Abstract
There is growing evidence that macroautophagic cargo is not limited to bulk cytosol in response to starvation, and can occur selectively for substrates including aggregated proteins. It remains unclear, however, if starvation-induced and selective macroautophagy share identical adapter molecules to capture their cargo. Here we report that Alfy, a phosphatidylinositol 3-phosphate binding protein, is central to the selective elimination of aggregated proteins. We report that the loss of Alfy inhibits the clearance of inclusions, with little to no effect on the starvation response. Alfy is recruited to intracellular inclusions and scaffolds a complex between p62(SQSTM1)-positive proteins and the autophagic effectors Atg5, Atg12, Atg16L and LC3. Alfy overexpression leads to elimination of aggregates in an Atg5-dependent manner, and likewise, to protection in a neuronal and Drosophila model of polyglutamine toxicity. We propose that Alfy plays a key role in selective macroautophagy, by bridging cargo to the molecular machinery that builds autophagosomes.
Introduction
The fundamental ability of a cell to function is tightly linked to its capacity for protein synthesis. Equally important is the cell's ability to eliminate proteins that are deemed no longer necessary or misfolded. In light of this, it is unsurprising that a common cytopathological feature of disease is the presence of intracellular inclusions. The pathological significance of aggregated proteins has been of particular interest in the area of neurodegeneration, as symptomatic reversal in mouse models of spinocerebellar ataxia 1 (SCA1) and Huntington's disease (HD) tightly correlates with the clearance of accumulated protein (Yamamoto et al., 2000; Zu et al., 2004).
Cytosolic proteins are degraded by the ubiquitin-proteasome and lysosome systems. Over the last several years, the latter has been brought to the forefront with macroautophagy- a process through which cytosolic constituents are taken up into a multimembranous structure known as the autophagosome, which upon fusion to endosomal (Berg et al., 1998; Filimonenko et al., 2007; Kochl et al., 2006) and lysosomal structures permits degradation (Klionsky, 2005). Predominantly known to nonspecifically engulf and degrade cytosol and long-lived proteins (LLP) in response to nutrient deprivation or inhibition of the kinase mTOR, macroautophagy also permits elimination of organelles such as mitochondria (Kanki et al., 2009; Klionsky, 2005; Okamoto et al., 2009). Recently, macroautophagy has been implicated in the elimination of aggregated proteins across various cell types including neurons (Boland and Nixon, 2006; Iwata et al., 2005b; Ravikumar et al., 2002; Yamamoto et al., 2006). These proteins are often polyubiquitinated and can be recognized by the ubiquitin- and LC3-binding proteins p62 and NBR1(Bjorkoy et al., 2005; Kirkin et al., 2009).
Alfy (Autophagy linked FYVE protein) is a 400kDa protein that contains a BEACH domain, WD-40 domain and a phosphatidylinositol 3-phosphate (PI3P) binding FYVE domain (Simonsen et al., 2004). We have previously shown that it is recruited to ubiquitin-positive protein inclusions under stress conditions (Simonsen et al., 2004). Although mammalian studies of Alfy have been limited, Drosophila lacking the Alfy homologue Blue Cheese (bchs) are adult viable, but have a reduced lifespan due to an accelerated accumulation of ubiquitin-positive inclusions and neuronal degeneration (Finley et al., 2003). In this study we demonstrate that aggregated proteins are sequestered into autophagic vesicles, and present evidence that Alfy is essential for their macroautophagic clearance. Despite, however, its role in selective autophagy, Alfy is not required for macroautophagy due to nutrient deprivation. Alfy co-localizes and interacts with p62 and NBR1 positive proteins, and directly interacts with Atg5. This interaction permits the creation of a greater complex with Atg12, Atg16L, and LC3, suggesting that Alfy acts as a scaffold that bridges its cargo to the macroautophagic machinery. Finally, we find that Alfy overexpression can decrease aggregated polyglutamine protein levels, and protect cells from expanded polyglutamine toxicity both a primary neuronal HD model and a Drosophila eye model of polyglutamine disease.
Results
Polyglutamine aggregates can be found in autophagosomes
Although macroautophagy has been implicated in several studies to eliminate aggregated proteins (Bjorkoy et al., 2005; Boland and Nixon, 2006; Iwata et al., 2005b; Kirkin et al., 2009; Ravikumar et al., 2004; Yamamoto et al., 2006), it is uncertain if oligomers or larger protein aggregates and inclusions are trafficked to autophagosomes. To examine this issue further, we turned to a model aggregation-prone protein, a short fragment of the protein huntingtin (exon1Htt) that carries an expanded polyglutamine (polyQ) mutation of greater than 37 glutamines (37Q). These proteins spontaneously aggregate in vitro, and are found as aggregated oligomers to large, aggresome-like inclusions in vivo. This is readily observed with transient transfection of a Flag-tagged exon1Htt construct expressing 68 polyQ (Flag-HttQ68, HeLa) (Figure S1A), as well as in previously described tetracycline (tet)-regulatable cell lines (HeLa and N2a) stably expressing the first 17 amino acids of Htt followed by a polyQ expansion (65Q or 103Q) tagged to monomeric CFP (HttPolyQ-mCFP) (Figure 1) (Yamamoto et al., 2006). As a control, we used cell lines that express 25Q, which is under the aggregation threshold of 37 repeats, and thus remains soluble (Yamamoto et al., 2006).
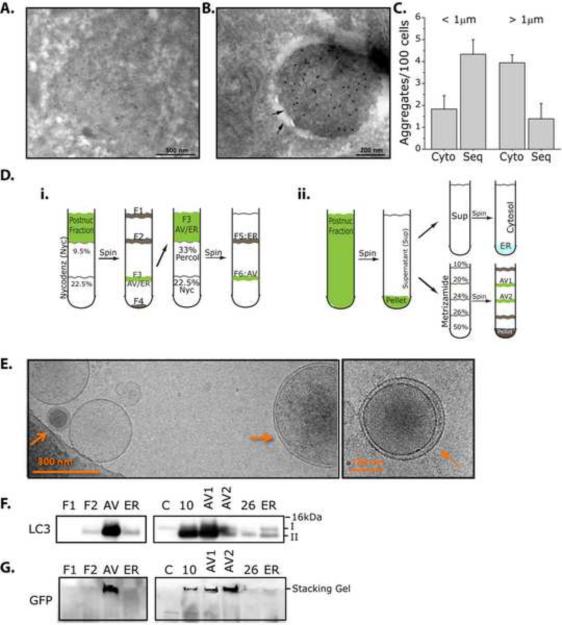
Figure 1.
Polyglutamine aggregates are found in autophagic vacuoles. A–G. Polyglutamine aggregates can be found in double membrane structures by immunoEM, and in the autophagosome fraction of cell fractionation experiments. A–C. ImmunoEM analysis of Htt103Q-mCFP cell lines. Cells were treated with 4h of BafA1 then fixed. Gold particles label structures positive for mCFP. A. Membrane-free aggregates. B. Aggregates sequestered within a double membrane. C. Quantification of Cytosolic (Cyto) vs Sequestered (Seq) aggregates. Aggregates positive for GFP were sorted based on diameter (< or > 1μm) then counted. < 1μm aggregates were more likely to be sequestered in autophagosomes (Cyto: 2.11+0.408; Seq: 4.10+0.689), while > 1μm aggregates were cytosolic (Cyto: 3.94+0.372; Seq: 1.40+0.700). 185 cells was counted across n=3 experiments. Data is shown as Mean+SEM. D–G. Cell fractionation experiments using 2 distinct protocols reveal that SDS-insoluble Htt103QmCFP positive proteins can be found in the autophagosome (AV) fractions. D. Summary of fractionation protocols used. E. CryoEM images generated from AV fractions using \Nycodenz protocol. F. Immunoblot analysis of cellular fractions. LC3 II is enriched in AV fractions. G. Immunoblotting of AV fractions reveals the presence of SDS-insoluble Htt103Q-mCFP in the loading stack of the gel, despite using denaturing conditions. 4–12% polyacrylamide gels were used for SDS-PAGE.
