Abstract
Targeting and down-regulating specific genes with antisense and decoy oligonucleotides, ribozymes or RNA interference (RNAi) offers the theoretical potential of altering a disease phenotype. This article reviews the molecular mechanism behind the in-vivo application of RNAi-mediated gene silencing, focusing on its application to the inner ear. RNAi is a physiological phenomenon in which small, double-stranded RNA molecules (small interfering RNA, siRNA) reduce expression of homologous genes. Notable for its exquisite sequence specificity, it is ideally applied to diseases caused by a gain-of-function mechanism of action. Types of deafness in which gain-of-function mutations are observed include DFNA2 (KCNQ4), DFNA3 (GJB2) and DFNA5 (DFNA5). Several strategies can be used to deliver siRNA into the inner ear, including cationic liposomes, adeno-associated and lentiviral vectors, and adenoviral vectors. Transduction efficiency with cationic liposomes is low and the effect is transient; with adeno-associated and lentiviral vectors, long-term transfection is possible using a small hairpin RNA (shRNA) expression cassette.
Introduction
The premise underlying many models of inner ear gene therapy is based on the transfer of DNA constructs into the cochlea using viral vectors with the dual aims of protecting existing inner ear anatomy and effecting the regeneration of sensory hair cells and neurons. Ultimately, these strategies will realize the aim of replacing inner ear hair cells, providing a habilitation option for deafness that obviates the need for hearing aids or cochlear implants. Perhaps closer on the clinical horizon, however, are therapeutic interventions that block the expression of specific genes thereby preventing damage mediated by toxic molecules or the dominant-negative mechanism of action of select mutant proteins (1, 2).
The Dominant-Negative Phenotype and Deafness
Major molecular mechanisms of autosomal dominant disease can be broadly classified as either loss-of-function or gain-of-function mutations. In the case of the former, the phenotype reflects the consequence of haploinsufficiency. Gain-of-function mutations, in contrast, predict a transcribed protein that interferes in a negative way with normal function of the native protein. The classic example is the multimeric protein in which function depends on oligomerization of several protein subunits (3). For example, in the inner ear GJB2 (Gap junction protein beta2) encodes the transmembrane protein Connexin 26 (CX26), which oligomerizes with five other connexins (CX26 or CX30) to form the component unit of gap junctions, the connexon. Several missense mutations [W44C (4, 5), R75W (5, 6), D66H (5)] and an in-frame deletion (delE42) (5) of GJB2 are known to cause dominant deafness at the DFNA3 locus. These mutations localize to the extracellular and transmembrane domains of CX26 and not only inhibit function of mutant GJB2 in cultured cells but also interrupt gap junction properties of co-transfected wild-type GJB2 (5–7).
Deafness at the DFNA2 locus also reflects a dominant-negative mechanism of action. It is caused by missense mutations of KCNQ4 (8, 9), which encodes KCNQ4, a member of the voltage-gated potassium channel family that forms heteromeric channels with KCNQ3 (10). Kubisch and colleagues have shown that the G285S mutation of KCNQ4 lies in the conserved channel pore region in exon 6 and abolishes ~ 90% of the potassium current of wild type KCNQ4 in the Xenopus oocyte system, reflecting a strong dominant-negative effect (8).
Dominant disease from a gain-of-function mechanism may also occur when the mutant protein acquires a novel and toxic function. Mutations in DFNA5, for example, segregate in deaf persons in three families with autosomal dominant nonsyndromic deafness at the DFNA5 locus. All of the described mutations are insertion/deletion or nucleotide substitutions in intron 7 that predict the skipping of exon 8 to generate a novel protein (11–13). In the yeast cell system, expression of this novel protein causes loss of cell viability as compared to wildtype DFNA5, which has no toxic effect (14), and in cultured mammalian cells, the novel DFNA5 protein localizes to the plasma membrane and induces non-apoptotic cell death. Van Laer and colleagues have postulated that the mutant DFNA5 protein has a deleterious new function, the phenotypic consequence of which is deafness (15) . The cellular function of DFNA5 remains to be determined.
The ability to prevent in an allele-specific manner the translation of these mutant proteins should theoretically mitigate the phenotype with which they are associated. In this review, we focus on strategies to down-regulate gene expression, highlighting steps that must be considered to move toward the goal of offering gene therapy as a habilitation option for some types of deafness.
Down-Regulating Gene Expression
Antisense oligonucleotides (ASOs)
An ASO is a single stranded DNA-like molecule that is complimentary to a specific target mRNA molecule. Generally 15–20 nucleotides in length, an ASO can be introduced into cells in vitro and in vivo using liposomes (16). Molecule base-pairing of an ASO with its cognate mRNA and pre-mRNA leads to nuclear RNase-H-mediated cleavage of the RNA-ASO heteroduplex and steric blockage of translation. An ASO may also interfere with transcription by forming triplexes with chromosomal DNA and modulating RNA splicing and mRNA transport (17).
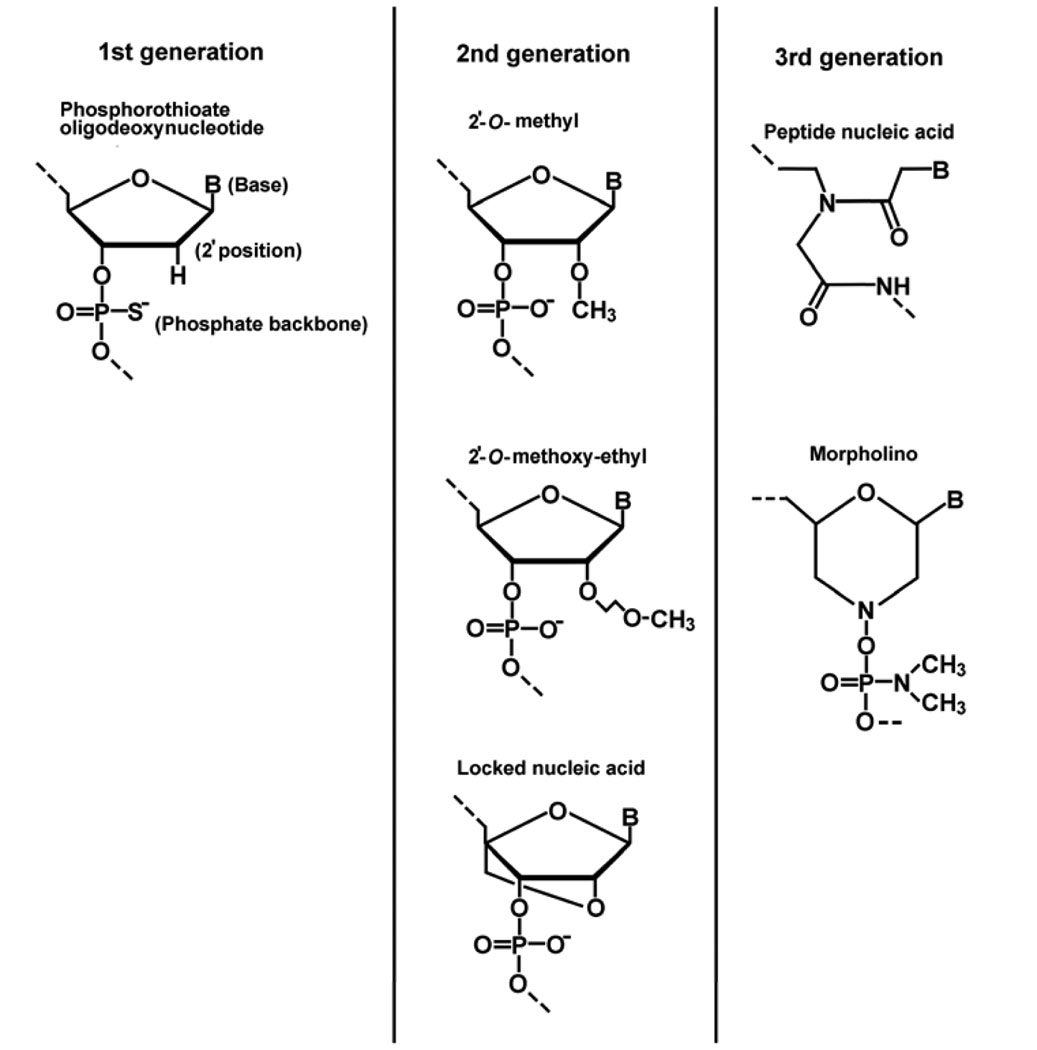
Several generations of ASOs have been developed, reflecting the limitation that endogenous nucleases rapidly degrade unmodified oligodeoxynucleotides, necessitating modifications to ASOs to make them resistant to nuclease activity while maintaining affinity and suppression potency for target mRNAs. First generation ASOs, for example, are phosphorothioate oligodeoxynucleotides with a nearly 10-fold greater serum half life as compared to conventional phosphodiester oligodeoxynucleotides. Second generation ASOs, with alkyl modifications at the 2’ position of the ribose, have enhanced affinity to complementary RNA when compared to first generation ASOs, however 2’-O-methyl and 2’-O-methoxy-ethyl RNA does not induce RNase-H-mediated cleavage of target RNA because of changes in the minor groove of the ASO-RNA duplex introduced with 2’- and backbone modifications (18). A central stretch of unmodified or phosphorothioate DNA between 5’- and 3’- 2’-O-methyl RNAs (so called “gapmers”) combines both nuclease resistance and RNase-H activation. Locked nucleic acids (LNA) are ribonucleotides containing a methylene bridge that connects the 2’ oxygen of the ribose and the 4’ carbon. Chimeric DNA·LNA gapmers possess enhanced stability and remarkable target affinity. Third generation ASOs, which include peptide nucleic acids and morpholino oligonucleotides are characterized by a deoxyribose backbone that has been replaced by either polyamide linkages or a morpholino moiety with phosphoroamidate linkages, respectively. This class of ASOs does not recruit RNase-H activity (19) (Fig. 1).
