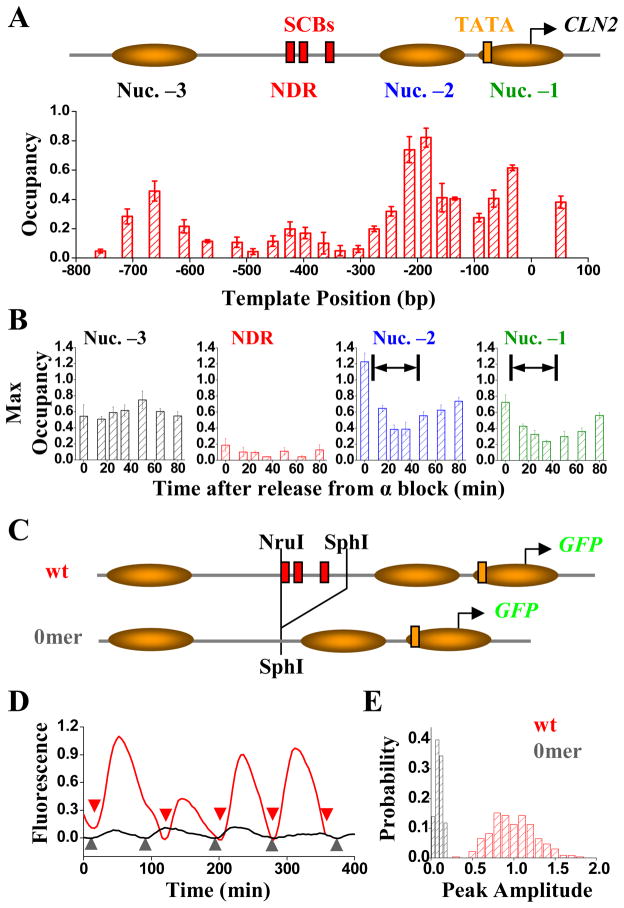
Figure 1.
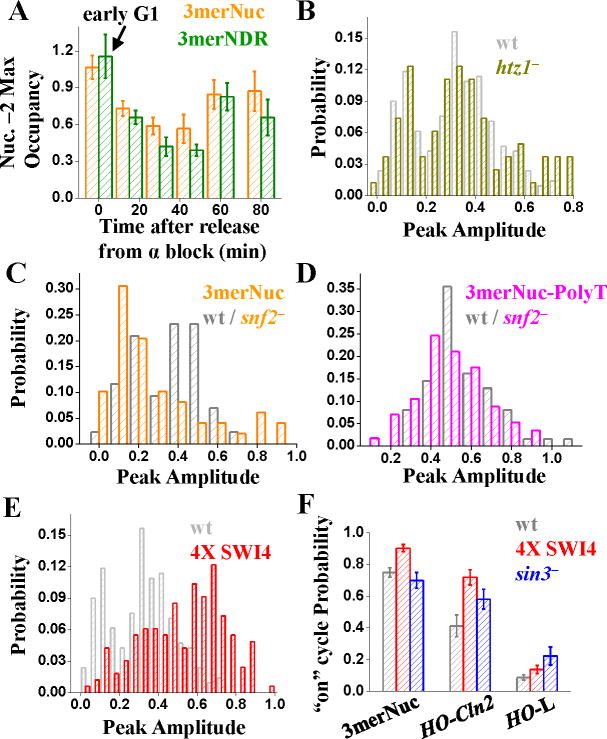
Nucleosome distribution and expression profile of wt CLN2pr. A) Genomic structure and nucleosome distribution on wt CLN2pr in asynchronized cells. The plot showed the measured nucleosome occupancy at different positions on the wt CLN2pr with the inferred locations of nucleosomes –1, –2 and –3 (shaded ovals) and NDR. The position 0 in the x axis is the CLN2pr TSS (chr16, 66788; Cross et al., 1994). The position of SCBs (red rectangles), TATA box (yellow rectangle) and TSS (arrow) were also shown. The error bars are the s.t.e from three independent measurements (same for below). B) The maximum occupancy of Nuc –3, NDR, Nuc –2 and Nuc –1 (left to right panels) in synchronized cells at different cell cycle time points after release from α factor block. The CLN2pr activation occurred within the marked time period (10–50 min after the release, including both mother and daughter cells; Figure S1B). C) The construct and the nucleosome distribution of 0mer promoter, where the NruI-SphI segment containing the SCBs were deleted. D) Typical GFP intensity vs time traces in a single cell driven by wt (red) or 0mer promoter (grey). The fluorescence signal was averaged over the cell area, corrected by subtracting a baseline connecting flanking troughs, and normalized by the average peak intensity per cell cycle of wt CLN2pr (same for the other figures unless specified). The colored arrows defined the cell division time marked by disappearance of Myo1 ring. E) The histogram of the peak-to-trough difference in the GFP signal per cell cycle for wt and 0mer promoters.
We mapped nucleosomes on the CLN2pr by MNase assay and qPCR (Kent and Mellor, 1995; Sekinger et al., 2005). Nucleosome occupancy was normalized to nucleosome –1 in PHO5pr, which was shown to be fully occupied (Jessen et al., 2006; Sekinger et al., 2005; Svaren and Horz, 1997). CLN2pr contained three positioned nucleosomes (–1, –2 and –3), and a ~300 bp NDR between nucleosome –2 and –3 (Figure 1A). The SCBs were located within the NDR, while the TATA box and TSS were buried inside nucleosome –1. Our data are consistent with two genome-wide nucleosome maps (Figure S1A; Lee et al., 2007; Mavrich et al., 2008a), which employed different methodologies: immuno-purification with H3 and H4 antibody and sequencing (Mavrich et al., 2008a), and hybridization with tiling microarray (Lee et al., 2007). Agreement between these data provides strong support to our nucleosome mapping data.
Next, we synchronized cells in G1 by α-factor block, and assayed nucleosome positioning at different times after release. Throughout a cell cycle, nucleosome –3 was constantly occupied and NDR remained nucleosome-free. In contrast, occupancy of nucleosomes –1 and –2 dropped upon activation of CLN2pr (Figure 1B, S1B; consistent with previous lower-resolution measurements, Hogan et al., 2006), probably allowing general transcription machinery to assemble onto the TATA box. Eviction of nucleosomes –1 and –2 required SBF binding, since it did not occur upon removal of SCBs (Figure S1C). After transcription was turned off, nucleosomes –1 and –2 were gradually reassembled (Figure 1B).
