Abstract
53BP1 is a well-known mediator of the cellular response to DNA damage. Two alternative mechanisms have been proposed to explain 53BP1’s interaction with DNA double-strand breaks (DSBs), one by binding to methylated histones and the other via an RNF8 E3 ligase–dependent ubiquitylation pathway. The formation of RNF8 and 53BP1 irradiation-induced foci are both dependent on histone H2AX. To evaluate the contribution of the RNF8-dependent pathway to 53BP1 function, we generated RNF8 knockout mice. We report that RNF8 deficiency results in defective class switch recombination (CSR) and accumulation of unresolved immunoglobulin heavy chain–associated DSBs. The CSR DSB repair defect is milder than that observed in the absence of 53BP1 but similar to that found in H2AX−/− mice. Moreover, similar to H2AX but different from 53BP1 deficiency, RNF8−/− males are sterile, and this is associated with defective ubiquitylation of the XY chromatin. Combined loss of H2AX and RNF8 does not cause further impairment in CSR, demonstrating that the two genes function epistatically. Importantly, although 53BP1 foci formation is RNF8 dependent, its binding to chromatin is preserved in the absence of RNF8. This suggests a two-step mechanism for 53BP1 association with chromatin in which constitutive loading is dependent on interactions with methylated histones, whereas DNA damage–inducible RNF8-dependent ubiquitylation allows its accumulation at damaged chromatin.
Protein ubiquitylation is emerging as an important posttranslational modification used to maintain genomic stability (Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007; Alpi and Patel, 2009; Doil et al., 2009; Panier and Durocher, 2009; Stewart et al., 2009). One component of this pathway is the E3 ubiquitin ligase RNF8, for which RNA interference–based studies have shown a role in the G2/M ionizing radiation (IR) cell-cycle checkpoint (Huen et al., 2007), homologous recombination (HR; Huang et al., 2009), and UV-induced nucleotide excision repair (Marteijn et al., 2009). RNF8-intiated protein ubiquitylation at DNA lesions is tightly coordinated with phosphatidylinositol 3-kinase–like kinase-dependent phosphorylations. Specifically, RNF8 recognizes ataxia telangiectasia mutated–mediated phosphorylated MDC1 bound to γ-H2AX, which permits it to catalyze ubiquitin-dependent recruitment of 53BP1 and Brca1 to DNA lesions via an interaction with the UBC13 E2 ligase (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). RNF8 and UBC13 also act in concert with another E3 ubiquitin ligase, RNF168, identified to be the gene mutated in the human RIDDLE immunodeficiency syndrome (Stewart et al., 2009). RIDDLE cells are characterized by IR sensitivity and impaired ability to form 53BP1 and Brca1 foci, and patients exhibit low levels of serum IgG, associated with impaired resolution of class switch recombination (CSR)–associated breaks (Stewart et al., 2007). Recent studies demonstrated that RNF168 acts to amplify RNF8-initiated protein ubiquitylation at sites of DNA double-strand breaks (DSBs; Doil et al., 2009; Stewart et al., 2009). Collectively, these results suggest that beyond phosphorylation, ubiquitylation might play an active role in the signaling of DSBs. However, the physiological relevance of such a response and the extent to which it modulates 53BP1 functions in vivo remain unexplored. In this paper, we report the effects of RNF8 deletion in mice. Our results reveal 53BP1-independent functions of RNF8 in meiotic recombination and, conversely, RNF8-independent functions of 53BP1 in CSR.
RESULTS AND DISCUSSION
To determine whether the ubiquitin-dependent arm of the DSB response affects CSR, we disrupted RNF8 in mice (Fig. S1 A). RNF8−/− mice were born at Mendelian frequencies, and the absence of RNF8 protein was confirmed by Western blotting with antibodies raised against full-length human RNF8 (Fig. S1 B).
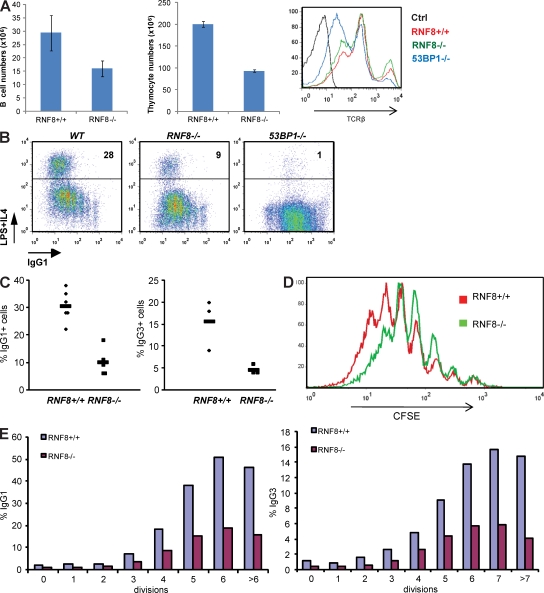
53BP1−/− and H2AX−/− mice exhibit a reduction in the number of mature lymphocytes (Celeste et al., 2002; Difilippantonio et al., 2008). Similarly, there was a 40–50% reduction in the number of thymocytes and B cells in the spleens of RNF8−/− mice (Fig. 1 A). 53BP1−/− thymocytes express low levels of TCRβ, which is associated with defective V(D)J recombination (Difilippantonio et al., 2008). Despite the decreased cellularity, RNF8−/−, like H2AX−/−, thymocytes showed normal TCRβ expression (Fig. 1 A; Celeste et al., 2002).
Figure 1.