Figure 1.
Three generations of antisense oligonucleotides (B, bases: adenine, guanine, cytosine or thymine)
Not all types of ASOs have been tested for in vivo gene suppression. The first generation phosphorothioate ASOs have been most intensively studied in animal experiments (16) and clinical trials (19) targeting cancer (e.g. BCL2, protein kinase Cα, HSP27) (20), viral infections like cytomegarovirus (21), Crohn’s disease (22) and asthma (23). With respect to the inner ear, Delprat and colleagues perfused the scala tympani of guinea pigs with phosphorothioate DNA to target endogenous Otos (otospiralin) expression and induced threshold elevation of cochlear compound action potentials. (24).
Decoy oligodeoxynucleotides (ODNs)
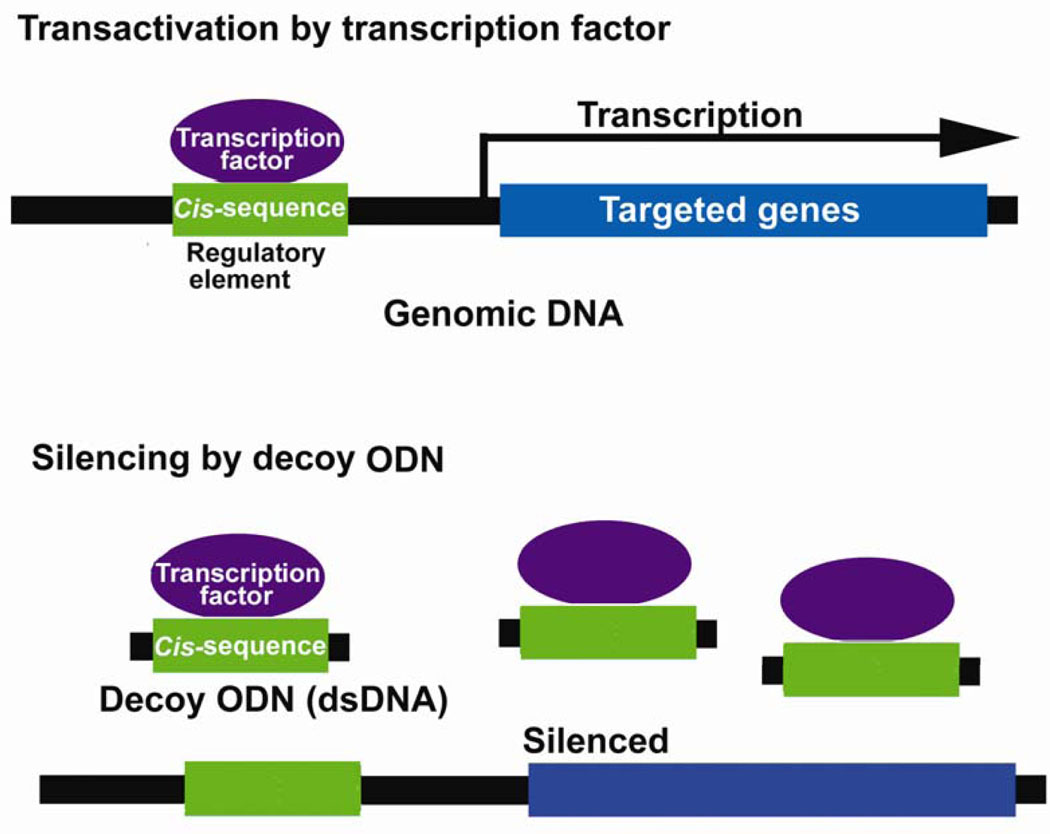
Exogenous double-stranded DNA (14–30 nt) corresponding to the cis sequence of a transcription factor can compete in the cis-trans interaction of a transcription factor with endogenous cis-elements, leading to regulation of gene expression at the transcriptional level (Fig. 2). Decoy ODNs can be transduced into cell nuclei using inactivated hemagglutinating virus of Japan (HVJ) complexed with liposomes (25). Examples include ODNs against the transcription factors E2F and NFκB.
Figure 2.
Scheme of decoy oligodeoxynucleotide (decoy ODN) strategy to down-regulate gene expression. Transcription factors transactivate transcription of target genes through binding to cis-consensus sequences of regulatory elements in genomic DNA. Decoy ODN (double-stranded DNA containing cis-consensus sequence) compete with this cis-trans interaction and regulate the expression of downstream targeted genes.
E2F plays a critical role in the coordinated transactivation of cell-cycle regulatory genes such as c-myc, cds2, and the proliferating-cell nuclear antigen gene. Transfection of decoy ODNs against E2F results in suppression of these genes, leading to inhibition of vascular smooth muscle cell proliferation in a rat model of carotid injury (26). NFκB may also mitigate the effects of ischemic myocardial damage. This transcription factor induces the coordinated transactivation of cytokine and adhesion molecule genes that are presumed to be involved in myocardial damage after ischemia and reperfusion. Consistent with this association, decoy ODNs against NFκB have an inhibitory effect on myocardial infarction in a rat model (27). Clinical trials of E2F- and NFκB- decoy ODNs are ongoing to treat patients after angioplasty, with clinical application of decoy ODNs against E2F being used to treat neointimal hyperplasia in vein bypass grafts (28).
In the inner ear of animal models, NFκB expression is up-regulated on induction of apoptosis by cisplatin (29), aminoglycoside (30) and explantation (31) in the cochlear lateral wall and organ of Corti . The latter two studies, by Schacht and colleagues and Bodmer and colleagues, respectively, found activation of NFκB protective against cell death in the organ of Corti. In vivo suppression experiments of either E2F or NFκB are yet reported in the inner ear
Hammerhead ribozymes
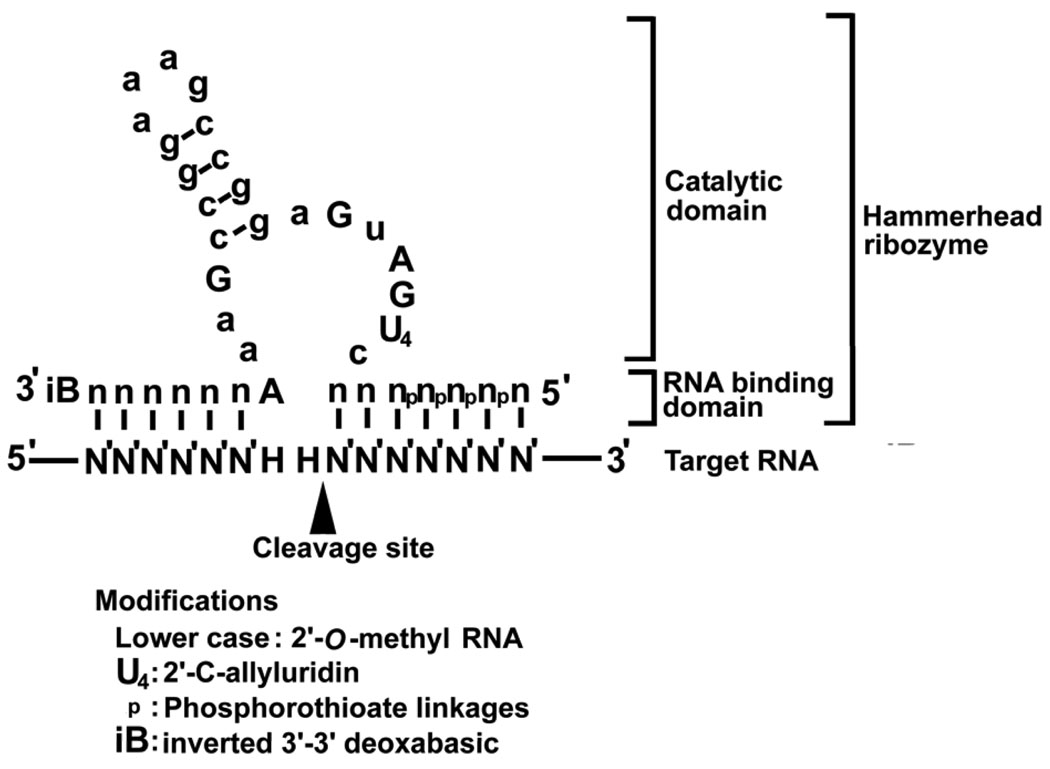
The hammerhead ribozyme is a 30–40 nucleotide-long RNA molecule so named for its hammer-like appearance. Like other ribozymes, the hammerhead ribozyme is an antisense RNA with two RNA binding domains and a catalytic domain that form covalent links with target RNA molecules, which are then cleaved at NHH triplets (H, any nucleotide except guanosine with U in the second position making the cleavage site most efficient) (32). The NHH triplets are flanked by sequence homologous to the RNA-binding domain, resulting in posttranscriptional down-regulation of specific gene expression (33) (Fig. 3).