CLN2pr activation occurs reliably once every cell cycle
We fused an unstable GFP reporter (Mateus and Avery, 2000) to the CLN2pr to evaluate its transcriptional activity in individual cells (Bean et al., 2006; Figure 1C). The promoter fusion only produces GFP; all strains for transcription analysis contain an intact, wt CLN2 gene. Myo1-mCherry, which forms a bud neck ring between bud emergence and cytokinesis, was used to accurately time cell birth (Di Talia et al., 2007). The time-lapse method allowed us to probe the average transcription level, cell-to-cell variability and activation kinetics, as well as correlations between different generations through cell pedigrees.
GFP driven by CLN2pr exhibited periodic change once per cell cycle (Bean et al., 2006; Figure 1D). The histogram of its peak expression per cell cycle was unimodal (Figure 1E; see Table S2 for the size of data set, same as below unless specified), with a coefficient of variation (mean divided by standard deviation) of 0.27. Deletion of a ~100 bp segment from the CLN2pr containing all three SCBs and two MCBs (“0mer promoter”; Figure 1C) almost eliminated transcription activation (Figure 1D,E). The remaining fluctuation in the fluorescence signal (0.1; normalized by wt CLN2pr level, same as below) mostly came from cell auto-fluorescence background (Figure S1D). The two histograms in Figure 1E were well-separated, showing SCB-dependent CLN2pr expression occurred reliably in every cell cycle.
CLN2pr variant with nucleosomal SCBs induces bimodal, “on/off” activation in individual cell cycles
To determine the functional significance of NDR localization of the CLN2pr SCBs, we engineered three closely spaced SCBs into the positioned nucleosome –2 in the 0mer promoter with the endogenous SCBs deleted (“3merNuc promoter”; Figure 2A). These synthetic SCBs were generated by mutagenesis with minimal perturbation to the original sequence (Figure S2A). MNase mapping confirmed that these SCBs were indeed covered by nucleosome –2 (Figure 2A, S2B).
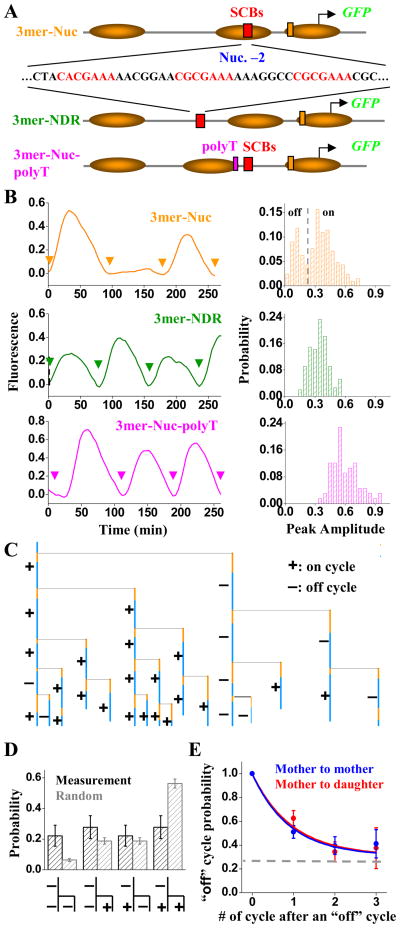
Figure 2.
Construct, nucleosome distribution and expression profile of 3merNuc, 3merNDR and 3merNuc-polyT promoters. A) The construct of the three promoters. The sequence of the SCB-containing region in these promoters was shown with the SCB consensus highlighted in red. The polyT sequence was shown in magenta. The inferred nucleosome distributions on these promoters were also shown (see Figure S2B for nucleosome occupancy data). B) The expression profile of GFP driven by 3merNuc, 3merNDR and 3merNuc-polyT promoters. Left panel showed a typical single-cell GFP intensity vs time trace driven by each promoter with the arrows representing the cell division time. Right panel was the corresponding histogram of the peak-to-trough difference in the GFP signal per cell cycle. C) One example of the 3merNuc cell pedigree with mapped “on” (+) and “off” (−) cycles. D) The measured fractions of different transcription profiles following an “off” mother cycle, and comparison with random probabilities. E) Propagation of the “off” cycle between different cell generations for 3merNuc promoter, i.e. given one “off” cycle, the probability of the “off” cycle in the subsequent cell cycles in both mother and daughter. The horizontal line represented the “baseline” of average “off” cycle probability.
The 3merNuc promoter could drive GFP expression at a level of 0.4, but only in ~75% of cell cycles. In the example shown in Figure 2B (top left panel), the promoter fired at cell cycle #1 and #3, but “skipped” cycle #2 (image data in Figure S2C). Accordingly, the expression from this promoter showed a clear bimodal, 2-Gaussian distribution (Figure 2B, S2D). The lower peak centered at ~0.1, identical to the 0mer promoter background, showing that SBF activation was effectively “off” in these cycles. The two Gaussian curves intersected at ~0.18, which we used as an empirical threshold to differentiate between “on” and “off” cycles.
The “on/off” pattern displays short-term memory from the previous mother cycle
To examine how the “on/off” transcription pattern propagated as cells divided, we mapped the “on/off” cycles across pedigrees (Figure 2C, more example in Figure S2E), and analyzed the correlation between adjacent cell generations. Interestingly, “on” and “off” cycles clustered within pedigrees: when a mother cycle was “off”, both the next mother and daughter cycle had an above-random probability to be “off” and conversely for “on” cycles (Figure 2D). For example, “off” cycles have an overall frequency of ~25%, but an “off” mother has two “off” descendants from a previous “off” mother cycle 22 ± 7% of the time, rather than the random expectation of 6% (P< 2 × 10−3).
To quantify the characteristic time of this generation-to-generation memory, we measured the “off” probability of sequential descendants of an individual cell following an initial “off” cycle. We observed an apparent exponential decay with a half life of 0.8 ± 0.3 cell cycle (Figure 2E). The transcription pattern was similarly propagated from mother to mother and from mother to daughter.
The “on” cycles of 3merNuc promoter had the same activation kinetics as the wt CLN2pr
We compared activation and repression kinetics between wild-type cycles and the “on” cycles of the 3merNuc promoter (Experimental Procedures; Figure S2FG). With wt CLN2pr, GFP induction occurred at 6.7 ± 0.4 min after cytokinesis in mother cells (N:163) and 22.2 ± 1.0 min in daughter cells (N:126). In both mother and daughter cells, the wt CLN2pr was activated for 24.8 ± 0.4 min before repression later in the cell cycle. The “on” cycles of the 3merNuc promoter had identical kinetics: the activation occurred at ~ 6.6 ± 0.4 min (mother cells, N:90) and 23.0 ± 1.5 min (daughter cells, N:60) after cytokinesis, and the activation period was 23.4 ± 0.5 min. Thus, despite frequent “off” cycles with 3merNuc promoter, the “on” cycles exhibit no defects in activation and repression kinetics and only a minor reduction in magnitude.