We previously reported that polyQ aggregates and inclusions can co-localize to transiently transfected LC3-YFP (Yamamoto et al., 2006). Transient expression of Flag-HttQ68 in cell lines stably expressing LC3-GFP leads to similar results (Figure S1A). To ensure that the co-localization of LC3 to inclusions is indeed indicative of the presence of autophagic membrane, we next used ultrastructural and biochemical approaches (Figure 1A–F). Lysosomal inhibitors increases the frequency with which LC3 co-localized to the mCFP-positive inclusion (Yamamoto et al., 2006), possibly by inhibiting the maturation of autophagosomes to autolysosomes. Htt103Q- and 65Q-mCFP stable cell lines were thus pre-treated with Bafilomycin A1 (BafA1) then processed for ultrastructural analyses. As expected, inclusions of differing sizes were detected by electron microscopy (EM). The majority of inclusions, especially larger inclusions of several microns in diameter, were found membrane-free (Figure 1A, S1B). In contrast, smaller polyQ inclusions up to 1 μm in diameter were preferentially detected within double membrane structures (Figure 1B, S1D,E). In rare cases we could observe larger aggregates (up to 5μm diameter) surrounded by a double membrane structure (Figure S1 double arrow in inset in C). Quantification of inclusions based on size (less than (<) or greater than (>) 1μm) and localization (cytosolic (cyto) or sequestered (seq)) is shown in Figure 1C.
To independently determine if inclusions are found in autophagic vesicles (AV), we next performed cell fractionation experiments with nycodenz (Stromhaug et al., 1998) (Figure 1Di) or metrizamide(Marzella et al., 1982) (Figure 1Dii) to isolate the AV fractions from Htt103Q-mCFP cells. These experiments were performed in the absence of lysosomal inhibitors. Cryo EM analysis revealed that multilamellar vesicles can be clearly detected in the AV fraction (Figure 1E). Moreover, the AV fraction was enriched for membrane bound LC3 (form II), confirming autophagosome enrichment (Figure 1F), while cytosolic fractions were positive for only soluble LC3 (form I). To determine if SDS-insoluble aggregates can be found in the AV fraction, we probed for Htt103QmCFP using an antibody against mCFP. In those fractions enriched for LC3-II, Htt103QmCFP remains in the stacking gel, indicative of the presence of SDS-insoluble aggregated Htt (Figure 1G)(Cornett et al., 2005). The same fractions were also associated with a high molecular weight polyubiquitin smear (Figure S1F,G)(Davies et al., 1997; DiFiglia et al., 1997). Taken together, these data indicate that SDS-insoluble aggregates can be found sequestered in double membrane structures and in an AV-enriched fraction.
Alfy shuttles from the nuclear membrane to co-localize with aggregated proteins
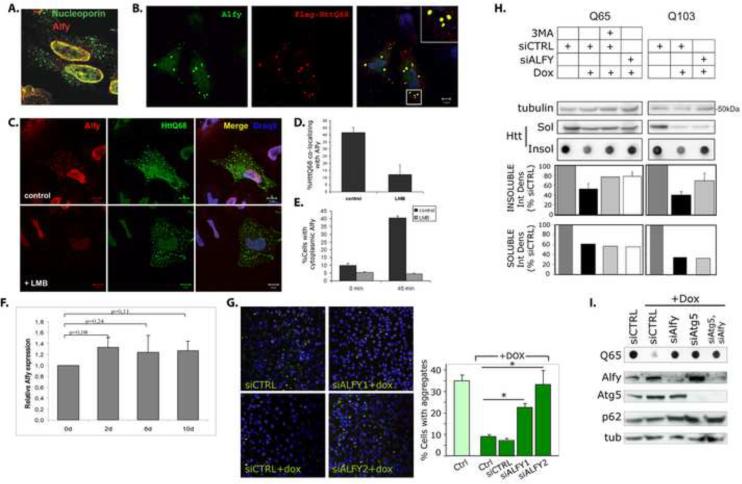
We have previously reported that Alfy is a predominantly nuclear protein that is recruited to cytoplasmic ubiquitinated structures in response to cellular stress such as proteasome inhibition (Simonsen et al., 2004). We therefore examined if expression of aggregated polyQ proteins also recruited Alfy. Immunofluorescence (IF) revealed that in the absence of aggregated proteins, Alfy is largely found within the nucleus along the nuclear membrane, co-localizing with nucleoporins (Figure 2A). The presence of the cytosolic polyQ inclusions led to the presence of Alfy in the cytosol, co-localized to the inclusions (Figure 2B). To determine if Alfy shuttled from the nucleus, cells were treated with leptomycin B (LMB), an inhibitor of nuclear export, and re-examined. Treatment with LMB significantly diminished the localization of Alfy to the inclusions, indicating that Alfy is actively exported from the nucleus in a CRM1-dependent manner (Figure 2C,D). LMB treatment also inhibited the recruitment of Alfy to cytoplasmic ubiquitin-positive aggregates due starvation-induced stress (Figure 2E). qRT-PCR analysis also showed that Alfy mRNA levels were unaffected by polyQ aggregation, indicating de novo synthesis of Alfy in response to aggregation is unlikely(Figure 2F). Interestingly, inhibition of nuclear export caused increased colocalization of Alfy with promyelocytic leukemia (PML) nuclear bodies (Figure S2A,B), intranuclear sites where misfolded proteins have been proposed to accumulate (Rockel et al., 2005). Consistent with this, Alfy co-localized with intranuclear inclusions of the nuclear protein Ataxin-1 (Figure S2C,D). Colocalization with other intranuclear structures was not detected (Figure S2B).
Figure 2.
Alfy translocates from the nucleus and is required to clear aggregated polyQ. A–F. Alfy translocates from the nucleus into cytoplasmic structures A. Alfy localizes to the nuclear membrane in untreated HeLa cells and co-localizes with nucleoporin. B. HeLa cells were transfected with Flag-HttQ68 (red), and probed for endogenous Alfy (green). Inset shows co-localization of Alfy and the polyQ inclusions (yellow). C,D. Flag-HttQ68 (green) transfected cells were treated 40h later with 5nM leptomycin B for 4h, fixed and stained with anti-Alfy antibodies (red). Draq5 (blue) labels nuclei. Scale bar = 10μm. LMB treatment inhibited recruitment of Alfy to cytoplasmic inclusions (30 cells, 2 independent experiments. Student t-test: p<0.005). E. LMB inhibits Alfy translocation under nutrient rich and nutrient starved conditions. HeLa cells were starved for 45min in HBSS±LMB. Cells containing cytoplasmic Alfy positive structures were counted (n>300 cells from n=3 experiments). F. Htt103Q-mCFP cells were treated with dox to abolish expression (7d). Dox was then removed to re-establish expression. Cells were collected at day 0, 2, 6 and 10. Relative Alfy mRNA levels were determined by qRT-PCR and normalized to d0. TBP and actin were used as controls. N=2 experiments were done in duplicate. G–I, Alfy is required for clearance of expanded polyQ aggregates. G. Htt103Q-mCFP cells were quantified for mCFP puncta per cell. 100μg/mL dox for 5d led to a significant decrease of `% Cells with aggregates' (p<0.001). siALFY1 and siALFY2 significantly inhibited clearance (*: p<0.001), while siCTRL did not (p=0.3218). Representative confocal images of each group generated on the INCA3000 shown. Nuclei are stained with Hoechst33342 (blue) and HttQ103-mCFP puncta are in green. H,I. Htt65Q- and Htt103Q-mCFP were transfected with siCTRL or siALFY and exposed 48h later to dox for 3d. H. Alfy KD inhibited clearance of SDS-insoluble protein for 65Q (p<0.05) and 103Q (p<0.05) cells (Student t-test). No detectable change was seen in SDS-soluble proteins as shown by SDS-PAGE. 5mM 3MA for 48h was used as a macroautophagy control. I. Alfy KD and Atg5 KD inhibited clearance of SDS-insoluble inclusions. Data shown as Mean+St.Dev. Complete statistics can be found in Supplemental Materials under Statistical Information for Figures
Alfy is required for macroautophagy-mediated clearance of aggregated protein
But why is Alfy recruited to these aggregating proteins? In previous studies, we and others have shown that macroautophagy is required to clear protein aggregates (Iwata et al., 2005a; Iwata et al., 2005b; Ravikumar et al., 2002; Yamamoto et al., 2006). To analyze aggregate clearance, HttPolyQ-mCFP cell lines were treated with doxycycline (dox) (Yamamoto et al., 2006), which led to inclusion clearance over several days in a macroautophagy-dependent manner (Figure S2, S3)(Yamamoto et al., 2006) (quantified as the number of mCFP puncta per cell). To determine if Alfy plays a role in clearance, we transfected siRNA sequences against Alfy (siALFY), which significantly impeded clearance of aggregated polyQ protein (Figure 2G). A control sequence (siCTRL) had no effect.