Figure 3.
Secondary structure of a hammerhead ribozyme. A hammerhead ribozyme is composed of two RNA binding domains and one catalytic domain. The target RNA is cleaved at NHH triplet (H, any nucleotide except for guanosine). Four types of modifications to achieve nuclease resistance are illustrated.
Hammerhead ribozymes were discovered originally as cis-acting ribozymes in plant viroids, but with engineering to minimize size and include substrate-specific domains, the hammerhead ribozyme has become a possible tool for gene therapy (34). Ribozymes can be introduced into cells using cationic liposome or plasmid and viral expression cassettes with RNA polymerase III (pol III) promoters. (35) To apply chemically synthesized hammerhead ribozymes in vivo, the RNA molecule must be stabilized against nucleolytic degradation by including 2’-O-methyl RNA and 2’-C-allyl uridine (36). The 5’ and 3’ ends can be protected, respectively, by phosphorothioate linkages and by an inverted 3’-3’ deoxyabasic sugar (37) (Fig. 3). These changes increase the serum half life to more than 5 h as compared to less than a few minutes for the unmodified ribozyme molecule. Zynzyme, a further modified ribozyme, has a half life of more than 100 h in human serum (38).
A stabilized hammerhead ribozyme against vascular endothelial growth factor receptor has been designed as a potential therapy to inhibit angiogenesis in neoplasms (39) and is expected to be used in combination with chemotherapy in the treatment of metastatic colorectal cancer. (40) Another potential therapeutic target for a hammerhead ribozyme is human epidermal growth factor-2 (HER2), which is overexpressed in types of breast and ovarian cancers (41).
Small interfering RNA
Small interfering RNA (siRNA) is a small-sized (21–23 nucleotides) double-stranded RNA molecule with 2 nucleotide 3’-overhangs which shares sequence identity with gene targets. siRNAs can be experimentally introduced into mammalian cells using liposomes or plasmid and viral vectors that express small hairpin RNA (shRNA), a substrate that induces gene-specific suppression as siRNA.
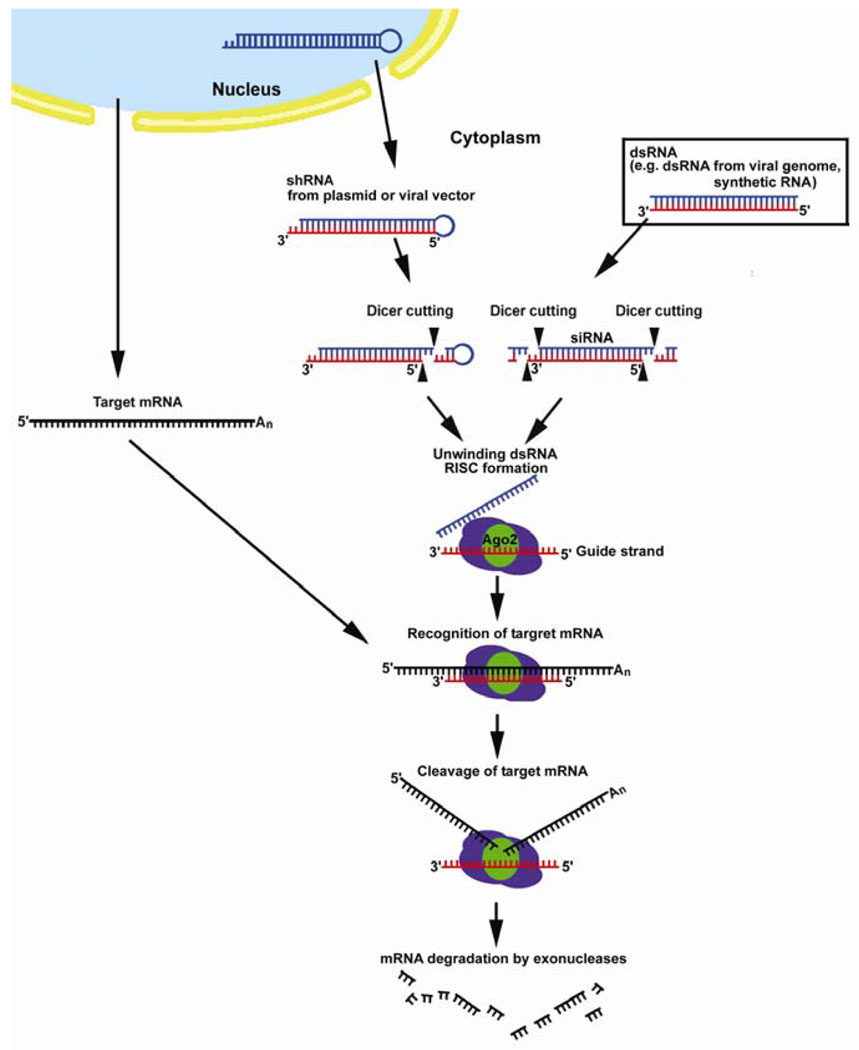
The complementary strand of siRNA is incorporated into a protein complex called RISC (RNA-induced silencing complex) where it serves as a cognate template for a specific mRNA in the cytoplasm. RISC catalyzes cleavage and degradation of the select mRNA, post-transcriptionally down-regulating gene expression (Fig. 4). siRNA also induces methylation of chromosomal DNA of homologous genes in plants, thus down-regulating expression at the level of transcription. In addition, it can direct the formation of repressive heterochromatin. (42).
Figure 4.
Cellular mechanism of RNA-interference . Synthetic double stranded RNA (dsRNA), dsRNA from viral genome and small hairpin RNA are processed to small interfering RNA (siRNA) by Dicer DNase III enzyme. SiRNAs are 21–23 nucleotide RNA duplexes with 2 nucleotide 3’ overhangs at both ends. The guide strand of siRNA is incorporated into the RNA-dependent silencing complex (RISC) in the cytoplasm and serves as the cognate template for the target mRNA. The target mRNA is cleaved by the enzyme Argonaute 2 (Ago2) in RISC and degraded by exonucleases.
The suppression efficiency of siRNA in cultured cells is at least comparable to ASOs that recruit RNase-H activity (43, 44). High sequence specificity is regarded as a feature of gene suppression by siRNA and, importantly, even single nucleotide mismatches against target sequence significantly affect suppression potency (45). Animal models of siRNA application have been developed as possible therapy for viral infections [i.e. HBV (46), HCV (47) , respiratory viruses (48, 49)), prostate cancer (50), spinocerebellar ataxia (51), Huntington’s disease (52)] and as we have shown, deafness associated with a dominant-negative allele of GJB2 (2). In our experiments, we were able to use siRNA to suppress efficiently and specifically transgene expression of the R75W allele of human GJB2 in cell cultures and in murine cochleae.
The Mechanism of RNA Interference
The first evidence that double stranded RNA (dsRNA) leads to post-transcriptional gene silencing in animals came from work on Caenorhabditis elegans. Interestingly, Fire and colleagues found double-stranded RNA to be substantially more effective at producing gene interference than either antisense or sense RNA individually (53). This phenomenon is referred to as RNA interference (RNAi), and is related to dsRNA-induced post-transcriptional gene silencing (PTGS) in plants, a presumed defense mechanism against viral infections and transposons (54).
As reconstructed in cell extract experiments in Drosophila melanogaster and Homo sapiens, dsRNAs from viral genome, microRNA (endogenous non-coding RNA) precursors, or artificial dsRNA are digested into 21–23 nucleotide fragments of siRNA by a member of the RNase III family of ATP-dependent, dsRNA-specific ribonucleases called Dicer . These siRNA duplexes bind to a nuclease complex to form RISC, with the antisense strand (guide RNA) serving as a cognate template for specific transcript recognition. Argonaute 2 (Ago2) is the slicer enzyme in RISC that cleaves target mRNA. When the guide RNA is extensively complementary to the target mRNA, RISC triggers rapid cleavage and degradation of the mRNA (55). As few as one or two base pair mismatches can significantly abolish the silencing function of siRNAs, especially when these mismatches are at the middle or 3’ end of the targeted sequence (56) (Fig. 4).