CLN2pr variants with NDR-localized synthetic SCBs induce unimodal activation
To test whether “on/off” transcription was specifically due to nucleosome positioning, we devised two control promoters with SCBs in the NDR. In the“3merNDR promoter”, the same synthetic 3mer-SCBs were inserted into the NDR of the 0mer promoter. Nucleosome analysis on this promoter confirmed that the inserted SCBs were unoccupied by nucleosomes (Figure 2A, S2B). In the “3merNuc-polyT” promoter, we introduced a polyT stretch into nucleosome –2 of the 3merNuc promoter. PolyT antagonizes nucleosome formation (Anderson and Widom, 2001; Field et al., 2008; Mavrich et al., 2008a; Yuan et al., 2005), and was shown to increase accessibility of the Gcn4 binding sites in chromatin in vivo (Iyer and Struhl, 1995). Indeed, nucleosome –2 in the 3merNuc-polyT promoter relocated upstream, leaving the 3mer SCBs nucleosome-free (Figure 2A, S2B).
These promoters expressed at a level comparable to the “on” cycles of the 3merNuc promoter (0.35 and 0.6 vs. 0.4). However, in striking contrast to 3merNuc promoter, their expression was unimodal, firing in every cell cycle (Figure 2B). Therefore, NDR localization of the SCBs correlated with high reliability and unimodality of expression in the CLN2pr variants.
A complementary case: HOpr
To check the generality of the observations above, we tested another SBF regulated promoter. Genome-wide, SBF binding was detected with high confidence at 50 promoters (Harbison et al., 2004), and 49 of these promoters have at least some of their candidate SCBs situated in NDR (Lee et al., 2007; Experimental Procedures). The one conspicuous exception was the well-characterized HO promoter, in which ~10 SCBs are packed within a positioned nucleosomal array (Figure 3A).
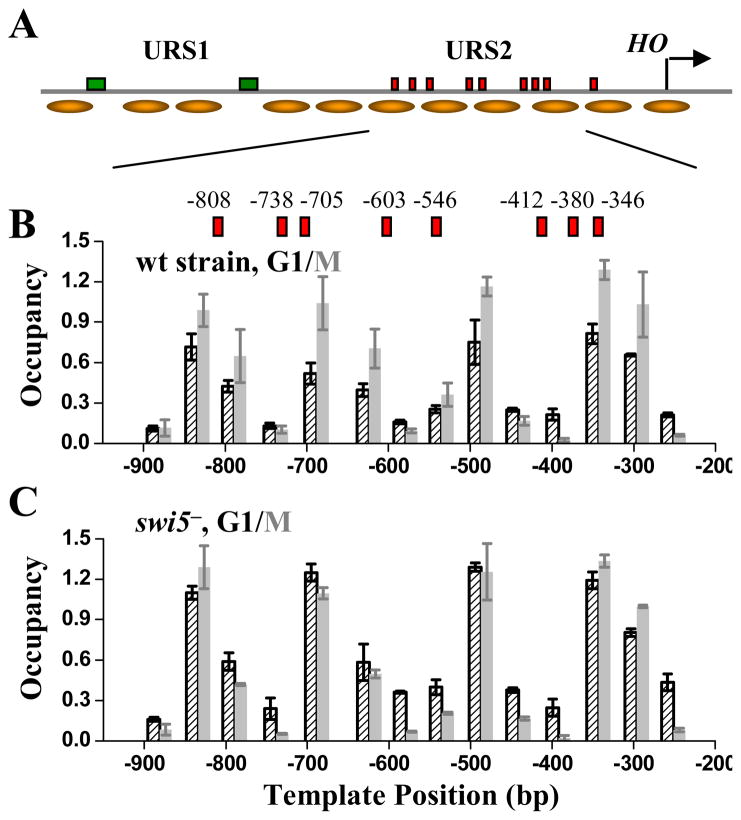
Figure 3.
Genomic structure and nucleosome distribution on wt HOpr. A) Genomic structure of the wt HOpr. The locations of URS1, URS2, Swi5 binding sites (green rectangle), SCBs (red rectangles) and TSS (arrow) were shown in the diagram. We measured nucleosome distribution over a ~600 bp segment in URS2 (Figure 3B), and the rest was obtained from global nucleosome positioning data (Lee et al., 2007). B) Nucleosome distribution on the wt HOpr in wt strain synchronized in G1 (black) and M (grey) phases. The x axis represents the HOpr position relative to its TSS (chr4, 48081; Nagalakshmi et al., 2008). There was significant decrease in nucleosome occupancy in G1 phase. The exact locations of the SCB binding sites (relative to the TSS) were also shown in the plot. C) Nucleosome distribution on the wt HOpr in swi5− strain synchronized in G1 (black) and M (grey) phases. There was no significant difference in nucleosome occupancy in the two cell cycle points.
Efficient SBF binding to HOpr requires prior upstream loading of Swi5, SWI/SNF and SAGA, while none of these factors contribute to the SBF binding on CLN2pr (Cosma et al., 1999). The reason for this difference was not well understood and we speculated that this could be due to nucleosomal localization of SCBs in the HOpr but not the CLN2pr. To test this idea, we examined nucleosome positioning on the ~600 bp SCB-containing region of HOpr (URS2) at different cell cycle stages. For cells blocked in early M phase by Cdc20 depletion, when Swi5 had not bound to the HOpr, we detected four positioned nucleosomes in this region with high occupancies. The occupancies were significantly lowered for cells blocked in G1 (Figure 3B) (this effect was clear but incomplete; this is likely due to the fact that Swi5 is only active in mother cells due to Ash1 inhibition in daughters (Cosma et al., 1999), so a two-fold difference is the maximum to expect). Nucleosome removal was Swi5-dependent (Figure 3C), suggesting Swi5 promotes SBF binding to the HOpr by evicting the URS2 nucleosomes. Interestingly, the Swi5 binding sites in URS1 of the HOpr are located in NDR (Figure 3A), which may allow Swi5, the most upstream factor for HO gene activation, to bind HOpr reliably without assistance from other factors. A recent publication also reported SWI5-dependent eviction of URS2 nucleosomes (Takahata et al., 2009).