To independently quantify the impact of Alfy knockdown (KD) on aggregate-clearance, we used a filter trap assay, which has been successfully used by several groups to examine the presence of SDS-insoluble aggregates (Bailey et al., 2002; Passani et al., 2000; Wanker et al., 1999). Alfy KD significantly inhibited clearance of insoluble Htt65Q- or Htt103Q-mCFP (Figure 2H, dot blots). The degree of inhibition due to Alfy KD was comparable to the effect of 3-methyladenine (3MA), a chemical inhibitor of macroautophagy. Similar Alfy-dependence was obtained in N2a Htt103Q-mCFP cells (Figure S3D). Co-depletion of Alfy and Atg5 led to no detectable further inhibition of Htt polyQ clearance, compared to depletion of each protein individually (Figure 2I, S3D), suggesting that Alfy eliminated the aggregated polyQ proteins by macroautophagy. Moreover, Alfy KD had no measureable effect on the chymotrypsin-like activity of the proteasome, suggesting that Alfy is not required for proteasome activity (Figure S3E). Interestingly, western blot analysis of the SDS-soluble Htt65Q- or 103Q-mCFP protein indicated that Alfy KD had little effect on the decrease of soluble polyQ protein (Figure 2H); however since it is unclear if this decrease is due to degradation of the SDS-soluble protein or conversion to an SDS-insoluble pool, we examined the clearance of non-aggregating Htt25Q-mCFP. Although 3MA and BafA1 treatment somewhat inhibited clearance of Htt25Q-mCFP, Alfy KD had no effect (Figure S2F,G), indicating that Alfy is not involved in lysosomal clearance of nonaggregated polyQ proteins.
Alfy is not required for starvation-induced macroautophagy
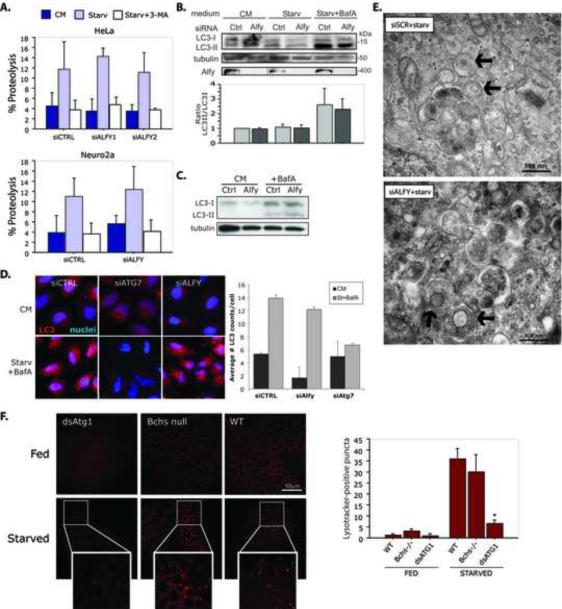
The requirement of Alfy for macroautophagic clearance of aggregated proteins implies that it may be required for general starvation-induced macroautophagy. We therefore determined the Alfy-dependence of macroautophagy in response to amino acid withdrawal (Klionsky et al., 2008)(Figure 3). We first examined the degradation of LLPs in both HeLa and N2a cells subjected to serum and amino acid withdrawal. As expected, a significant increase in LLP proteolysis that was 3MA dependent was observed in both cell types (Figure 3A, Starv+3MA). Although KD of Beclin1 also inhibited LLP degradation (Figure S2C), depletion of Alfy had no significant effect (Figure 3A). Constitutive gene knockdown in cell lines stably expressing shRNA against Alfy also showed no deficits in LLP degradation (Figure S2H).
Figure 3.
Alfy is not required for starvation-induced autophagy. A. Alfy is not required for LLP degradation. Amino acid withdrawal leads to a significant effect of amino acid withdrawal (Starv) in both HeLa and N2a cells. Knockdown of Alfy had no significant effect on LLP degradation (HeLa: p=0.6991; N2a: p=0.3505). To ensure that LLP degradation was due to macroautophagy, starvation experiments were also performed +10mM 3MA. Cells were transfected with siCTRL or siALFY, then monitored for LLP degradation 72h later (n = 3 experiments in duplicate or triplicate). B, C. Alfy depletion has no effect on starvation-induced or basal (C) LC3-lipidation (p=0.7641; n=4). HeLa cells transfected with siALFY or siCTRL were cultured in complete medium (CM) or starved (Starv) in the absence or presence of BafA1 for 4h (B) or 2h (C). D. Alfy depletion has no effect on LC3 puncta formation. Cells transfected with siCTRL, siATG7 or siALFY were starved, fixed and stained for endogenous LC3. In starved cells, there is significantly greater number LC3-positive puncta in siCTRL and siALFY cells, whereas siATG7 cells have significantly fewer LC3-positive puncta upon starvation between siCTRL and siATG7 (p<0.0001) and siALFY and siATG7 (p<0.0001). Bars represent mean+St.Dev (n=8). E. EM reveals no detectable difference in autophagosome (black arrows) formation due to Alfy depletion. siALFY or siCTRL transfected cells were starved for 4h before fixation and imaging. F. Autolysosome accumulation in Drosophila fat body cells is not dependent on Bchs. In response to starvation (3h sucrose, n=3), WT larvae (n=3) show an increase in autolysosome numbers, while loss of Bchs (bchs3/Df(2L)clot7, n=3) has no significant impact on this response (p=0.2151). As a control of suppressed macroautophagy, knockdown of Atg1 (CG-Gal4;UAS-dsAtg1i, n=3) leads to a significant inhibition of autolysosome accumulation (p=0.0005). Scale bar is 50μM. All bars represent Mean+St.Dev. Complete statistics can be found in Supplemental Materials under Statistical information for Figures.
We next explored whether Alfy is required for LC3-lipidation. The conversion of cytosolic LC3 (form I) to membrane bound LC3 (form II) has been correlated with autophagosome production (Kabeya et al., 2000). Neither starvation-induced (Figure 3B) nor basal lipidation (Figure 3C) was affected by Alfy depletion. In a complementary experiment, we examined endogenous LC3 puncta formation in response to starvation. siCTRL, siALFY and Atg7-depleted (siAtg7) cells were starved for 4h in the presence of BafA1, and stained for endogenous LC3 (Iwata et al., 2005b) (Figure 3D). Starvation led to a significant increase of LC3 puncta in control cells, which was inhibited in the absence of Atg7. In contrast, Alfy KD had no significant impact on LC3 puncta formation, consistent with the LC3 conversion data (Figure 3B). Finally, EM analyses of cells after starvation also showed no observable difference in autophagosome formation after Alfy KD(Figure 3E).