The relevance of this phenomenon in mammals was initially questioned because infection of mammalian somatic cells with long dsRNA (>50bp) was known to arrest protein synthesis through an interferon response and protein kinase activation. However, with the realization that shorter siRNAs of 21–23 nucleotides circumvent this response and cause targeted gene suppression (57), siRNA has became one of the most intensively studied tools for possible gene therapy, and in murine models, in vivo RNAi has been achieved using naked synthetic siRNA, liposomes, plasmid and viral vectors (Table 1).
Table 1.
In vivo models of RNAi-based gene therapy
| In vivo RNAi model | Targeted gene | Vehicle for siRNA | Delivery route | Reference |
|---|---|---|---|---|
| Spinocerebellar ataxia type1 | Ataxin-1 | shRNA from AAV | Injection to cerebellum | (51) 2004 |
| Huntington's disease | Huntingtin | shRNA from AAV | Stereotaxic injection to brain | (52) 2005 |
| ALS | SOD1 | shRNA from LV | Intraspinal injection | (75) 2005 |
| ALS | SOD1 | shRNA from LV | Intamuscular injection | (76) 2005 |
| Hereditary deafness | GJB2 | Synthetic siRNA with liposome | Intratympanic administration | (2) 2005 |
| Neovascularization | VEGF | Synthetic siRNA | Subretinal injection | (77) 2003 |
| Chronic pain | P2X3 cationic channel | Chemically stabilized siRNA | Cannulation to spinal cord | (78) 2004 |
| Chronic pain | NMDA-R2B receptor subunit | Synthetic siRNA with polyethileneimine polymer | Cannulation to spinal cord | (79) 2005 |
| Optic nerve axotomy | c-Jun, Bax, Apaf-1 | Synthetic siRNA with liposome | Injection to optic nerve stump | (80) 2005 |
| Prostate cancer | PI 3-kinase | shRNA from plasmid with lipid complex | Stable transfection of plasmid | (50) 2003 |
| Glioblastoma | MMP-9, cathepsin B | shRNA from plasmid | Injection to tumor | (81) 2004 |
| Germ cell tumor | FGF-4 | Synthetic siRNA with atelocollagen | Injection to tumor | (82) 2004 |
| Pancreatic tumor, metastatic tumor | CEACAM6 | Synthetic siRNA | Injection to tail vein | (83) 2004 |
| Small cell carcinoma | Skp-2 | siRNA from Ad | Injection to tumor | (84) 2005 |
| Ovarian carcinoma | HER-2 receptor | Synthetic siRNA with polyethileneimine polymer | Intraperitoneal injection | (85) 2005 |
| Autoimmune hepatitis | Fas | Synthetic siRNA | Hydrodynamic injection to tail vein | (86) 2003 |
| Hepatitis B | Viral HBsAg gene | Synthetic siRNA | Hydrodynamic injection to tail vein | (87) 2003, (88) 2003 |
| Hepatitis B | Viral genome (multiple target) | shRNA from plasmid | Hydrodynamic injection to tail vein | (46) 2003 |
| Influenza virus | Viral genome (multiple target) | Synthetic siRNA with polyethileneimine polymer shRNA from plasmid |
Injection to retroorbital vein intratnasal administration |
(89) 2004 |
| Influenza virus | Viral genome (multiple target) | Synthetic siRNA Synthetic siRNA with liposome |
Hydrodynamic injection to tail vein Intranasal administration |
(90) 2004 |
| Hypercholesterolemia | apoB | Chemically stabilized siRNA | Injection to tail vein | (91) 2004 |
| Renal tubular apoptosis | Fas | Synthetic siRNA | Hydrodynamic injection to tail vein | (92) 2004 |
| Parainfluenza virus Respiratory syncytial virus |
viral RSV P and PIV P gene | Synthetic siRNA with and without lipid | Intranasal administration | (48) 2005 |
| Respiratory syncytial virus | viral NS1 gene | siRNA from plasmid with nanochitosan polymer | Intranasal administration | (49) 2005 |
Abbreviations: Ad, adenovirus; AAV, adeno-associated virus; ALS, Amyotrophic lateral sclerosis; Apaf-1, apoptotic protease activating factor 1; apoB, apolipoprotein B; CEACAM6, carcinoembryonic antigen-related cell adhesion molecule 6; FGF-4 , fibroblast growth factor-4; GJB2, Gap junction protein, beta 2; HbsAg, hepatitis B surface antigen; LV, lentivirus; MMP-9, matrix metalloproteinase-9; NMDA, N-methyl-D-aspartate; NS1, non-structural protein 1; PI 3, phosphatidylinositol 3; PIV, parainfluenza virus; RSV, respiratory syncytial virus; RNAi, RNA interference; Skp-2, S phase kinase-associated protein; SOD1, superoxide dismutase 1; VEGF, vascular endothelial growth factor.
Delivering Gene Therapy into the Inner Ear
In vivo drug delivery to introduce exogenous DNA or RNA into the inner ear can be achieved using a variety of techniques that can be broadly divided into local or systemic approaches. The inner ear is a particularly suitable organ for the latter as its bony capsule within the temporal bone provides relative isolation from the rest of the body but is accessible through the middle ear. Local delivery, in turn, can directly or indirectly access the perilymphatic or endolymphatic systems, and it is reasonable to expect that different approaches will result in different cochlear effects. Vehicles used in transfection will also impact outcome and include cationic liposomes complexed with nucleic acid constructs and viral vectors like adenovirus (Ad), adeno-associated virus (AAV) herpes simplex type I (HSV-1) and lentivirus (LV) (Table 2).
Table 2.
Models of in vivo gene transfer into inner ear of rodents
| Animal | Delivery route | Expression in cochlea |
Vector | Dose | Duration | Others | Ref |
|---|---|---|---|---|---|---|---|
| Mouse | Lateral canalostomy | (Sensory cells in crista ampullaris) | Ad.CMV-lacZ (β-galactose gene), serotype5 (E1-, E3-) | 8×1011 pfu/ml, 0.5–1.0 µℓ | 28 d | (58 | |
| Cochleostomy | Inner hair cells, Deiter’s cells Scala tympani, Reissner’s membrane |
ABR threshold increase 13 d post injection |
|||||
| Neonatal rat (P5) |
Cochleostomy | Scala tympani, Reissner’s membrane Organ of Corti (sporadic) |
Ad.CMV-lacZ (E1-) | 5×106 pfu/ml, 2 µℓ | 7d | (93) | |
| Guinea pig |
Cochleostomy | Ad.CMV-lacZ (E1-, E3-) | 5×108 pfu/cochlea, infusion | 8 d | DPOAE lost | (65) | |
| Inner and outer hair cells | Ad.CMV-lacZ (E1-, E3-, pol-) | 5×108 pfu/cochlea, infusion | DPOAE intact | ||||
| Guinea pig |
Round window Membrane damaged by local anesthetics |
Spiral limbus, Spiral ligament Inner and outer hair cells, Supporting cells ,Spiral ganglion, Scala tympani |
Ad..lacZ | 1×109 p/ml, 20 µℓ | 3 d | (70) | |
| Guinea pig |
Endolymphatic sac Widening of endolymphatic duct |
Reissner’s membrane, Marginal cells of stria vascularis, Spiral ligament, supporting cells (Endolymphatic duct, Transitional epithelium surrounding the sensory area of utricle and saccule) |
Ad.CMV-lacZ, serotype 5 (E1-, E3-) | 1×1010 p/ml,10–15 µℓ | 4 d | (63) | |
| Guinea pig |
Cochleostomy | Inner hair cells, Pillar cells | Ad.CMV-lacZ, serotype 5 (E1-, E3-) | 5×108 pfu/cochlea, infusion | 8d | DPOAE lost | (94) |
