To the editor:
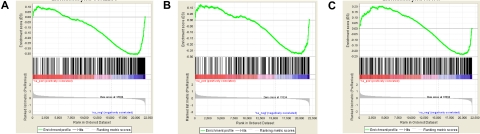
Leukemias induced by MLL-fusion genes are associated with poor prognosis and with overexpression of HOXA9 and its cofactor MEIS1. We previously reported in Blood that MEIS1 was essential for maintenance of both human and mouse cell lines bearing MLL-fusion genes.1 In a knockin mouse MLL-AF9 cell line, we found that 594 genes (contained in 1053 Affymetrix probe sets) showed significant inhibition at 48 hours with short-hairpin RNA (shRNA) mediated Meis1 knockdown (raw data can be found on the Gene Expression Omnibus under accession number GSE14101).1 A recent search for overlaps with other gene signatures in the Molecular Signatures Database2 (MSigDB) revealed that the Meis1-dependent gene signature in murine leukemia significantly overlapped with those associated with neural and embryonic stem cells (p = 3.27 e−28 and p = 3.71 e−13, respectively, hypergeometric distribution).3,4 The malignant potential of cancers has been linked to the expression of embryonic stem cell (ESC)–like gene signatures.5,6 This led us to postulate that in MLL-fusion leukemia, MEIS1 is necessary for maintenance of an ESC-like molecular profile. However, in a recent paper Sommervaille et al report that while expression of ESC-like signatures correlated with leukemia stem cell (LSC) frequency in retrovirally generated murine MLL-fusion gene leukemias, expression of ESC-like genes and high LSC frequency were independent of Meis1 expression.7 Subsequently, we obtained the compiled ESC-associated gene sets from 2 publications—the core ESC-module map by Wong et al and the ESC-like gene set by Ben Porath et al (the same ones used by Somervaille et al), and used Gene Set Enrichment Analysis (GSEA) to compare the expression of these sets in our dataset of control and Meis1-knockdown MLL-AF9 leukemia cells.3,5,6 We found that both the ESC-associated gene sets were significantly inhibited in Meis1 sh-RNA transduced murine MLL-AF9 leukemia cells compared with vector controls (Figure 1). This was true even when the proliferation associated genes were removed from the ESC-module map. Instead of GSEA, if we used simple intersects, we similarly found that our set of Meis1-dependent genes (described above) significantly overlapped with the ESC-like gene sets (P < e−7, hypergeometric distribution). Thus, our data indicate that at least in the knockin model of MLL-AF9 mouse leukemia, Meis1 is required for the maintenance of an ESC-like gene signature. The reasons for the differences between our results and those of Sommervaille et al are not clear but there are differences in experimental design. In our MLL-AF9 knock-in model, the expression of the oncogene is at physiologic levels.8 On the other hand, the supra-physiologic expression of the MLL-fusion gene by retroviral expression (as used by Sommervaille et al) might render Meis1 redundant upon transformation. It is also notable that MYB, included in the ESC-like gene list that correlated with LSC frequency in the study by Sommervaille et al and in our list of Meis1-dependent genes, was previously shown to be up-regulated by Hoxa9/Meis1 in a murine MLL-ENL model.9 Retroviral models have provided valuable fundamental data on the transforming abilities of oncogenes.10 However, for more intricate problems such as the role of downstream targets like MEIS1 in MLL-fusion gene leukemias, studies with models that use gene expression at physiologic levels might be better suited.
Figure 1.
Significant inhibition of ESC-like gene sets with Meis1 knockdown in murine MLL-AF9 leukemia. Expression of gene sets composed of core-ESC module, core-ESC module without proliferation-associated genes,4,6 and another ESC-like gene set7 were compared across the dataset of control and Meis1-shRNA transduced murine MLL-AF9 leukemia cells using GSEA. Shown are the enrichment plots of GSEA analysis illustrating that the gene sets composed of core ESC module (A), core ESC module without the genes associated with proliferation (B), and the ECS-like signature (C) were all significantly inhibited with Meis1 knockdown (preranked analysis using Log2 fold change statistic, 1000 permutations, P < .001 for all analyses; results were similar when the paired t test statistic was used).
Authorship
Acknowledgments: The authors thank Kevin Silverstein, PhD, Bioinformatics Research Scientist at the Masonic Cancer Center, University of Minnesota, for help with the analysis of microarray results.
This research was supported by grants from the National Institutes of Health (R01-CA087053 to J.H.K., and K08-CA122191 to A.R.K.), the Leukemia Research Fund (A.R.K.), and by the Children's Cancer Research Fund (J.H.K. and A.R.K.).
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ashish R. Kumar, Division of BMT and Immune Deficiency, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave, Cincinnati, OH 45236; e-mail: ashish.kumar@cchmc.org.
References
- 1.Kumar AR, Li Q, Hudson WA, et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113(8):1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molecular Signatures Database. [Accessed January 5, 2010]. http://www.broad.mit.edu/gsea/msigdb/index.jsp.
- 3.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298(5593):597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 5.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2(4):333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Somervaille TC, Matheny CJ, Spencer GJ, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Kumar AR, Hudson WA, et al. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13(5):432–440. doi: 10.1016/j.ccr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess JL, Bittner CB, Zeisig DT, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108(1):297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCormack E, Bruserud O, Gjertsen BT. Review: genetic models of acute myeloid leukaemia. Oncogene. 2008;27(27):3765–3779. doi: 10.1038/onc.2008.16. [DOI] [PubMed] [Google Scholar]