To further clarify the role of Alfy in starvation under physiologic conditions, we examined the role of its Drosophila homologue bchs in the starvation response of the larval fat body. Previous studies have shown that rapid onset of starvation-induced autophagy can be reliably followed in fat body cells by following autolysosomal content using the marker Lysotracker Red (LR) or a fluorescent autophagy marker, GFP-Atg8a (Lindmo et al., 2006; Rusten et al., 2004; Scott et al., 2004). Fat body tissue isolated from wild type (WT), two different bchs mutants (bchs3, bchs7) (Finley et al., 2003) and Atg1 depleted (CG-GAL4;UAS-dsAtg1-RNAi) fed or starved second instar larvae were incubated with LR and autolysosome accumulation was examined (Figure 3F, S4). Fat bodies dissected from fed larvae of all genotypes showed diffuse staining, with little to no detectable punctate structures. In contrast, starvation for 3h caused wildtype and bchs mutant larvae to display intense, punctate LR staining in fat body cells (Figure 3F, bchs3 S4A, bchs7). Larvae expressing dsAtg1-RNAi in the fat body had significantly fewer puncta, indicating that starvation-induced autophagy can be inhibited at this developmental stage. Similar patterns were also detected using LR in combination with a GFP-Atg8a (Figure S4B). These data provide further evidence that the Alfy/Bchs is not an essential component for a response to cellular starvation.
Alfy directly interacts with Atg5 and can be found in a complex with Atg5, Atg12 and Atg16L
We next sought to determine how Alfy contributes to the degradation of aggregation prone proteins. The C-terminus of Alfy contains protein-protein interaction domains, such as a Beige and Chediak Higashi (BEACH) domain and WD-40 repeats, and a PI3P-binding FYVE-domain (Figure 4A). In light of the potential of Alfy to interact with both protein and lipid, we hypothesized that Alfy may act as a scaffold for the macroautophagic machinery onto the aggregation-prone protein.
Figure 4.
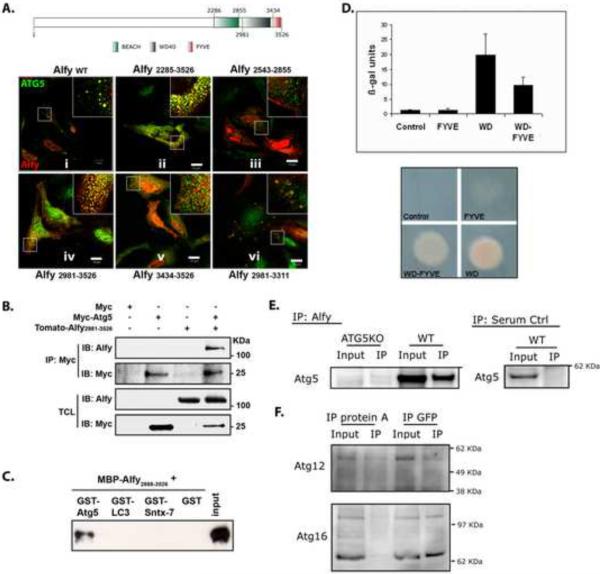
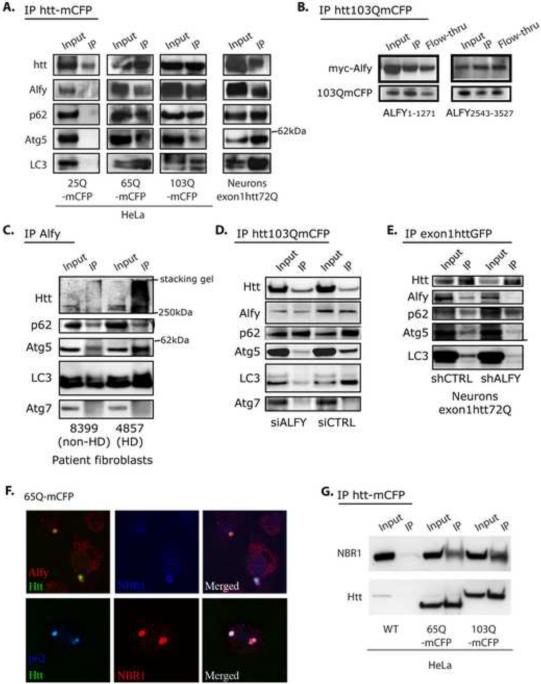
Alfy interacts directly with Atg5 A. Schematic figure of Alfy noting the location of its BEACH domain, WD-40 repeats and FYVE domain. Numbers indicate amino acids. Endogenous Alfy (red) co-localizes with endogenous Atg5 (green, panel i) in non-starved HeLa cells. Constructs containing the WD-40 repeats (amino acids 3095–3436) co-localized with Atg5 (ii, iv, vi). Expression of neither BEACH (iii) nor FYVE domains (v) alone led to Atg5 colocalization. B. Alfy C-terminus and Atg5 can be found in the same complex in cell lysates. HeLa cells were cotransfected with the indicated plasmids and subjected to IP using anti-myc antibodies, then probed with anti-Alfy or anti-myc antibodies. C. Atg5 directly interacts with the WD-40 repeats of Alfy as determined by GST-pull down. GST, GST-Atg5, GST-LC3 or GST-Syntaxin-7 were immobilized on pre-washed beads and then incubated with MBP-Alfy2888–3526. Bound proteins were eluted and analyzed by immunoblotting against Alfy. D. Atg5-Alfy interaction revealed by Y2H. The yeast reporter strain L40 was co-transformed with pLexa-Atg5 and the indicated pGAD-Alfy plasmids. As a negative control, the strain was transformed with the plasmids pLexa-Atg5 and pGAD-GH. A positive interaction was detected in yeast co-expressing Gal4 activation domain fused to the WD40-repeats of Alfy together with the reporter activation sequence binding LexA-Atg5 fusion, as shown by β-galactosidase activities (upper) and HIS3 activities (lower). Reporter activity in yeast expressing the FYVE domain alone was similar to controls. The values of β-galactosidase units are represented as Mean+Std. Dev. (n=4). E. Endogenous Alfy can be found in a complex with endogenous Atg5–Atg12 in WT mouse embryonal fibroblasts (MEFs) but not in ATG5 knockout MEFs. Whole cell lysates generated from MEFs were subject to IP using Rb anti-Alfy antibody serum and probed for Atg5. Serum controls in WT MEFs were also negative for Atg5. F. Atg12 and Atg16L can also be found in a complex with Alfy. Whole cell lysates generated from Htt103Q-mCFP cells were immunoprecipitated with GFP or with protein A beads as a negative control.
Using IF, we found that endogenous Alfy co-localizes with endogenous Atg5 (Figure 4A, panel i), and they both localize to the polyQ inclusion in the stable Htt103Q-mCFP cells (Figure S5A). To determine which region of Alfy is required for the co-localization with endogenous Atg5, different regions of Alfy tagged to the fluorophore tdTomato were over-expressed in non-starved cells. We found that the Atg5-Alfy co-localization required the C-terminal region of Alfy encoding the WD40 repeats (Figure 4A, panel ii, iv, vi) whereas the BEACH domain (Figure 4A, panel iii) and FYVE domain (Figure 4A, panel v) were not required. There was no colocalization of the N-terminal part of Alfy with Atg5 (Figure S5B).