Hybrid HO/CLN2 promoters with different localization of SCBs also affect expression variability
We further pursued the role of NDR localization by constructing CLN2 and HO hybrid promoters. We inserted three short SCB-containing segments (each <120 bp) from HO URS2 into the 0mer promoter (“HO-S1”, “-S2”, “-S3”; Figure 4A). Although these segments were nucleosome-bound in the context of the HO promoter, they were nucleosome-free upon transplantation (Figure 4B, S3A). In contrast, insertion of a ~550 bp segment of HO URS2 (including S1,2,3) into the same site in the 0mer promoter (“HO-L”) yielded largely nucleosome-covered SCBs, although the nucleosomes seemed to be less well-positioned than in the wt HOpr (Figure 4AB, S3A). Thus, this set of promoters allowed comparison of HO SCBs transplanted to the same site in the CLN2pr, either in NDR or nucleosome-bound.
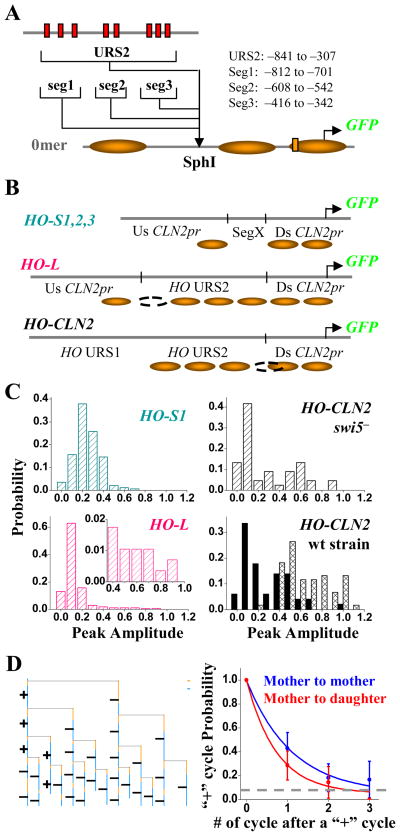
Figure 4.
Construct and expression profile of HOpr variants. A) The construct of HO-S1,2,3 and HO-L promoters. These promoters were constructed by inserting HO URS2 (or part of it) into the SphI site in the 0mer promoter, and integrated into the CLN2 locus. The range of the HOpr inserts was shown on the side (relative to the HO TSS). B) The composition and nucleosome distribution on the HO-S1,2,3, HO-L and HO-CLN2 promoters (see Figure S3A for nucleosome occupancy data). Us: upstream; Ds: downstream. The short vertical bars represented SphI sites, and the arrows were the TSS. C) The expression profile of GFP driven by HO-S1, HO-L (left panels) and HO-CLN2pr in wt and swi5− strain background (right panels). The inset in the HO-L panel zoomed in the histogram of HO-L expression with high level. Note this long “tail” did not exist in the histogram of 0mer promoter expression (Figure 1E). HO-CLN2pr also shows “on/off” expression in wt daughter cells and both mother/daughter cells in swi5− strain (see text for details). D) One example of the HO-L cell pedigree with mapped “on” (+) and “off” (−) cycles (left panel). The right panel shows the propagation of the “on” cycle between different cell generations for HO-L promoter from mother to mother (blue) and mother to daughter (red). The analysis is identical as in Figure 2E.
The HO-S1, -S2, and -S3 promoters all generated a low (~0.25) but unimodal expression every cell cycle (Figure 4C, S3B). In contrast, the HO-L promoter induced highly variable expression. The histogram for the HO-L promoter expression showed mostly “off” cycles, with a long tail (~8%) of “on” cycles (Figure 4C) and an overall average of ~0.14. Note that HO-L promoter contains all the SBF binding sites in HO-S1, 2, 3, but has lower and more variable expression. This initially counterintuitive observation could be well explained by different nucleosome positioning on these promoters.
As was observed with the 3merNuc promoter, the “on/off” cycles on the HO-L promoter exhibited memory across generations (Figure 4D). Interestingly, the memory half life (0.7 ± 0.2 cycle to mother and 0.5 ± 0.1 cycle to daughter; Figure 4D) was similar to that of the 3merNuc promoter, despite large differences in average expression level and “on” frequency. Also similar to the 3merNuc promoter, the “on” cycle of the HO-L promoter had the same activation kinetics as the wt CLN2pr: the activation time after cytokinesis was 7.6 ± 1.0 min in mother (N:15) and 21.3 ± 3.0 min in daughter (N:11), and the activation period was 23.8 ± 1.0 min.
To make sure the “on/off” transcription pattern was not specific to the CLN2 locus, we constructed a hybrid HOpr/CLN2pr at the HO locus. This “HO-CLN2pr” contained the CLN2pr TATA box and TSS (Figure 4B), allowing a direct comparison to CLN2pr variants. Endogenous Swi5-dependent HO regulation would create an NDR over the SCBs in mothers, and Ash1 inhibition of Swi5 would leave the SCBs nucleosome-bound in daughters. Therefore on HO-CLN2pr, the NDR localization of SCBs was manipulated by altering trans-factors rather than by cis mutation of the sequence. This promoter was fully activated in every mother cell cycle, while daughter cells exhibited bimodal expression, with ~40% of “on” cycles (Figure 4C, lower right panel). In a swi5− background, the HO-CLN2pr became bimodal in both mother and daughter cells, firing in 41% of all cell cycles (Figure 4C, upper right panel). In summary, across all the promoters we examined, uniform vs. bimodal expression correlated with NDR vs. nucleosomal localization of SCBs, strongly supporting the notion that nucleosomal localization causes variable expression.
Bimodal expression is unlikely due to nucleosome partial occupancy
We next probed the mechanism for the “on/off” transcription. The simplest explanation for bimodal expression from promoters with nucleosomal SCBs is that the nucleosome(s) covering SCBs is only present in some cell cycles. For instance, 25% occupancy of the nucleosome –2 in the 3merNuc promoter could account for a 75% “on” cycle probability. However, inconsistent with this idea, in early G1 before SBF activation, we found that nucleosome –2 was fully occupied (Figure 5A). This suggested that SBF could gain access to nucleosomal SCBs, at least in some cell cycles. Subsequent transcriptional activation of the 3merNuc promoter was associated with eviction of nucleosome –2, similar to the wt and 3merNDR promoters (Figure 5A), which could explain why the “on” cycle has comparable activation level as that from promoter with NDR-localized SCBs.
Figure 5.