| (No expression) | AAV. CMV-lacZ, promoter | 5×108 ip/cochlea, infusion | 8d | DPOAE intact | |||
| Nerve fibers | NSE promoter | ||||||
| Spiral limbus | EF-1α promoter | ||||||
| Epithelium of blood vessels | PDGF promoter | ||||||
| (No expression) | β-actin promoter | ||||||
| Guinea Pig |
Rupture of round window membrane |
Spiral ganglion, Reissner’s membrane Epithelial cells in basilar membrane |
Ad.RSV-lacZ (E1-,E3-) | 1×1011-2×1011 pfu/ml | 56 d | (68) | |
| Spiral ganglion, Stria vascularis, | AAV.CMV-GFP | 1.5×10^8 ip/ml | 28 d | ||||
| Guinea pig |
Cochleostomy | Spiral limbus, Spiral ligament Reissner’s membrane, Organ of Corti Spiral neurons |
AAV.lacZ AAV.CMV-GFP |
1×10^5 ip/ cochlea, infusion | 14 d | (95) (59) |
|
| Guinea pig |
Cochleostomy | Scala tympani, Scala vestibuli | VSV G-pseudotyped LV.CMV-GFP | 1×107 p/ml, infusion | 14 d | (60) | |
| Guinea pig |
Rupture of round window membrane |
Spiral ligament, Reissner’s membrane Supporting cells Scala vestibuli |
HSV-1.lacZ | 1×1010 pfu/ml, 2 µℓ | 4 d | Reduction in gene expression after 6 days. | (67) |
| Spiral ligament, Reissner’s membrane Inner and outer hair cells Scala vestibuli, Scala tympani |
Vaccinia virus.lacZ | 6×109 pfu/ml, 2 µℓ | |||||
| Guinea pig |
Rupture of round window membrane |
Spiral ligament, Spiral limbus, Organ of Corti, Reissner’s membrane Spiral neurons |
Liposome-plasmid.CMV-lacZ | 0.2µg/µℓ 10µℓ (injection) or infusion |
14 d | (66) | |
| Mouse | Intact round window membrane | Spiral limbus, Reissner’s membrane Spiral neurons |
Liposome-plasmid.CMV-GFP | 0.2 µg/µℓ | 3 d | (61) | |
| Cochleostomy | |||||||
| Mouse | Intact round window membrane |
Spiral limbus, Spiral ligament Organ of Corti, Reissner’s membrane Spiral neurons |
Liposome-plasmid.CMV-eGFP | 0.5µg/µℓ, 2µℓ | 3 d | (62) | |
| Scala tympani, Reissner’s membrane | Ad.CMV-eGFP | 6×1010 pfu/ml, 2 µℓ | 7 d | ||||
| (No expression) | AAV.CMV-GFP, AAV.CMV-lacZ | 2×1011 pfu/ml, 2 µℓ | 7 d | ||||
| Mouse | Intact round window membrane |
Spiral limbus, Spiral ligament Outer hair cells, Inner and outer pillar cells, |
Liposome-plasmid.CMV-eGFP | 0.5 µg/µℓ, 5 µℓ | 3 d | (2) | |
| Mouse | Rupture of round window membrane |
Stria vascularis, Spiral ligament, Outer hair cells, Deiter’s cells |
Ad.CMV-GFP | 1×108−1×1010 pfu/ml,1 µℓ | 3 d | (64) | |
| Stria vascularis, Supporting cells, Spiral neurons |
HSV-1.CMV-lacZ | 2×107,1×108 pfu/ml, 1 µℓ | |||||
| Stria vascularis, Spiral neurons, Scala tympani |
Liposome-plasmid.CMV-GFP |
Abbreviations: Ad., adenovirus; pol, Ad polymerase gene; CMV, cytomegalovirus promoter; d, day; EF-1α, elongation factor 1α; pfu, plaque forming unit; E1, E1 region of Ad genome; E3, E3 region of Ad genome; eGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; HSV-1, herpes simplex virus type1; ip, infectious particle; LV, lentivirus; NSE, neuron-specific enolase; p, particle; PDGF, platelet-derived growth factor; RSV, Rous sarcoma virus promoter; VSV, vesicular stomatitis virus.
Direct approaches to the perilymphatic system
A number of studies have been conducted on rodents in which the perilymphatic system was approached by direct microinjection through the round window membrane (RWM), via a cochleostomy or through the posterior semicircular canal. Because of the anatomical continuity of membranous perilymphatic space, these approaches primarily allow transgene transfection in fibrocytes and mesothelial cells lining the scala tympani, although in some experimental conditions transfection is also seen in the area surrounding the scala media.
Differences in the anatomical sites of expression may be due to differences in rodent models, titers and doses of vector (single injection vs continuous infusion using an osmotic minipump), particle size, presence or absence of viral receptors, and time points of sacrifice of the drug-treated animals. For example, after cochleostomy and injection of Ad vector, Kawamoto and colleagues detected LacZ transgene expression in the inner hair cells and supporting cells as well as Reissner’s membrane and lining cells of the scala tympani. The lateral canalostomy approach also resulted in the transgene expression in the sensory cells of the organ of Corti. (58) Lalwani and colleagues demonstrated AAV-mediated GFP transgene expression in the spiral limbus, spiral ligament, organ of Corti , Reissner’s membrane and spiral neurons after cochleostomy (59), however a single injection of AAV vector through the RWM, resulted in eGFP transgene expression that was limited to the spiral ganglion and stria vascularis. Direct infusion of LV vectors resulted in eGFP transgene expression only in the lining cells of the scala tympani (60).
Indirect approaches to perilymphatic systems
A more atraumatic method has been studied by Jero and colleagues who demonstrated the feasibility of diffusion of liposome-complexed plasmid and AV vector through the intact RWM of mice (61). These investigators were able to observe eGFP and β-gal transgene expression in the spiral limbus, spiral ligament, sensory and supporting cells of the organ of Corti, Reissner’s membrane and spiral ganglion cells 3–7 days after placing gelfoam soaked with a lipocomplexed plasmid directly on the RWM (62).
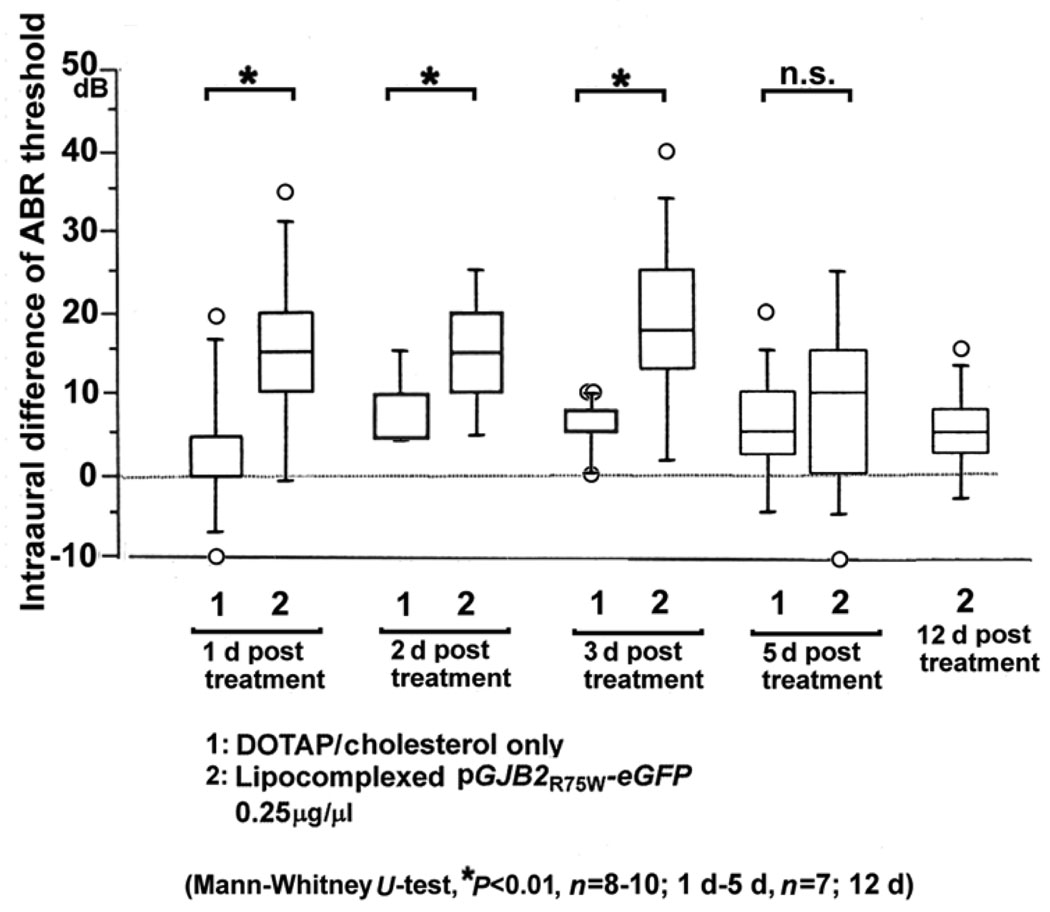
We used a similar approach to introduce a CMV-driven, dominant-negative GJB2 mutant construct into the inner ear and also observed expression in the spiral limbus, spiral ligament, epithelial cells in the basilar membrane, outer hair cells, inner and outer pillar cells and Claudius cells in the organ of Corti (2). The hearing loss associated with the mutant transgene expression was significant at 1–3 days posttransfection, but as expected not at 5 days posttransfection (Fig. 5)
Figure 5.
Hearing loss associated with dominant-negative GJB2 mutant expression in mice.(n=7–10, for each group). A plasmid vector expressing the dominant-negative GJB2R75W-eGFP (0.5 µg/µℓ) was complexed with DOTAP liposome, soaked in gel foam and applied to the intact round window membrane (1–5 µℓ). The ABR threshold in the treated ear was expressed as the intraaural difference from untreated ear to minimize variation between animals. Transient hearing loss was induced in the plasmid-applied ear on 1d, 2d and 3d after treatment but hearing levels were nearly normal by 5d. After 12 d, hearing levels were equivalent in both treated and control animals (whiskers, 1.5 SD from the median; circles, outliers> 1.5 SD from the median).