To further resolve the nature of this co-localization, we next performed co-immunoprecipitation (co-IP) experiments using over-expressed and recombinant deletion mutants of Alfy. As shown in Figure 4B, overexpression of myc-tagged Atg5 co-immunoprecipitated with the same fragment of tdTomato-tagged Alfy that co-localized in Figure 4A, panel iv (Alfy2981–3526). In contrast, overexpression of the myc tag alone did not co-IP Alfy. To determine if this interaction was direct, recombinant GST-tagged Atg5 was incubated with the C-terminal WD40-containing region of Alfy fused to maltose binding protein (MBP-Alfy2888–3526). MBP-Alfy2888–3526 was pulled down with GST-Atg5, indicating that this region of Alfy interacts directly with Atg5. To ensure the specificity of this interaction, GST-LC3 and GST-Syntaxin-7 were used as negative controls (Figure 4C). Next, yeast two hybrid (Y2H) confirmed a positive interaction between the WD40-repeats of Alfy and Atg5 in both a beta-galactosidase liquid assay and a growth selection assay scoring for the HIS3 reporter (Figure 4D, S5C). No growth was detected with the FYVE domain alone or a control (Figure 4D). These results are consistent with the data from our co-localization studies, and strongly indicate that Alfy directly interacts with Atg5 through the WD-40 domain. Since overexpression can yield false-positive results, we also examined if this interaction can be found with endogenous proteins (Figure 4E). Whole cell lysates from immortalized WT and ATG5KO mouse embryonic fibroblasts (MEFs) (Hosokawa et al., 2006) were incubated with anti-Alfy or Ctrl serum, and immunoprecipitated using proteinA beads. Atg5 can be found in a complex with endogenous Alfy in WT cells, confirming that Alfy and Atg5 interact. No precipitation was observed in either Atg5KO MEFs or serum control IP in WT MEFs.
While the monomeric form of Atg5 is approximately 35kDa, the co-IP experiment revealed an approximately 56kDa protein. This higher molecular weight species is indicative of the Atg5-Atg12 complex, which is the predominant form at which Atg5 is found in vivo. Indeed probing for Atg12 confirmed the presence of the Atg5-Atg12 complex, since Atg12 was also found at 56kDa (Figure 4F). Moreover, probing for Atg16L also revealed its presence (Figure 4F), strongly suggesting that in cells, Alfy can be found in a complex with the Atg5-Atg12-Atg16L E3-like ligase for the lipidation of LC3(Fujita et al., 2008).
Alfy scaffolds the aggregation prone protein with the macroautophagic machinery
We have thus far determined that Alfy is required for macroautophagy-mediated clearance of aggregated polyQ protein and that it can form a complex with Atg5-Atg12-Atg16L. We next asked if Alfy forms a complex with aggregated polyQ to potentially recruit the autophagic machinery to the inclusions. IF showed that endogenous, full length Alfy co-localized with the Htt inclusions (Figure 2B). To determine if this is indicative that Alfy and aggregated mutant Htt is in a complex, we performed co-IP experiments. The polyQ protein tag was immunoprecipitated by its fluorophore from lysates generated from either the HttpolyQ-mCFP stable cell lines or a primary neuronal lentiviral model that expresses an exon1 fragment of Htt with 72Q (exon1Htt72Q) tagged to GFP (Figure 5A). Alfy co-immunoprecipitated with Htt65Q- and 103Q-mCFP, as well as with exon1Htt72Q. Interestingly, Alfy did not co-IP with Htt25Q-mCFP (Figure 5A), consistent with our findings that Alfy is not required for the elimination of soluble polyQ proteins (Figures 2H and S3).
Figure 5.
Alfy is in a complex with the aggregation prone mutant huntingtin protein and endogenous Atg5, p62, NBR1 and LC3. A. Co-IP experiments from whole cell lysate from stable cell lines or primary neuronal model of HD. Lysates from stable cell lines Htt25Q-, Htt65Q- and Htt103Q-mCFP were first immunoprecipitated via the CFP-tag of the exon1Htt protein, then probed for endogenous proteins with antibodies against Alfy, p62, Atg5, Atg7 and LC3, and reprobed for Htt. Primary neurons transduced with lentivirus carrying exon1Htt72Q-GFP were first immunoprecipitated for Htt, then probed with antibodies against Alfy, p62 and LC3, and reprobed for Htt. B. Both a myc-tagged N-terminal and C-terminal fragment of Alfy co-IPs with exon1Htt-mCFP. 103QmCFP was IPed with an antibody against GFP which also recognizes the mCFP tag. C. Co-IP experiments with anti-Alfy serum from whole cell lysates from control or HD patient fibroblasts. In HD patient samples, Alfy co-IP with the mutant huntingtin protein, p62, Atg5 and LC3, but not Atg7. Alfy still co-IPs p62, Atg5 and LC3 in unaffected patient samples, suggesting that there are non-HD aggregates that may be recognized by Alfy. D–E. Alfy is required for bringing Atg5 and LC3 to the complex. Experiments were performed similarly to B, but after transfection with siRNA or shRNA against Alfy or a Ctrl sequence. D. Htt103Q-mCFP cell line. E. Primary rat cortical neurons. Immunoblotting (with antibodies against GFP, Alfy, p62, Atg5 and LC3) were all performed on the same blot. Based on the Htt input levels, transduction efficiency for 5F was not as efficient as in 5B. All IP experiments are representative images from at least 3 independent experiments. F. Endogenous Alfy, p62 and NBR1 all localize to 65Q-mCFP aggregates G. NBR1 forms a complex with the aggregation prone mutant huntingtin protein. Htt65Q- and Htt103Q-mCFP were IP'ed and the blot probed for NBR1
To delineate which portion of Alfy may interact with the aggregating protein, an N-terminal fragment (Alfy1–1271), a central fragment (Alfy2285–2981) and a C-terminal fragment (Alfy2981–3526)) tagged to tdTomato, were over-expressed in Htt103Q-mCFP cells. Two different patterns emerged (Figure S12A): The N-terminal fragment completely co-localized with the Htt aggregate, suggesting it was distributed throughout or decorated the outside of the inclusion. In contrast, the C-terminal fragment was only found to decorate the outside of the inclusion. The internal Alfy fragment or tdTomato alone neither co-localized with nor coated the inclusion (Figure S6A). To determine if the co-localization indicated that the proteins were in a complex, we performed co-IP experiments (Figure 5B). A myc-tagged Alfy1–1271 was found to complex with Htt103Q-mCFP. Under the same stringency conditions, the myc-tagged Alfy2981–3526 was also bound to Htt103Q-mCFP. Taken together, both the N- and C-terminal fragments of Alfy can interact with the polyQ protein.
We next sought to determine if Alfy can recruit Atg5 or any other autophagic proteins to the polyQ protein aggregates (Figure 5A). Further immunoblotting revealed that endogenous Atg5, LC3 (form I and II) and p62 also co-IP with the aggregated HttpolyQ protein, both in the stable cell lines and in the neuronal lentiviral model (Figure 5A). The interaction appeared specific, since another macroautophagic protein, Atg7, failed to co-IP with the mutant polyQ protein (Figure 5D,E). Moreover, while Alfy was required for recruitment of Atg5 to the inclusion, interaction of Alfy with the polyQ protein did not require the presence of Atg5 (Figure S6B). Finally consistent with Alfy recruitment, Atg5 did not co-IP with Htt25Q-mCFP. LC3 (form I and II) and p62 were also absent (Figure 5A).
To further ascertain whether Alfy is central to recruiting Atg5 to the aggregates, we used RNAi to deplete Alfy in Htt103Q-mCFP cells (Figure 5D) and in neurons (Figure 5E) and re-examined the complex. Decreased Alfy levels diminished the interaction with Atg5 to a degree consistent with KD efficiency. The interaction of p62 with Htt103QmCFP, however, was not Alfy dependent, consistent with p62 interacting with polyQ proteins in an ubiquitin-dependent manner(Bjorkoy et al., 2005). Unexpectedly, Alfy KD led to a diminished amount of LC3 present in the complex (Figure 5D,E). Since there was no direct interaction of C-terminus Alfy with LC3 (Figure 4C), this data implies that Alfy-mediated recruitment of Atg5 to the inclusion might aid the stable association of LC3 with p62 or that LC3 interacts through another region of Alfy.