Mechanism of “on/off” activation. A) Nucleosome –2 occupancy as a function of time in synchronized cell population on both 3merNuc and 3merNDR promoters. Note that at time 0 (before SBF activation), nucleosome –2 had ~100% occupancy in both promoters. B) The expression profile of 3merNuc promoter in wt (gray) vs htz1− strain (dark yellow). The two profiles are essentially identical. C,D) The expression profile for the 3merNuc (C) and 3merNuc-PolyT (D) promoters in the CY337 (wt)/CY407 (snf2–) background. 3merNuc promoter still activates in an “on/off” fashion in the absence of SWI/SNF, but the probability of “on” cycle decreases. In contrast, the expression of 3merNuc-PolyT promoter is not significantly affected. E) The expression profile of 3merNuc promoter in wt (1X SWI4) (gray) vs 4X SWI4 strain (red). Note the increase of the “on” cycle probability in the 4X SWI4 strain. F) The “on” cycle probability for 3merNuc, HO-CLN2 and HO-L promoters in the wt, 4X SWI4 and sin3− strains.
H2A.Z is not responsible for bimodal expression
A histone variant, H2A.Z, is thought to reduce nucleosome stability and promote more rapid activation for some genes (Abbott et al., 2001; Santisteban et al., 2000; Suto et al., 2000). H2A.Z is preferentially distributed in promoter regions flanking NDR, including the CLN2pr nucleosome -2 (Albert et al., 2007; Jin et al., 2009). In addition, H2A.Z is required for epigenetic memory of transcription for some yeast genes (Brickner et al., 2007). It is thus possible that a H2A/H2A.Z mixed population in nucleosome –2 is responsible for the bimodal expression of 3merNuc promoter.
Deletion of the H2A.Z-encoding gene HTZ1 did not affect the nucleosome distribution on the wt CLN2pr, or on the 3merNuc promoter (Figure S4A). Consistent with previous results (Dhillon et al., 2006), SBF activation and repression was mildly slower in the htz1− strain. However, it is not clear whether this is a specific effect, since the strain in general grows more slowly than wt.
Importantly, HTZ1 deletion had no effect on the expression profile of 3merNuc promoter: it was still bimodal and the maximum expression level per cell cycle remained essentially the same (Figure 5B). The memory of the “on/off” profile was not affected in the htz1− strain either (data not shown).
SWI/SNF is not strictly required for but contributes to the activation of 3merNuc promoter
The activation of wt CLN2pr is not SWI/SNF dependent (data not shown). However, nucleosomal SCBs could have elevated dependence on SWI/SNF, as has been demonstrated for Gal4 binding sites (Burns and Peterson, 1997). SWI/SNF was also implicated in memory of gene activation (Kundu et al., 2007). To test whether stochastic recruitment of SWI/SNF leads to the “on/off” activation, we carried out the time-lapse analysis of the 3merNuc and 3merNuc-polyT promoters driving unstable GFP in the snf2− background (CY407).
Similar to the wt CLN2pr, the activation of 3merNuc-PolyT promoter is not significantly affected by the absence of SWI/SNF (Figure 5D). GFP expression from 3merNuc promoter could also be detected in a small subset of cell cycles in the snf2− strain (Figure 5C), therefore SWI/SNF is not strictly required for its activation. However, the probability of “on” cycle decreases significantly in the snf2− background (~30%, comparing with ~65% in the wt CY337 background), indicating that SWI/SNF may contribute to the activation of the 3merNuc promoter, probably by destabilizing/evicting nucleosome –2.
“On” cycle probability is increased with elevated [SBF] and histone acetylation level
To investigate whether the “on/off” transcription could be rescued by high [SBF], we increased SBF level by integrating 3 extra copies of SWI4 (likely to be the limiting factor in the SBF complex; Ghaemmaghami et al., 2003). The increased Swi4 had little effect on the expression profile of wt promoter, suggesting that Swi4 may be saturating for wt. It also had no effect on the 0mer promoter, as expected since this promoter lacks Swi4 binding sites. Interestingly, extra copies of SWI4 increased the “on” cycle probability of all the “on/off” promoters (Figure 5EF, S4B). Reversely, when we reduced the [SBF/MBF] by constructing diploid strain with single copy of SWI4/MBP1 gene (JB22d), the “on” probability of the 3merNuc promoter was reduced comparing to that from a diploid strain with two copies of SWI4/MBP1 (JB14d; Figure S4C).
Histone acetylation is thought to weaken the interaction between neighboring nucleosomes and help recruit/stabilize SWI/SNF on target nucleosomes, therefore may allow more accessibility of buried TFBS (for review, see Hansen, 2002). Previous bulk experiments showed that deletion of histone deacetylase Sin3p increased HO expression in the absence of Swi5 and Gcn5 (Krebs et al., 1999; Mitra et al., 2006). Interestingly, our single cell assay revealed that the average expression level in the “on” cycles of HO-CLN2 (in swi5− strain) and HO-L promoters was similar with or without Sin3p (both ~0.5–0.6), but the “on” probability were significantly increased by sin3− deletion (Figure 5D, S4D). In contrast, this deletion did not affect the expression of 3merNuc promoter (probably not a Sin3p target, Figure S4D). Therefore, the increase of “on” cycle probability in the sin3− strain was not a global effect, but likely correlated with local changes in acetylation levels. These results indicate that the “on/off” transcription is related to the lowered accessibility of SBF to nucleosomal SCBs.
Biological consequences of NDR
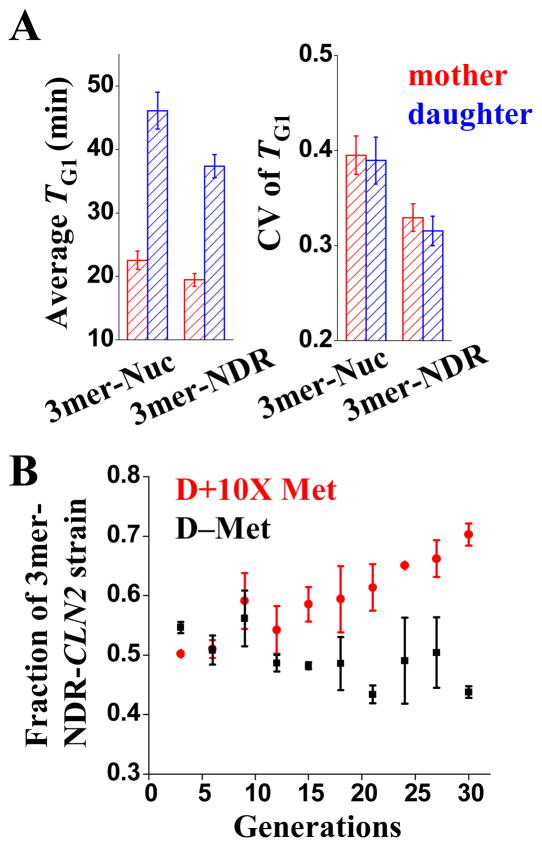
To determine if increased transcriptional noise due to nucleosomal localization of SCBs has functional consequences, we fused the 3merNuc and 3merNDR promoters to CLN2 coding sequence instead of to GFP, and placed them in a background where both wt CLN1 and CLN2 were deleted (cln1−, MET3pr-CLN2, cln2−::3merNuc/3merNDR -CLN2). These two promoters are directly comparable because they have similar structure and almost identical average transcription level (0.32 ± 0.01 vs 0.35 ± 0.01), but the 3merNuc promoter is much more variable (Figure 2B). When growing in D+10X Met media where the MET3pr-CLN2 is repressed, the G1 time (from cell division to budding) was longer and more variable in the 3merNuc-CLN2 strain than the 3merNDR-CLN2 strain for both mother and daughter cells (Figure 6A). Qualitatively similar effects could be detected even in the presence of CLN1 (data not shown).