Direct approaches to endolymphatic system
Surgical approaches to the endolymphatic sac allow drug delivery to the endolymphatic system, resulting primarily in transgene expression in the endolymphatic duct epithelium and transitional epithelium in the utricle and saccule. Yamasoba and colleagues have reported that after high-dose injection of Ad vector (1×1010 particles/ml, 10–15µl), LacZ expression could be detected in cells bordering the scala media, including endothelial cells in Reissner’s membrane, marginal cells in the stria vascularis and supporting cells in the organ of Corti (63).
Advantages and disadvantages of these approaches
There are many potential complications associated with gene delivery into the inner ear irrespective of the route of delivery, including iatrogenic cell damage from hydropressure and ototoxicity of the delivery vehicle. In rodent experiments, after cochleostomy and injection to scala tympani, damage of inner ear cells and an inflammatory response are occasionally seen (64), although it is relatively uncommon to observe inner ear cytoarchitectural damage and inflammation at the light microscopic levels. The damage that is observed is usually confined to the basal turn of the cochlea or the site of injection at the RWM.
In experiments in which Ad vectors have been perfused into the scala tympani by osmotic pumps, replication-deficient AV (E1-, E3-) vectors presumably affected the function of transfected outer hair cells as evidenced by compromised distortion product otoacoustic emissions, while replication defective AV (E1-, E3-, pol-) vectors did not (65). After cochleostomy and infusion of AAV (59), LV vectors (60), or liposome vectors (66), the cytoarchitecture of the cochlea was intact and free from inflammation, with the exception of mild fibrosis at the injection site. Dervy and colleagues observed a lymphocytic infiltration in the cochlea after injection of HSV vector or vaccinia virus vectors into scala tympani (67), highlighting the importance of transgene selection in circumventing complications like loss of cell viability and inflammation.
The duration of transgene expression is also an important factor that must be studied under comparative experimental conditions. Using direct injection of AV vectors driven by CMV (Cytomegalovirus) or RSV (Rous sarcoma virus) promoters, transgene expression in cochleae was detected after 28 and 56 days posttransfection in mice and guinea pigs, respectively (58, 68). AAV, HSV and LV vectors can potentially offer permanent transgene expression by chromosomal integration, which would be beneficial for “one-time” therapies to correct hereditary deafness. These vectors, however, show a more limited cell-type tropism in the inner ear when compared to Ad vectors, with most published data using direct injection of Ad into the perilymphatic system of rodents showing substantial transduction of sensory and supporting cells in the organ of Corti.
Clearly, an indirect approach through the intact or chemically permeabilized RWM reduces iatrogenic damage to a minimum, however transgene expression is relatively low and primarily in the basal turn of the cochlea. The round window membranes of various species differ in their thickness (70 µm in humans, 40–60 µm in rhesus monkeys, 10–14 µm in chinchillas) but share a similar composition consisting of outer and inner epithelial layers between which lies a connective tissue core. Passage of substances from the middle to the inner ear via round window membrane is an active process by the epithelia as demonstrated with cationic ferritin, horseradish peroxidase and 1 µm latex spheres in experiments using rodents and monkeys (69). In studies of guinea pigs, Suzuki and colleagues were able to enhance permeability of the outer epithelium of the round window membrane by phenol-containing local anesthetics. Middle ear application of adenovirus vector into the middle ear of these animals was followed by transgene expression in the sensory epithelium of the cochlea (70).
Vectors to Deliver siRNAs In Vivo
In rodent experiments, in vivo transfection of siRNA has been achieved using a number of different vectors in a number of different organs. Surprisingly, even naked, synthetic siRNA has been successfully introduced and functioned in the cells of organs such as liver and kidney following hydrodynamic injection in mouse tail veins. However, naked synthetic RNA is degraded in serum unless it is modified for stabilization making this transfection route infeasible for clinical use. Cationic liposomes, polyethylenimine (PEI) vectors and atelocollagen are alternate efficient methods to deliver synthetic siRNAs by either topical or systemic injection. However, if synthetic siRNAs are used, RNAi will remain a transient phenomenon.
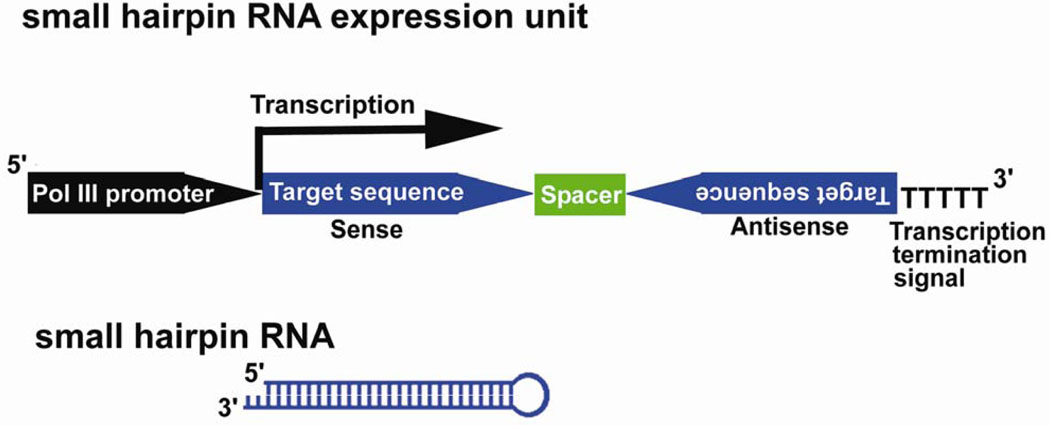
To achieve long-term, stable expression of siRNA, plasmid vectors with RNAi expression units driven by RNA polymerase III (Pol III) promoters U6 and H1 have been developed by several researchers (71–73). Pol III transcribes significantly shorter RNAs than those transcribed by RNA polymerases I and II (pol II), with a short stretch of about 5 thymidine residues serving as the termination signal. The expression cassette contains an shRNA sequence of an inverted repeat of 19–29 nt separated by a short spacer sequence. The transcribed RNA molecule forms short hairpin RNA that is indistinguishable from siRNAs in terms of RNAi efficiency and mechanism (Fig. 6).
Figure 6.
Scheme of RNA polymerase III (Pol III) driven, small hairpin RNA (shRNA) expression cassette. Pol III promoter (H1 or U6 promoter) is followed by DNA template that corresponds to the target sequence of the gene to be suppressed. The motif of inverted repeats is separated by a short spacer sequence and followed by transcriptional termination signal of five thymidines. The transcript folds back on itself to form shRNA.
By using a RNA polymerase II promoter, Xia and colleagues developed an shRNA-expression plasmid with a modified CMV promoter, a downstream shRNA template with inverted repeat and a minimal polyA cassette. In this expression unit, the hairpin sequence was placed immediately adjacent to the transcription initiation site of the promoter (74).
Ad and AAV vectors with a pol II or modified CMV shRNA expression unit have been demonstrated to induce in vivo RNAi in neural tissues. In one study, an Ad vector with a modified CMV expression cassette of shRNA against eGFP was directly injected to the basal ganglia of transgenic mice expressing eGFP. Western blot analysis showed that eGFP expression was diminished in the injected hemisphere 5 d post injection (74). The same group also injected an AAV vector with the H1-shRNA expression cassette into the cerebellum of a transgenic murine model of spinocerebellar ataxia type 1. Expression of shRNA targeting human ataxin1 was verified 10 d after the injection (51). When they injected AAV.U6-shRNA vectors into the basal ganglia of a murine model of Huntington’s disease, shRNA expression against the human Huntington’s disease gene was detected 21 d post injection (52). In these experiments using AAV vectors, a CMV-driven, eGFP expression unit was included in the chimeric viral constructs, and eGFP expression was detected in the injected sites up to 5 months later.
Lentiviral vectors with H1-shRNA expression units have been used to treat a mouse model of amyotrophic lateral sclerosis (ALS) that expresses a human SOD1 mutant. The SOD1 mutant causes a dominantly inherited form of ALS through a gain-of-function mechanism, and the lentiviral-mediated shRNA selectively targets the expression of the mutant gene. In two studies using these animals, lentiviral vectors were injected intraspinally (75) or intramuscularly (utilizing retrograde axonal transport to motoneurons) (76). Western blotting and immunofluorescence demonstrated a reduction in SOD1 mutant protein expression 15–50 d after spinal injection and an improvement was observed in motor neuron survival and motor ability 80–100 d after the spinal and intramuscular injection.
Conclusion
The development of technologies to induce specific gene suppression by small DNA or RNA molecules is outpacing the development of appropriate drug delivery systems that are applicable to the inner ear. The AV vector with poI II or modified CMV promoters that expresses shRNA has the potential to induce RNAi in the inner ear for relatively short periods of time (up to 2 months). Future studies focusing on methods for efficient and substantial long-term transfection into the inner ear sensory epithelium are crucial if RNAi-based gene therapy of hereditary deafness caused by gain-of-function mechanisms is to become a clinical reality.