Although the Htt fragment used in heterologous systems can effectively model protein aggregation, the full length Htt protein is also known to aggregate in HD. We therefore determined if Alfy plays a similar role under physiologic levels of Htt protein, using fibroblasts collected from HD and unaffected patients. Indeed, immunoprecipitation with Alfy revealed that endogenous Htt comes down with Alfy only in HD patient samples. Interestingly, however, Atg5, LC3 and p62 are found in a complex with Alfy in both samples, indicating that the Alfy complex exists independent of Htt aggregation, and thus may generalize across different aggregating proteins. To explore this further, we turned to a non-polyQ protein known to form cellular aggregates, α-synuclein (α-Syn) with and without its more aggregation-prone mutation A53T (Figure S6C). Although the soluble form of α-Syn can be degraded by chaperone mediated autophagy(Cuervo et al., 2004; Vogiatzi et al., 2008), its aggregated form may be degraded by macroautophagy (Sarkar et al., 2007). Confocal microscopy and filter trap analysis revealed that in these cell lines, both α-Syn-mCFP and α-Syn(A53T)-mCFP form aggregates, although more frequently in the latter (Figure S6C, D). Consistent with polyQ aggregate clearance, filter trap analysis revealed that Alfy KD inhibited the ability of the cells to eliminate the SDS-insoluble α-Syn proteins, and co-IP analysis found that Alfy and Atg5 precipitated with the synuclein proteins(Figure S6E). Taken together, these data suggest that Alfy is required for the elimination of different types of aggregated proteins.
Recently, NBR1 was found to cooperate with p62 in the sequestration of misfolded ubiquitinated proteins for degradation by autophagy (Kirkin et al., 2009). Both proteins have an ubiquitin-binding UBA domain and LC3 interacting regions (LIR). Since we found that Alfy makes a complex p62-positive polyQ proteins (Figure 5A,D,E), we also pursued NBR1. NBR1 colocalized with Alfy and polyQ inclusions and was found to co-IP with mutant Htt polyQ proteins (Figure 5F,G). Overall this suggested that polyQ protein can interact with p62 and NBR1 and Alfy may target these proteins for degradation.
Alfy overexpression can enhance aggregate clearance in a primary neuronal model of HD
We have found that Alfy is required for aggregate-clearance, and it may do so by scaffolding the macroautophagy machinery through its C-terminus. Interestingly, we generally noticed fewer polyQ inclusions upon ectopic expression of the C-terminal fragment of Alfy and we therefore speculated that by increasing Alfy expression, we may increase mutant Htt elimination and in turn abrogate toxicity of mutant Htt. Htt103QmCFP cells were transiently transfected with the C-terminus of Alfy containing the WD repeats and the FYVE domain (Alfy2981–3526). Quantification of the number of cells containing inclusions revealed that overexpression of this fragment indeed enhanced clearance of polyQ aggregates, but not in cells depleted of Atg5 (Figure S7A). Moreover, mutagenesis throughout the WD-40 repeat of this construct (4 W mutated to A) also inhibited inclusion clearance, indicating that clearance is dependent on Atg5 binding to Alfy (Figure S7A).
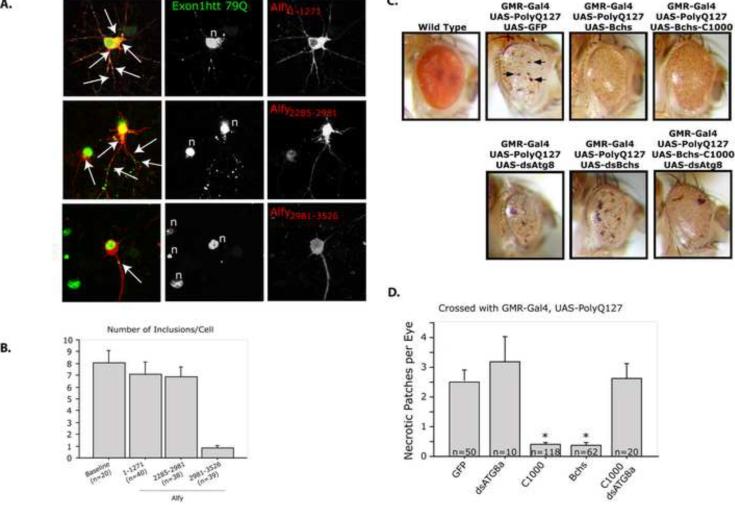
To determine if overexpression of Alfy can diminish the level of mutant Htt aggregates in a non-dividing cell, we used the neuronal model of HD we described for Figure 5. Neurons were transduced at day in vitro (DIV) 2 and transfected with different Tomato-tagged Alfy constructs 5d later, after the appearance of inclusions. Cells were then fixed 72h post transfection and examined for the presence or absence of inclusions. As shown in Figure 6A, transduced neurons demonstrated robust staining of exon1Htt72Q-GFP, and inclusions were found in the soma as well as within the processes of the neurons. Protein also accumulated within the nucleus, as has previously been reported; however due to the intensity of the staining it was unclear if discreet puncta could be observed. Although transfection of an N terminal (Alfy1–1271) or internal (Alfy2285–2981) portion of Alfy had no effect on aggregation, overexpression of the C-terminal fragment of Alfy (Alfy2981–3526) led to a significant reduction of protein aggregates in the neurons (Figure 6A,B). Similar to our finding in HeLa cells, co-localization of Alfy1–1271 with Htt inclusions could also be observed (Figure S7B). Overexpression of the full length 400kDa Alfy was very difficult to achieve, but in the few cells that were successfully transfected, the frequency of inclusions was fewer (Figure S7C). These data indicate that overexpression of Alfy or the C-terminal of Alfy can diminish protein accumulation, further supporting that Alfy is required for clearance of aggregates by acting as a scaffold for the macroautophagic machinery.
Figure 6.
Alfy/Bchs overexpression leads to a disappearance of inclusions in a primary neuronal model of HD and diminished neurotoxicity in a Drosophila eye model of polyQ toxicity. A,B. Alfy overexpression in a lentiviral model of HD leads to fewer inclusions in rat primary cortical neurons. A. IF against exon1Htt (green) with MAB5492 reveals that transduced neurons exhibit robust aggregation throughout the cell, including the soma and processes (white arrows), and nuclei (n). Transfection of Alfy1–1271 (p=0.4241) and Alfy2285–2981 (p=0.3325) had no noticeable effect on the number of inclusions/neuron. In contrast, expression of Alfy2981–3526 led to a significant reduction in the aggregate load in the neurons (p<0.001). The visible green puncta in the field that do not co-localize with red are attributable to neuronal processes from non-Alfy transfected neurons. B. Number of inclusions per cell was counted in the number of cells indicated. All visible puncta within a 100 m radius from the center of the nucleus were counted. Cells were considered within the cell if the puncta (green) co-localized to cytosol expressing the tomato fluorophore (red). C,D. Alfy/Bchs overexpression in the Drosophila eye leads to protection against polyQ toxicity. C. Canton-S control flies (wild type) show the normal pigmentation profiles and ommatidial features of the adult Drosophila eye. Expression of polyQ127 peptide (UAS-PolyQ-127, n=20) using the pGMR-Gal4 driver leads to distinctive defects throughout the eye including necrotic regions (pGMR-Gal4/UAS-PolyQ-127, arrows, n=20). Co-expression of full-length Bchs (pGMR-Gal4, UAS-PolyQ-127/UAS-EP-bchs, n=20) reduced the level of observable necrosis. Co-expression of PolyQ127 peptide with the C-terminal region of Bchs (pGMR-Gal4, UAS-PolyQ-127/UAS-bchs-C1000, n=20) leads to suppress this external toxic phenotypes. Co-expression of a dsAtg8a (pGMR-Gal4, UAS-PolyQ-127/UAS-bchs-C1000/UAS-dsAtg8a, n=15) RNAi transgene blocked this protective effect. D. The number of necrotic regions was determined for C. While co-expression of dsAtg8a further reduced eye size it did not significantly alter necrosis levels (p=0.2391). Both full-length and C1000-Bchs significantly reduced fly eye necrosis (p<0.0001). This decrease was abrogated when C1000 was co-expressed with a dsRNA against ATG8a (p=0.9977), which significantly inhibits macroautophagy in the developing eye. Complete statistics can be found in Supplemental Materials under Statistical information for Figures.