Figure 6.
Biological consequence induced by 3merNuc-CLN2 vs 3merNDR-CLN2. A) The average TG1 (time from cell division to budding) and its coefficient of variation (CV) for both mother (red) and daughter cells (blue) in cln1− strains with either 3merNuc and 3merNDR promoters driving CLN2 expression. B) The fraction of 3merNDR-CLN2 strain in a growth competition assay with 3merNuc-CLN2 strain. 3merNDR-CLN2 strain gradually out competed 3merNuc-CLN2 strain in D+10X Met media (red), but not in D-Met where Met3pr-CLN2 was expressed.
The prolonged and more variable G1 period in the 3mer-Nuc-CLN2 strain could potentially serve as a driving force favoring low variability in the Cln2p expression. To test this idea, we carried out a growth competition assay (Experimental Procedures) where the two strains above were grown together and the composition of the culture were monitored over 30 generations. The 3merNDR-CLN2 strain successfully out-competed 3merNuc-CLN2 strain (Figure 6B). This effect was specifically due to defects in CLN2 expression, since no competitive advantage was observed when the ectopic MET3-CLN2 was also expressed (Figure 6B). This is a direct demonstration that low variability of house-keeping gene driven by NDR-localized TFBSs increases cell fitness.
Discussion
NDR localization of TFBS reduces gene expression variability
Conflicting results have been obtained with respect to the effect of nucleosome positioning on transcription in vivo. The abundance of NDR-localized TFBSs genome-wide suggested that an “open” chromatin domain is important for activators to gain access; on the other hand, nucleosome-embedded binding sites still allow efficient transcription factor binding and activation.
Based on genome-wide correlation between transcription noise and nucleosome occupancy, a hypothesis was proposed that promoter chromatin structure could affect variability in gene expression (Choi and Kim, 2009; Field et al., 2008; Tirosh and Barkai, 2008). This intriguing hypothesis still required direct experimental testing, for several reasons: 1) correlation does not demonstrate causality; low expression variability and NDR could both be consequences of other structural or regulatory features, so that NDR could correlate with but not directly cause low expression variability. The correlations display a substantial P-value because of the sample size, yet the information conveyed for any particular gene is low. 2) The noise dataset used in these papers is from snapshots of GFP-tagged strain in asynchronous culture (Newman et al., 2006), therefore does not apply to cell-cycle regulated genes. For instance, wt CLN2pr would be scored as highly variable in this dataset, but the actual expression noise is low once cell cycle timing is taken into account (see above). 3) Two of these papers (Choi and Kim, 2009; Tirosh and Barkai, 2008) analyzed the nucleosome occupancy relative to transcription and translation start sites, which lowers sensitivity for detecting specific effects of TFBS localization.
Here, through direct experimental manipulation of nucleosomal occupancy over TFBS in individual promoters, we demonstrate a key role of NDR in suppressing transcriptional variability. On both CLN2 and HO promoter variants, we found that SBF activation could still occur even with nucleosome-embedded SCBs. However, this activation is highly variable, firing only in a subset of cell cycles. Such an “on/off” transcription profile was not observed for any wild-type cell-cycle regulated promoter so far assayed at the single-cell level (Skotheim et al., 2008), suggesting a strong evolutionary pressure for suppressing such variable expression, at least partially through NDR localization of TFBS (see below).
Some origins of DNA replication only fire in some cell cycles, and there is evidence that origin efficiency correlates with its nucleosome density (Field et al., 2008; Gerbi and Bielinsky, 2002). These results are potentially related to our observations. However, the high density of replication origins on chromosomes means that failure of initiation from a given origin has relatively minor consequence of passive replication from adjacent origins – replication will not fail altogether. In contrast, failure to express a gene is more problematic; in particular, failure to express an unstable protein, such as many cell cycle regulators, is not correctable within a cell cycle. This could be why NDRs have been efficiently selected on promoters but not replication origins.
The mechanism of the “on/off” cell cycles and its memory
The lowered accessibility of the nucleosomal SCBs could lead to “on/off” expression through two, non-exclusive mechanisms: 1) Nucleosomal SCBs could stochastically delay activation onset, resulting in sporadic “skips” of a transient pulse of SBF activity. 2) There could be structural heterogeneity in the structure of promoters with nucleosomal SCBs, resulting in different accessibility in different cell cycles. Evidence so far excluded the heterogeneity in nucleosome occupancy and H2A.Z, but there are other possibilities, such as variation in SBF concentration, variation in factors bound to the promoter, and heterogeneity in nucleosome positioning and/or modifications. We can divide these variations into “intrinsic” component, such as the heterogeneity in local chromatin structure, and “extrinsic” component, such as the variation in [SBF] (Elowitz et al., 2002; Raser and O’Shea, 2005). These two components can be differentiated by using multiple copies of the same promoter driving different fluorescence reporters in the same cell (Elowitz et al., 2002), which will be an interesting direction to pursue in the future. Some of these variations likely occur at the wt CLN2pr as well, but NDR localization of SCBs renders the promoter robust to these environmental noises.
We also observed partial inheritance of “on/off” transcription. Heritable chromatin structure/nuclear localization can provide memory in several systems, such as telomeric transcriptional silencing (Laurenson and Rine, 1992; Xu et al., 2006), and rapid reactivation of GAL1-10 (Brickner et al., 2007; Kundu et al., 2007). If the “memory” we observed is also related to local chromatin/nuclear structure, our results would reflect how these structures are inherited across generations in single cell during continuous growth, a property hard to measure directly. The ~1 cell cycle half life of memory in our system is consistent with the idea that at division, the prior transcription pattern is “remembered” in one of the descendants, while the pattern in the other one is randomly acquired. Further experiments are required to test such a model.