Acknowledgments
This research was supported by NIH grant DC03544 (RJHS).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of financial interests.
References
- 1.Van De Water TR, Lallemend F, Eshraghi AA, et al. Caspases, the enemy within, and their role in oxidative stress-induced apoptosis of inner ear sensory cells. Otol Neurotol. 2004;25:627–632. doi: 10.1097/00129492-200407000-00035. [DOI] [PubMed] [Google Scholar]
- 2.Maeda Y, Fukushima K, Nishizaki K, Smith RJ. In vitro and in vivo suppression of GJB2 expression by RNA interference. Hum Mol Genet. 2005;14:1641–1650. doi: 10.1093/hmg/ddi172. [DOI] [PubMed] [Google Scholar]
- 3.Wilkie AO. The molecular basis of genetic dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denoyelle F, Lina-Granade G, Plauchu H, et al. Connexin 26 gene linked to a dominant deafness. Nature. 1998;393:319–320. doi: 10.1038/30639. [DOI] [PubMed] [Google Scholar]
- 5.Rouan F, White TW, Brown N, et al. trans-dominant inhibition of connexin-43 by mutant connexin-26: implications for dominant connexin disorders affecting epidermal differentiation. J Cell Sci. 2001;114:2105–2113. doi: 10.1242/jcs.114.11.2105. [DOI] [PubMed] [Google Scholar]
- 6.Richard G, White TW, Smith LE, et al. Functional defects of Cx26 resulting from a heterozygous missense mutation in a family with dominant deaf-mutism and palmoplantar keratoderma. Hum Genet. 1998;103:393–399. doi: 10.1007/s004390050839. [DOI] [PubMed] [Google Scholar]
- 7.Marziano NK, Casalotti SO, Portelli AE, Becker DL, Forge A. Mutations in the gene for connexin 26 (GJB2) that cause hearing loss have a dominant negative effect on connexin 30. Hum Mol Genet. 2003;12:805–812. doi: 10.1093/hmg/ddg076. [DOI] [PubMed] [Google Scholar]
- 8.Kubisch C, Schroeder BC, Friedrich T, et al. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999;96:437–446. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- 9.Coucke PJ, Van Hauwe P, Kelley PM, et al. Mutations in the KCNQ4 gene are responsible for autosomal dominant deafness in four DFNA2 families. Hum Mol Genet. 1999;8:1321–1328. doi: 10.1093/hmg/8.7.1321. [DOI] [PubMed] [Google Scholar]
- 10.Kharkovets T, Hardelin JP, Safieddine S, et al. KCNQ4, a K+ channel mutated in a form of dominant deafness, is expressed in the inner ear and the central auditory pathway. Proc Natl Acad Sci U S A. 2000;97:4333–4338. doi: 10.1073/pnas.97.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Laer L, Huizing EH, Verstreken M, et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet. 1998;20:194–197. doi: 10.1038/2503. [DOI] [PubMed] [Google Scholar]
- 12.Yu C, Meng X, Zhang S, Zhao G, Hu L, Kong X. A 3-nucleotide deletion in the polypyrimidine tract of intron 7 of the DFNA5 gene causes nonsyndromic hearing impairment in a Chinese family. Genomics. 2003;82:575–579. doi: 10.1016/s0888-7543(03)00175-7. [DOI] [PubMed] [Google Scholar]
- 13.Bischoff AM, Luijendijk MW, Huygen PL, et al. A novel mutation identified in the DFNA5 gene in a Dutch family: a clinical and genetic evaluation. Audiol Neurootol. 2004;9:34–46. doi: 10.1159/000074185. [DOI] [PubMed] [Google Scholar]
- 14.Gregan J, Van Laer L, Lieto LD, Van Camp G, Kearsey SE. A yeast model for the study of human DFNA5, a gene mutated in nonsyndromic hearing impairment. Biochim Biophys Acta. 2003;1638:179–186. doi: 10.1016/s0925-4439(03)00083-8. [DOI] [PubMed] [Google Scholar]
- 15.Van Laer L, Vrijens K, Thys S, et al. DFNA5: hearing impairment exon instead of hearing impairment gene? J Med Genet. 2004;41:401–406. doi: 10.1136/jmg.2003.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dass CR. Liposome-mediated delivery of oligodeoxynucleotides in vivo. Drug Deliv. 2002;9:169–180. doi: 10.1080/15227950290097606. [DOI] [PubMed] [Google Scholar]
- 17.Juliano RL, Dixit VR, Kang H, Kim TY, Miyamoto Y, Xu D. Epigenetic manipulation of gene expression: a toolkit for cell biologists. J Cell Biol. 2005;169:847–857. doi: 10.1083/jcb.200501053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamaratski E, Pradeepkumar PI, Chattopadhyaya J. A critical survey of the structure-function of the antisense oligo/RNA heteroduplex as substrate for RNase H. J Biochem Biophys Methods. 2001;48:189–208. doi: 10.1016/s0165-022x(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 19.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 20.Gleave ME, Monia BP. Antisense therapy for cancer. Nat Rev Cancer. 2005;5:468–479. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 21.Marwick C. First “antisense” drug will treat CMV retinitis. Jama. 1998;280:871. [PubMed] [Google Scholar]
- 22.Dove A. Isis and antisense face crucial test without Novartis. Nat Biotechnol. 2000;18:19. doi: 10.1038/71852. [DOI] [PubMed] [Google Scholar]
- 23.Sandrasagra A, Leonard SA, Tang L, et al. Discovery and development of respirable antisense therapeutics for asthma. Antisense Nucleic Acid Drug Dev. 2002;12:177–181. doi: 10.1089/108729002760220770. [DOI] [PubMed] [Google Scholar]
- 24.Delprat B, Boulanger A, Wang J, et al. Downregulation of otospiralin, a novel inner ear protein, causes hair cell degeneration and deafness. J Neurosci. 2002;22:1718–1725. doi: 10.1523/JNEUROSCI.22-05-01718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morishita R, Gibbons GH, Horiuchi M, et al. Molecular Delivery System for Antisense Oligonucleotides: Enhanced Effectiveness of Antisense Oligonucleotides by HVJ-liposome Mediated Transfer. J Cardiovasc Pharmacol Ther. 1997;2:213–222. doi: 10.1177/107424849700200308. [DOI] [PubMed] [Google Scholar]
- 26.Morishita R, Gibbons GH, Horiuchi M, et al. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc Natl Acad Sci U S A. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morishita R, Sugimoto T, Aoki M, et al. In vivo transfection of cis element "decoy" against nuclear factor-kappaB binding site prevents myocardial infarction. Nat Med. 1997;3:894–899. doi: 10.1038/nm0897-894. [DOI] [PubMed] [Google Scholar]
- 28.Morishita R, Aoki M, Ogihara T. Does gene therapy become pharmacotherapy? Exp Physiol. 2005;90:307–313. doi: 10.1113/expphysiol.2005.030403. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe K, Inai S, Jinnouchi K, et al. Nuclear-factor kappa B (NF-kappa B)-inducible nitric oxide synthase (iNOS/NOS II) pathway damages the stria vascularis in cisplatin-treated mice. Anticancer Res. 2002;22:4081–4085. [PubMed] [Google Scholar]
- 30.Jiang H, Sha SH, Schacht J. NF-kappaB pathway protects cochlear hair cells from aminoglycoside-induced ototoxicity. J Neurosci Res. 2005;79:644–651. doi: 10.1002/jnr.20392. [DOI] [PubMed] [Google Scholar]
- 31.Nagy I, Monge A, Albinger-Hegyi A, Schmid S, Bodmer D. NF-kappaB is Required for Survival of Immature Auditory Hair Cells In Vitro. J Assoc Res Otolaryngol. 2005:1–9. doi: 10.1007/s10162-005-0006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kore AR, Vaish NK, Kutzke U, Eckstein F. Sequence specificity of the hammerhead ribozyme revisited; the NHH rule. Nucleic Acids Res. 1998;26:4116–4120. doi: 10.1093/nar/26.18.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Citti L, Rainaldi G. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr Gene Ther. 2005;5:11–24. doi: 10.2174/1566523052997541. [DOI] [PubMed] [Google Scholar]
- 34.Haseloff J, Gerlach WL. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 35.Michienzi A, Rossi JJ. Intracellular applications of ribozymes. Methods Enzymol. 2001;341:581–596. doi: 10.1016/s0076-6879(01)41178-5. [DOI] [PubMed] [Google Scholar]
- 36.Beigelman L, McSwiggen JA, Draper KG, et al. Chemical modification of hammerhead ribozymes. Catalytic activity and nuclease resistance. J Biol Chem. 1995;270:25702–25708. doi: 10.1074/jbc.270.43.25702. [DOI] [PubMed] [Google Scholar]
- 37.Usman N, Blatt LM. Nuclease-resistant synthetic ribozymes: developing a new class of therapeutics. J Clin Invest. 2000;106:1197–1202. doi: 10.1172/JCI11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zinnen SP, Domenico K, Wilson M, et al. Selection, design, and characterization of a new potentially therapeutic ribozyme. RNA. 2002;8:214–228. doi: 10.1017/s1355838202014723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciafre SA, Niola F, Wannenes F, Farace MG. An anti-VEGF ribozyme embedded within the adenoviral VAI sequence inhibits glioblastoma cell angiogenic potential in vitro. J Vasc Res. 2004;41:220–228. doi: 10.1159/000077777. [DOI] [PubMed] [Google Scholar]