Alfy/Bchs overexpression is neuroprotective in a fly-eye model of HD
To determine if enhanced Alfy/Bchs expression can be neuroprotective, we assessed the impact of overexpressing Bchs in an established Drosophila eye model of polyglutamine toxicity (Kazemi-Esfarjani and Benzer, 2000). For this study we used the bipartate Gal4/UAS system to drive expression of a UAS-polyQ127 transgene in the developing fly eye (pGMR-Gal4). As previously shown, this genotype (Gal4,UAS-polyQ127) generates a phenotype that includes a reduced eye size, pigmentation loss, and ommatidial disorganization and the formation of necrotic regions (Figure 6C, arrows). This model has successfully identified genetic modifiers of polyglutamine toxicity (Bilen and Bonini, 2005; Kazemi-Esfarjani and Benzer, 2000; Lam et al., 2006; Pandey et al., 2007; Steffan et al., 2001).
Quantification of the number of necrotic areas revealed a significant degree of protection due to co-expression of full-length bchs (UAS-FL-Bchs) or C-terminal Bchs (UAS-bchs-C1000) with UAS-polyQ127 (Figure 6D). This included the reduction or absence of necrosis and an overall improvement in eye size, morphology and pigmentation. Interestingly, overexpression of Bchs-C1000 produces a higher level of protection than the full-length protein. This may in part be due to the fact that enhanced expression of the large and complex 400kDa Bchs protein produces its own external eye phenotype as well as subcellular axonal trafficking defects within the eye (GMR-Gal4,UAS-bchs) (Figure S7D) (Simonsen et al., 2007).
To determine if the Bchs-mediated protection required macroautophagy, we suppressed the pathway through depletion of Atg8a (UAS-dsAtg8a-RNAi). We have previously shown that the loss of Atg8a was sufficient to inhibit macroautophagy in adult Drosophila head, and that Atg8b is not expressed in this tissue (Simonsen et al., 2008). When the C-terminal Bchs was expressed together with Atg8a KD using a UAS-dsAtg8a transgene, the protection against polyQ127 generated by higher Bchs-C1000 levels was no longer observed (Figure 6C,D). Western analysis of polyQ127 peptide levels prepared from the different fly genotypes showed that changes in eye phenotypes were not due to altered expression of the transgene (Figure S7E). This set of experiments indicates that the Alfy/Bchs proteins have a significant role in suppressing the in vivo cytotoxicity of aggregation-prone proteins, in large part mediated through the macroautophagic pathway.
DISCUSSION
In this study we have found that Alfy is required for the macroautophagic elimination of aggregated proteins, but not for macroautophagic elimination of bulk cytosol in response to starvation. We propose that Alfy functions as a scaffold which brings together the E3-like ligase, Atg5-Atg12-Atg16L and LC3 to the target substrate to permit the degradation of expanded polyglutamine proteins in different cell types, including neurons. Finally, we have found that its overexpression can enhance the elimination of toxic polyglutamine proteins in a primary neuronal model, and permit protection in a Drosophila eye model of polyQ toxicity.
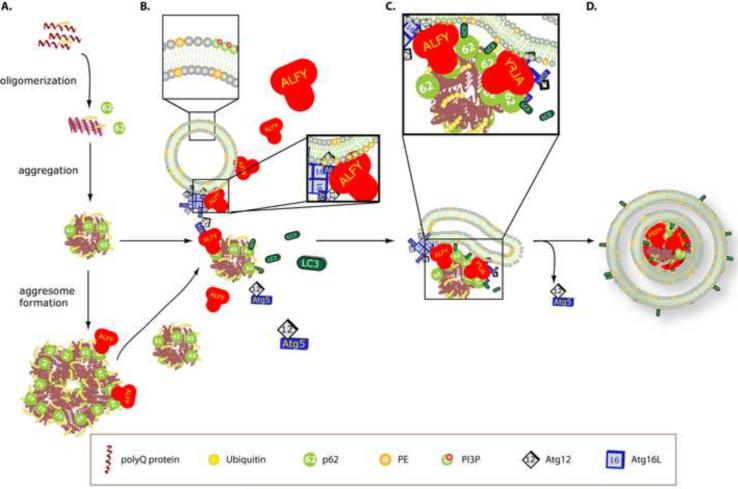
The role of the lysosome and its capacity to eliminate aggregated proteins of expanded polyQ proteins has been explored extensively (Iwata et al., 2005a; Ravikumar et al., 2002; Yamamoto et al., 2006). It has remained a question, however, how they are targeted to the autophagosome for degradation. Our findings demonstrate that Alfy plays an important role in this process, and we suggest the following model (Figure 7): As the polyQ protein is expressed and accumulates, it evolves from oligomers to increasingly larger aggregates to inclusions. The polyubiquitinated forms of the accumulating mutant protein are recognized by p62, which may aid aggregate-growth via self-polymerization of its PB1 domain (Bjorkoy et al., 2005), together with previously implicated microtubule- and HDAC6-dependent mechanisms (Iwata et al., 2005b; Johnston et al., 1998; Kawaguchi et al., 2003). Meanwhile, in the presence of aggregation-prone proteins, Alfy is transported from the nucleus to the cytoplasm where it may interact with PI3P-containing membranes through its FYVE domain (Simonsen et al, 2004) and Atg5-Atg12 through its WD-40 domain. Although drawn as a vesicle, the membrane source for the autophagosome is still unclear, although the ER(Axe et al., 2008; Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009) has been implicated. Once at the membrane, Atg5-Atg12 can interact with Atg16L (Fujita et al., 2008; Mizushima et al., 2003). Alfy also interacts with aggregated polyQ proteins most likely through its interaction with p62 (Høyevarde et al, 2010). In this manner, we speculate that Alfy brings a membrane source and Atg5-Atg12-Atg16L to the inclusion, so that they can act as part of a greater E3 ligase-like complex for LC3 (Hanada et al., 2007). The presence of p62 may stabilize the lipid bound LC3, thus permitting the autophagosome to build directly around the inclusion. Although giant (diameter > 1um) inclusions were not found frequently in double membrane structures by immuno-EM, the large inclusions contained were much larger than the usual 100nm size AVs. Clearly further in vitro and in vivo experiments must be performed to test this model directly.
Figure 7.
Proposed model of Alfy-mediated clearance of protein aggregates. A. Expression of expanded polyQ proteins lead to their polyubiquitination, accumulation and aggregation. The ubiquitinated protein can be recognized by p62. B. In the presence of aggregating protein, Alfy translocates from the nucleus into the cytosol, where it interacts with PI3P containing membranes through its FYVE domain, Atg5–Atg12 through its WD-40 domain and the ubiquitin- and p62- positive protein. p62, which interacts directly with LC3, may bring unbound LC3 to the inclusion as well. C. Alfy brings the different components together, permitting Atg5-Atg12-Atg16L to aid the conversion of LC3 to its PE bound form. It is uncertain if p62 binds free LC3, PE-bound LC3 or both; however interaction with PE-bound LC3 may help stabilize the membrane around the aggregate. Atg5–Atg12 possibly leaves as LC3 conjugation occurs along the membrane. D. Aggregates are packaged into autophagosomes to permit eventual degradation upon fusion to the lysosome. Not to scale.