NDR mechanism
Although the mechanism of generating and maintaining NDR is not the focus of our work, some of our observations are relevant to this question. First, SBF is neither necessary nor sufficient for generating NDR. During a cell cycle, the NDR on CLN2pr is constitutive even though SBF binding and transcriptional activation only occur for a short period of time. In a swi4 mbp1 double deletion strain where there is no SBF or MBF to bind the sites, and CLN2pr activation is completely abolished (Koch et al., 1993), the CLN2pr NDR remains intact (unpublished data). These observations are consistent with other controllable promoters such as GAL1-10 and PHO5, which contain constitutive NDRs irrespective of the transcriptional status of the gene (Lohr, 1997; Svaren and Horz, 1997). Conversely, ~10 copies of SCBs are not sufficient to evict the nucleosome on HOpr URS2, even when this sequence is transplanted to the NDR region of the CLN2pr. Second, although H2A.Z is enriched in nucleosome –2 of CLN2pr (Albert et al., 2007), its deletion has no effect on the nucleosome distribution on the promoter. This is consistent with a recent finding (Hartley and Madhani, 2009) that H2A.Z disposition is dispensable for NDR formation. Third, NDR is resistant to short sequence deletion/insertion (0mer, 3merNDR, and HO-S1,2,3 promoter), but not long sequence insertion (HO-L promoter). This result suggests that the factors responsible for NDR formation have local effects on nucleosome positioning, and are not able to compete with nucleosome-positioning sequences that are longer than the nucleosome repeat length.
Direct evidence that NDR-localized TFBSs could be selected through evolution
NDRs are in general energetically unfavorable due to the high affinity between histone and DNA (Thastrom et al., 2004). Indeed, we have found that the NDR sequence of CLN2 efficiently forms nucleosomes in vitro using salt dialysis, indicating extra energy is required for nucleosome depletion (unpublished data). Nevertheless, many natural promoters contain NDRs, suggesting an important biological function.
We demonstrated that NDR localization of transcription factor binding sites limits transcriptional noise, and we tested the biological consequences of NDR- or nucleosomal-localized SCB sites driving Cln2 expression. Cln2 is responsible for transcriptional positive feedback that ensures the sharp activation of SBF at Start, which is important for coherent expression of the G1/S regulon (Skotheim et al., 2008), and therefore unreliable CLN2 expression is likely to be detrimental. Indeed, 3merNuc-Cln2 strain showed reduced cell fitness. This argument could likely be extended to other SBF-regulated genes (as noted above, 49/50 SBF-bound promoter contain NDR-localized SCBs). Although high expression variability in stress-response genes was indicated to be beneficial for the cell fitness (e.g. Blake et al., 2006), noisy expression of most genes, especially house-keeping genes such as CLN2, is likely to be harmful and therefore minimized by natural selection (Fraser et al., 2004). Especially in higher multi-cellular organisms, the development and patterning relies on the coordination of the expression of many genes over multiple cells. Many of these genes are under control of complicated regulation network, where gene expression noise tends to propagate and amplify. The robustness of gene regulation in these organisms indicates highly evolved mechanisms to suppress noise. Since NDR reduces gene expression noise, these results provide an explanation for the abundance and conservation of NDR.
Experimental Procedures
Strains and Plasmids
Standard methods were used to construct the strains and plasmids. All strains are W303-congenic. To introduce CLN2pr variation, we started from plasmid pLB02 (containing wt CLN2pr-GFP-CLN2pest with CaURA3 marker), mutated the wt CLN2pr as desired, digest at BbsI site in the wt/mutant CLN2pr, and integrated into the strain GC46-03 (MATa, MY01::MY01-mCherry-SpHIS5, ADE2) at the CLN2 locus. The 0mer promoter is constructed by deletion of the NruI-SphI segment from wt CLN2pr (Figure 1C). 3merNuc, 3merNDR and 3merNuc-polyT promoters are all mutations from the 0mer promoter, each only differ by a few bases; their detailed construction methods are shown in Figure S2A. For the hybrid promoters of HO and CLN2, we inserted the HO URS2 (from −841 to −307 of HOpr relative to its TSS) into the SphI site in the 0mer promoter (Figure 4A), and integrate the plasmid either into the CLN2 locus to form HO-L promoter, or into the HO locus to form HO-CLN2pr. The change in integration site is achieved by digesting the plasmid either in the CLN2pr region upstream the SphI site (at BbsI site), or inside the HO URS2 insert (at AflII site). For the HO-S1,2,3 promoters, we inserted short segments from HO URS2 (see Figure 4A for the range of the segments) into the SphI site in the 0mer promoter, and integrated into the CLN2 locus.
For nucleosome analysis, the ura– “popout” strain containing the varied CLN2pr was selected so that the entire CLN2pr region is kept single copy. sin3–::KanMx strain was obtained by one-step gene replacement. Extra copies of SWI4 were introduced by integrating multiple pRS404-SWI4 (derived from pTOW-SWI4; Moriya et al., 2006) into W303α, and crossing with strains containing CLN2pr variants driving GFP. The SWI4 copy number was estimated by qPCR. See Table S1 for the complete strain list.
Nucleosome mapping
For the MNase assay, we used the protocol described in Kent and Mellor (Kent and Mellor, 1995). In brief, we first grew 10ml cell to OD ~ 0.15, harvested the cell, and washed in 0.5ml water. Then we re-suspended the cells in 0.5ml of sphaeroplasting solution (1M sorbitol, 0.5mM 2-mercaptoethanol, 0.18mg/ml zymolyase), incubated at room temperature for ~5min with gentle stir. We harvested the cell, wash in 1ml 1M sorbitol, then re-suspend the pellet in 200ul of digestion buffer (1M sorbitol, 50mM NaCl, 100mM Tris-Cl (pH 7.4), 5mM MgCl2, 1mM CaCl2, 1mM 2-mercaptoethanol, 0.5mM spermidine, 0.075% NP-40, micrococcal nuclease with final concentration 1–10 unit/ml) for ~8 min in 37°C. After terminating the MNase digestion by adding 20ul quench buffer (250mM EDTA, 5% SDS), we extracted the DNA with phenol/chloroform, and proceeded with the qPCR analysis with stacking PCR primer pairs as described by Sekinger et al., 2005. The PCR products were all ~100bp in length, and the distances between adjacent primers were typically 30–50 bp. We used the nucleosome –1 on the PHO5pr as the standard to scale the occupancy from 0 to 1 (Sekinger et al., 2005).