- 40.Wright L, Kearney P. Current status of ribozymes as gene therapy agents for cancer. Cancer Invest. 2001;19:495–509. doi: 10.1081/cnv-100103848. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki T, Anderegg B, Ohkawa T, et al. Adenovirus-mediated ribozyme targeting of HER-2/neu inhibits in vivo growth of breast cancer cells. Gene Ther. 2000;7:241–248. doi: 10.1038/sj.gt.3301065. [DOI] [PubMed] [Google Scholar]
- 42.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 43.Bertrand JR, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296:1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- 44.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 45.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. Embo J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCaffrey AP, Nakai H, Pandey K, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- 47.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 48.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Yang H, Kong X, et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat Med. 2005;11:56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
- 50.Czauderna F, Santel A, Hinz M, et al. Inducible shRNA expression for application in a prostate cancer mouse model. Nucleic Acids Res. 2003;31:e127. doi: 10.1093/nar/gng127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia H, Mao Q, Eliason SL, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 52.Harper SQ, Staber PD, He X, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 54.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 55.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 56.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 58.Kawamoto K, Oh SH, Kanzaki S, Brown N, Raphael Y. The functional and structural outcome of inner ear gene transfer via the vestibular and cochlear fluids in mice. Mol Ther. 2001;4:575–585. doi: 10.1006/mthe.2001.0490. [DOI] [PubMed] [Google Scholar]
- 59.Lalwani AK, Han JJ, Walsh BJ, Zolotukhin S, Muzyczka N, Mhatre AN. Green fluorescent protein as a reporter for gene transfer studies in the cochlea. Hear Res. 1997;114:139–147. doi: 10.1016/s0378-5955(97)00151-2. [DOI] [PubMed] [Google Scholar]
- 60.Han JJ, Mhatre AN, Wareing M, et al. Transgene expression in the guinea pig cochlea mediated by a lentivirus-derived gene transfer vector. Hum Gene Ther. 1999;10:1867–1873. doi: 10.1089/10430349950017545. [DOI] [PubMed] [Google Scholar]
- 61.Jero J, Tseng CJ, Mhatre AN, Lalwani AK. A surgical approach appropriate for targeted cochlear gene therapy in the mouse. Hear Res. 2001;151:106–114. doi: 10.1016/s0378-5955(00)00216-1. [DOI] [PubMed] [Google Scholar]
- 62.Jero J, Mhatre AN, Tseng CJ, et al. Cochlear gene delivery through an intact round window membrane in mouse. Hum Gene Ther. 2001;12:539–548. doi: 10.1089/104303401300042465. [DOI] [PubMed] [Google Scholar]
- 63.Yamasoba T, Yagi M, Roessler BJ, Miller JM, Raphael Y. Inner ear transgene expression after adenoviral vector inoculation in the endolymphatic sac. Hum Gene Ther. 1999;10:769–774. doi: 10.1089/10430349950018526. [DOI] [PubMed] [Google Scholar]
- 64.Staecker H, Li D, O'Malley BW, Jr, Van De Water TR. Gene expression in the mammalian cochlea: a study of multiple vector systems. Acta Otolaryngol. 2001;121:157–163. doi: 10.1080/000164801300043307. [DOI] [PubMed] [Google Scholar]
- 65.Luebke AE, Steiger JD, Hodges BL, Amalfitano A. A modified adenovirus can transfect cochlear hair cells in vivo without compromising cochlear function. Gene Ther. 2001;8:789–794. doi: 10.1038/sj.gt.3301445. [DOI] [PubMed] [Google Scholar]
- 66.Wareing M, Mhatre AN, Pettis R, et al. Cationic liposome mediated transgene expression in the guinea pig cochlea. Hear Res. 1999;128:61–69. doi: 10.1016/s0378-5955(98)00196-8. [DOI] [PubMed] [Google Scholar]
- 67.Derby ML, Sena-Esteves M, Breakefield XO, Corey DP. Gene transfer into the mammalian inner ear using HSV-1 and vaccinia virus vectors. Hear Res. 1999;134:1–8. doi: 10.1016/s0378-5955(99)00045-3. [DOI] [PubMed] [Google Scholar]
- 68.Li Duan M, Bordet T, Mezzina M, Kahn A, Ulfendahl M. Adenoviral and adeno-associated viral vector mediated gene transfer in the guinea pig cochlea. Neuroreport. 2002;13:1295–1299. doi: 10.1097/00001756-200207190-00016. [DOI] [PubMed] [Google Scholar]
- 69.Goycoolea ML, Lundman L. Round window membrane. Structure function and permeability: a review. Microscop Res Tech. 1997;36:201–211. doi: 10.1002/(SICI)1097-0029(19970201)36:3<201::AID-JEMT8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki M, Yamasoba T, Suzukawa K, Kaga K. Adenoviral vector gene delivery via the round window membrane in guinea pigs. Neuroreport. 2003;14:1951–1955. doi: 10.1097/00001756-200310270-00014. [DOI] [PubMed] [Google Scholar]
- 71.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 75.Raoul C, Abbas-Terki T, Bensadoun JC, et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 76.Ralph GS, Radcliffe PA, Day DM, et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 77.Reich SJ, Fosnot J, Kuroki A, et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- 78.Dorn G, Patel S, Wotherspoon G, et al. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 2004;32:e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan PH, Yang LC, Shih HC, Lan KC, Cheng JT. Gene knockdown with intrathecal siRNA of NMDA receptor NR2B subunit reduces formalin-induced nociception in the rat. Gene Ther. 2005;12:59–66. doi: 10.1038/sj.gt.3302376. [DOI] [PubMed] [Google Scholar]
- 80.Lingor P, Koeberle P, Kugler S, Bahr M. Down-regulation of apoptosis mediators by RNAi inhibits axotomy-induced retinal ganglion cell death in vivo. Brain. 2005;128:550–558. doi: 10.1093/brain/awh382. [DOI] [PubMed] [Google Scholar]
- 81.Lakka SS, Gondi CS, Yanamandra N, et al. Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene. 2004;23:4681–4689. doi: 10.1038/sj.onc.1207616. [DOI] [PubMed] [Google Scholar]
- 82.Minakuchi Y, Takeshita F, Kosaka N, et al. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004;32:e109. doi: 10.1093/nar/gnh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duxbury MS, Matros E, Ito H, Zinner MJ, Ashley SW, Whang EE. Systemic siRNA-mediated gene silencing: a new approach to targeted therapy of cancer. Ann Surg. 2004;240:667–674. doi: 10.1097/01.sla.0000140755.97224.9a. discussion 75–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sumimoto H, Yamagata S, Shimizu A, et al. Gene therapy for human small-cell lung carcinoma by inactivation of Skp-2 with virally mediated RNA interference. Gene Ther. 2005;12:95–100. doi: 10.1038/sj.gt.3302391. [DOI] [PubMed] [Google Scholar]
- 85.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 86.Song E, Lee SK, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 87.Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8:769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 88.Klein C, Bock CT, Wedemeyer H, et al. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9–18. doi: 10.1016/s0016-5085(03)00720-0. [DOI] [PubMed] [Google Scholar]
- 89.Ge Q, Filip L, Bai A, Nguyen T, Eisen HN, Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc Natl Acad Sci U S A. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tompkins SM, Lo CY, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci U S A. 2004;101:8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 92.Hamar P, Song E, Kokeny G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dazert S, Aletsee C, Brors D, Gravel C, Sendtner M, Ryan A. In vivo adenoviral transduction of the neonatal rat cochlea and middle ear. Hear Res. 2001;151:30–40. doi: 10.1016/s0378-5955(00)00189-1. [DOI] [PubMed] [Google Scholar]
- 94.Luebke AE, Foster PK, Muller CD, Peel AL. Cochlear function and transgene expression in the guinea pig cochlea, using adenovirus- and adeno-associated virus-directed gene transfer. Hum Gene Ther. 2001;12:773–781. doi: 10.1089/104303401750148702. [DOI] [PubMed] [Google Scholar]
- 95.Lalwani AK, Walsh BJ, Reilly PG, Muzyczka N, Mhatre AN. Development of in vivo gene therapy for hearing disorders: introduction of adeno-associated virus into the cochlea of the guinea pig. Gene Ther. 1996;3:588–592. [PubMed] [Google Scholar]