This selective sorting of cargo in macroautophagy is reminiscent of protein trafficking, from endocytosis to secretion, for which particular cargo are directed to unique trafficking fates. Recently in yeast, Atg32 has been shown to confer selectivity during mitophagy(Kanki et al., 2009; Okamoto et al., 2009). In mammalian systems, several proteins that link the autophagic machinery to its substrates have been identified. Termed `autophagy receptors', they are involved in degradation of ubiquitinated bacteria (NDP52) (Thurston et al., 2009), mitochondria (NIX)(Novak et al.,; Sandoval et al., 2008), as well as protein aggregates (p62 and NBR1)(Kirkin et al., 2009; Pankiv et al., 2007). All proteins contain an LIR and substrate recognition domain. Since Alfy interacts with p62 (Høyevarde et al, 2010), it will be interesting to determine how Alfy influences the degradation of other autophagy receptor proteins and their cargo.
Our findings indicate that an innate cellular process to package protein aggregates into autophagic vacuoles exists in mammalian cells. Several results echo the Cvt (cytosol to vacuole targeting) pathway in yeast (Nair and Klionsky, 2005). Our ultrastructural studies show that relatively large aggregated structures can be found sequestered in double membrane vesicles, devoid of bulk cytosol. Further, the clearance of aggregated proteins clearly requires the macroautophagic machinery, but like Cvt, also requires additional unique factors to acts as a scaffold between the cargo aggregate and the membrane building machinery. Aggregation of the cargo may therefore be one of the first steps towards degradation, and hence inhibition thereof may confer toxicity, since the protein can no longer be eliminated. This may explain studies indicating the `protective' nature of inclusions in neurons(Arrasate et al., 2004; Taylor et al., 2003). Nonetheless, if indeed a Cvt-like process exists, why is there a prevalence of protein accumulation in human disease? Do levels of Alfy relate to the susceptibility to protein aggregation? We have previously shown that Alfy is ubiquitously expressed in mouse tissue, with highest levels found in brain (Simonsen et al., 2004). In Drosophila, expression level of the Alfy homologue bchs is highest in neurons. Alfy/bchs may be especially important in non-dividing cells which require `more help' to clear aggregate-prone or aggregated proteins to prevent accumulation of protein aggregates. Consistent with this are previously published studies demonstrating that loss of Bchs leads to the accumulation of ubiquitinated proteins in fly neurons (Finley et al., 2003). Our complimentary experiment using overexpression of Alfy in neurons shows that increased levels of Alfy or its C-terminus permits clearance (Figure 6, S7A), suggesting that Alfy may be rate limiting to the degradation of aggregated proteins. This may be linked to the prospect that Alfy can corral key members of the macroautophagy machinery, and thus its constitutive presence may limit the starvation response, leading to cell death. The nuclear localization of Alfy may limit its bioavailability, to avoid this possibility. This hypothesis and how cellular stress permits Alfy to exit the nucleus is under current investigation.
Materials and methods
Complete methods can be found in Supplemental Materials.
Human fibroblasts were obtained with permission from Coriell Cell Repositories.
Promoter Shutdown Experiments and Filter-Trap Assay
48h after siRNA transfection, HttPolyQ-mCFP cells were exposed to 100ng/ml dox for 3d to permit more than 50% of clearance. The effect of siRNAs on aggregate clearance was analyzed by confocal quantification as described (Yamamoto et al., 2006) or by membrane filter trap assay as previously published (Bailey et al., 2002; Passani et al., 2000; Wanker et al., 1999).
Long-lived protein degradation assay
Cells were labeled overnight with [14C]valine, chased in fresh media with excess valine overnight, then incubated for 4h with complete media (CM) or starvation media (HBSS/EBSS+10mM HEPES, Starv). To ensure that protein degradation during nutrient deprivation was due to macroautophagy, experiments were also performed in the presence of 3MA. LLP degradation was monitored 72h post-transfection.
Autolysosome assay in Drosophila fat body cells
Performed as previously described (Rusten et al., 2004). Briefly, second instar larvae were fed or starved on limited medium (3hr sucrose only medium). Fat bodies from individual larvae were dissected, stained with LR(Molecular probes) and immediately imaged by confocal microscopy. The number of autolysosomes in fat tissue was quantified from replicate 50μm2 areas. Information on Drosophila lines and genetic crosses can be found in supplemental materials.
Primary neuronal model of HD
Primary rat cortical neurons were plated at E18. Neurons were transduced with lentiviri carrying exon1Htt with 72Q on DIV2. Alfy constructs were transiently introduced at DIV7. Neurons were fixed and stained 72h later.
Quantification of necrotic eye regions in Drosophila eye model
Crosses were maintained at 25 °C and replicate eyes from individual genotypes were examined and the average number of necrotic regions counted and SEM calculated. Multiple representative digital images were taken from each genotype using a Leica MZ6 dissecting microscope and Nikon Coolpix 990 camera system. Information on lines and genetic crosses can be found in supplemental materials
Statistical analyses were performed using Statview 5.0 (SAS Institute). Images were processed using Adobe Photoshop 7.0 and 8.0.
Supplementary Material
Acknowledgements
This study was supported by the NINDS RO1 NS050199 (MA, KM, AY), NINDS RO1 NS063973 (TJM, AY), Parkinson's Disease Foundation (KMM, AY), Hereditary Disease Foundation (AY), NIH/NIA R21 AG030187 (KDF), the FUGE programme of the Research Council of Norway (PI, TL, AB, HS, AS) and the Norwegian Cancer Society (AS). We would like to thank Drs. R. Kopito and T. Yoshimori for their anti-LC3 antibodies, and Drs. N. Mizushima for the ATG5 KO MEFs and S.A.Tooze for HEK293 LC3-GFP cells. The UAS-PolyQ127 line was a gift from Seymour Benzer. The cryo EM was performed at the New York Structural Biology Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annual review of genetics. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27:503–519. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Cornett J, Cao F, Wang CE, Ross CA, Bates GP, Li SH, Li XJ. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat Genet. 2005;37:198–204. doi: 10.1038/ng1503. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase K, Davies S, Bates G, Vonsattel J, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, Hwang CE, Benedetti M, McKeown M. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohusmi Y. The ATG12–ATG5 conjugate has a novel e3-like activity for protein lipidation in autophagy. J Biol Chem. 2007 doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci U S A. 2005a;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005b doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Molecular cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy. Curr Biol. 2005;15:R282–283. doi: 10.1016/j.cub.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun ED, Duvick LA, Orr HT, Botas J, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- Lindmo K, Simonsen A, Brech A, Finley K, Rusten TE, Stenmark H. A dual function for Deep orange in programmed autophagy in the Drosophila melanogaster fat body. Exp Cell Res. 2006;312:2018–2027. doi: 10.1016/j.yexcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12–Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Passani LA, Bedford MT, Faber PW, McGinnis KM, Sharp AH, Gusella JF, Vonsattel JP, MacDonald ME. Huntingtin's WW domain partners in Huntington's disease post-mortem brain fulfill genetic criteria for direct involvement in Huntington's disease pathogenesis. Hum Mol Genet. 2000;9:2175–2182. doi: 10.1093/hmg/9.14.2175. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Rockel TD, Stuhlmann D, von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J Cell Sci. 2005;118:5231–5242. doi: 10.1242/jcs.02642. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nature chemical biology. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Lindmo K, Galaviz V, Cheng S, Rusten TE, Finley KD. Genetic modifiers of the Drosophila blue cheese gene link defects in lysosomal transport with decreased life span and altered ubiquitinated-protein profiles. Genetics. 2007;176:1283–1297. doi: 10.1534/genetics.106.065011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Stromhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335(Pt 2):217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, Fischbeck KH. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nature immunology. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283:23542–23556. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanker EE, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Membrane filter assay for detection of amyloid-like polyglutamine- containing protein aggregates. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- Zu T, Duvick LA, Kaytor MD, Berlinger MS, Zoghbi HY, Clark HB, Orr HT. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J Neurosci. 2004;24:8853–8861. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.