Error bars
The error bars on all figures and supplementary figures represent the s.e. in the measurements. For nucleosome occupancy, the error bars represent the s.e. from three independent measurements. For the fluorescence measurements, see Table S2 for the size of data set.
Timelapse fluorescence microscopy and data analysis
Sample preparation for the time-lapse assay was performed as previously described (Bean et al., 2006; Charvin et al., 2008). The instrumentation of the timelapse microscopy and the Matlab software for data acquisition and analysis have been described in Charvin et al (Charvin et al., 2008). Images were acquired every 4 minutes for ~8 hours. Within this period, the majorities of the cells remained in focus (we do not analyze the cells if they are out of focus). Occasionally we observed cells arrested in mitosis due to photo damage (<5% of the whole population), and these cells were discarded in the analysis. The GFP intensity vs time curves as shown in Figure 1D were smoothed, then corrected by subtracting a baseline connecting flanking troughs, and finally normalized by the average peak GFP intensity of wt CLN2pr.
Analysis of activation kinetics
We setup a simplified model that during one cell cycle from t = 0 to T (division to division time), SBF activation occurred within ton to toff. In this window, GFP protein was produced at a constant rate kp, and meanwhile degraded at a rate kd. Outside the window, the [GFP] simply decayed exponentially at the rate kd. To observe GFP fluorescence signal, the GFP must fold first, which is assumed to be a first order process with folding rate kf (Charvin et al., 2008):
in which
We used the function above to fit the experimental data of GFP intensity vs time during each single cell cycle (from cell division to division). We assumed the kf = 0.1 min−1, and the fitted parameters were ton, toff, kp and kd. For examples of fitted curve and fitted parameters, see Figure S2F-H.
Nucleosome positioning at SBF binding sites in genome-wide scale
Based on the global nucleosome distribution data (Lee et al., 2007), NDR were recognized with the following steps: first, we picked out region where nucleosome density was lower than −0.6 (log scale); second, to account for the fluctuations in the density measurement, the neighboring low density regions were lumped together if the distance between them was smaller than 80 bp; finally, the low density region was considered as a NDR if its length was longer than 80 bp. Then for each of the ~100 SBF binding sites detected with the highest confidence (distributed on 50 promoters) (Harbison et al., 2004), we examined whether they fall into a NDR.
Growth competition assay
First, individual strains were grown to saturation in D-MET medium, then we mixed approximately equal number of cells from both strains into 5ml of D-MET or D+10X MET media to a final OD ~ 0.1. After OD reaches 0.8 (3 generations), we harvested most of the culture, stored in 4C and diluted the remaining cells into the same 5 ml medium to OD ~ 0.1 and let it grow to OD ~0.8 again. After 30 generations, we extracted the gDNA from the cultures collected at different time points, and analyzed the culture composition using rtPCR with PCR primer pairs specific to each strain.
Supplementary Material
Acknowledgments
The authors acknowledge Dr. Jon Widom and Dr. Irene Moore for advice on the MNase assay; Dr. Hisao Moriya for the pTOW-SWI4 plasmid; Dr. Craig Peterson for CY337 and CY407 strain; Lea Schroeder.and Ying Lu for help with strain construction and all the members of the F. Cross laboratory for insightful comments. This work was supported by the National Institute of Health (E.D.S. and F.R.C.), the Damon Runyon cancer research fellowship (L.B.), Human Frontier Science Program Organization (G.C.) and the National Science Foundation (E.D.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J Biol Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol. 2006;4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- Balasubramanian B, Morse RH. Binding of Gal4p and bicoid to nucleosomal sites in yeast in the absence of replication. Mol Cell Biol. 1999;19:2977–2985. doi: 10.1128/mcb.19.4.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR. Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell. 2006;21:3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns LG, Peterson CL. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE. 2008;3:e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Kim YJ. Intrinsic variability of gene expression encoded in nucleosome positioning sequences. Nat Genet. 2009;41:498–503. doi: 10.1038/ng.319. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Cross FR, Hoek M, McKinney JD, Tinkelenberg AH. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol Cell Biol. 1994;14:4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Dhillon N, Oki M, Szyjka SJ, Aparicio OM, Kamakaka RT. H2A.Z functions to regulate progression through the cell cycle. Mol Cell Biol. 2006;26:489–501. doi: 10.1128/MCB.26.2.489-501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA, Bielinsky AK. DNA replication and chromatin. Curr Opin Genet Dev. 2002;12:243–248. doi: 10.1016/s0959-437x(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen WJ, Hoose SA, Kilgore JA, Kladde MP. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat Struct Mol Biol. 2006;13:256–263. doi: 10.1038/nsmb1062. [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. Nucleosome transactions on the promoters of the yeast GAL and PHO genes. J Biol Chem. 1997;272:26795–26798. doi: 10.1074/jbc.272.43.26795. [DOI] [PubMed] [Google Scholar]
- Mateus C, Avery SV. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 2000;16:1313–1323. doi: 10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008a;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008b;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol Cell Biol. 2006;26:4095–4110. doi: 10.1128/MCB.01849-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Shimizu-Yoshida Y, Kitano H. In vivo robustness analysis of cell division cycle genes in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e111. doi: 10.1371/journal.pgen.0020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi N, Nakajima K, Kurihara D, Mukai Y, Mitchell AP, Shimizu M. A nucleosome positioned by alpha2/Mcm1 prevents Hap1 activator binding in vivo. Biochem Biophys Res Commun. 2007;364:583–588. doi: 10.1016/j.bbrc.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RH. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1563–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- Morse RH. Transcription factor access to promoter elements. J Cell Biochem. 2007;102:560–570. doi: 10.1002/jcb.21493. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T, Workman JL. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol Cell Biol. 1994;14:4788–4801. doi: 10.1128/mcb.14.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- Svaren J, Horz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell. 2009;34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrom A, Lowary PT, Widom J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR. Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell. 2006;23:219–229. doi: 10.1016/j.molcel.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Xu M, Simpson RT, Kladde MP. Gal4p-mediated chromatin remodeling depends on binding site position in nucleosomes but does not require DNA replication. Mol Cell Biol. 1998;18:1201–1212. doi: 10.1128/mcb.18.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zawadzki KA, Morozov AV, Broach JR. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:3503–3513. doi: 10.1091/mbc.E09-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.