Lymphopenia and reduced CSR in RNF8−/− mice. (A, left) Mean number of CD43− cells isolated from spleens of RNF8+/+ and RNF8−/− mice. (middle) Mean number of thymocytes in RNF8+/+ and RNF8−/− mice (error bars = SD; n ≥ 3). (right) TCRβ surface expression in freshly isolated RNF8+/+, RNF8−/−, and 53BP1−/− thymocytes (one representative out of three independent experiments). (B) Two-color flow cytometric analysis of IgG1 expression on CFSE-labeled B cells that were stimulated with LPS plus IL-4 for 4 d (percentages are shown). (C) Frequency of IgG1 and IgG3 expression in B cells that were stimulated for 4 d with LPS plus IL-4 or LPS alone in five or three independent experiments, respectively. Horizontal bars indicate means. (D) Representative example of CFSE incorporation profiles of B cells stimulated with LPS plus IL-4 for 4 d. (E) Percentage of cells expressing IgG1 (left) or IgG3 (right) that had undergone a given number of cell divisions. One representative out of at least three independent experiments is shown in D and E.
To further compare 53BP1 and RNF8 deficiencies, we examined the efficiency of CSR in the mutant mice. RNF8 deficiency caused a significant reduction in the frequency of IgG1 or IgG3 surface expression in response to activation with LPS/IL-4 or LPS alone, respectively (Fig. 1, B and C). However, this reduction in CSR was less severe than the one observed in 53BP1-deficient B cells (Fig. 1 B).
To assess whether loss of RNF8 affects cellular proliferation, we labeled LPS/IL-4–activated B cells with CFSE. In contrast to the normal proliferation of 53BP1 mutant B cells (Ward et al., 2004), CFSE dye dilution histograms revealed that RNF8−/− B cells underwent fewer cell divisions than WT cells (Fig. 1 D). One potential explanation for the proliferation defect (Fig. 1 D) and lymphopenia (Fig. 1 A) is that RNF8 regulates HR (Huang et al., 2009), which is required for DNA replication. For example, loss of RNF8 leads to a defect in the ability to relocalize Brca1 and Rad51 to sites of DNA damage (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). Consistent with this notion, we found that IR-induced Rad51 focus formation was partially impaired in RNF8−/− B cells (Fig. S2 A). Impaired Rad51 focus formation was not caused by alterations in cell-cycle distribution, as determined by cellular DNA content (Fig. S2 B). Because proliferation defects might affect CSR, we measured surface IgG1 and IgG3 levels as a function of cell division (Fig. 1 E). Even in cells that had undergone several rounds of cell division, CSR was significantly reduced in the absence of RNF8 (Fig. 1 E).
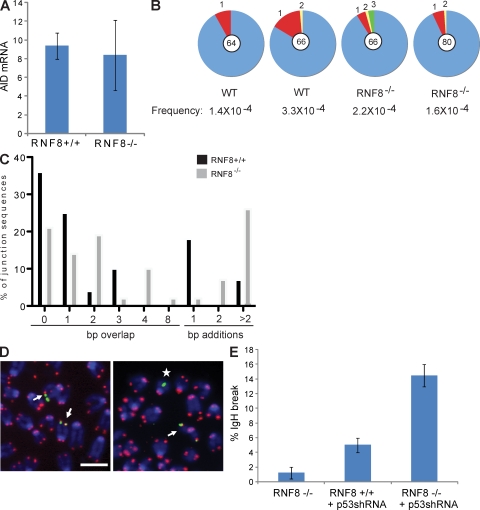
To determine at which level RNF8 deficiency affects CSR, we measured activation-induced cytidine deaminase (AID) expression and determined whether AID targeting to the switch region was affected by analysis of Sμ mutation in B cells stimulated to switch with LPS and IL-4. The level of AID mRNA in stimulated B cells was not affected by RNF8 deletion (Fig. 2 A). Moreover, we found similar levels of Sμ mutation in IgM+ RNF8−/− and RNF8+/+ B cells (Fig. 2 B). These results indicate that RNF8 is dispensable for AID accessibility to the Sμ switch region.
Figure 2.
AID targeting and genomic instability in RNF8−/− B cells undergoing CSR. (A) AID mRNA levels measured by quantitative real-time PCR performed on cDNA from B cells stimulated with LPS plus IL-4 for 48 h. The abundance of AID transcripts relative to GAPDH is shown. Error bars represent means ± SD. (B) Sµ mutations in B cells stimulated with LPS plus IL-4 and labeled with CFSE. The analysis was performed on sorted IgM+ cells that had undergone five divisions. The frequency of mutations per basepair sequenced and the total number of independent sequences analyzed are indicated underneath and in the center of each chart, respectively. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated in the periphery of the charts. (C) RNF8+/+ and RNF8−/− B cells were stimulated with LPS and IL-4 for 72 h, IgG1-positive cells were sorted, genomic DNA was amplified by PCR, and Sμ–Sγ junctions were sequenced. The percentage of junctions with the indicated nucleotide overlap or insertion is indicated (42 RNF8−/− and 28 RNF8+/+ sequences were analyzed). (D) LPS plus IL-4–stimulated B cells were retrovirally transduced with p53 shRNA (see Materials and methods). At 72 h after infection, the cells were sorted and metaphase spreads were hybridized with a combination of painting probes for IgH Cα (green) and telomere-specific peptide nucleic acid probes (red), and counterstained with DAPI (blue). (left) Example of a normal metaphase spread (arrows). (right) Example of an IgH lesion (star). Bar, 5 µm. (E) The percentage of metaphases with abnormalities specifically associated with chromosome 12 is quantified. At least 100 metaphases for each group were analyzed in two independent experiments. Error bars represent means ± SD.
Sμ–Sγ1 switch region junctions cloned from 53BP1−/− B cells show a high frequency of DNA insertions because of defective processing of CSR DSBs (Reina-San-Martin et al., 2007). To ascertain whether switch junctions are also abnormal in the absence of RNF8, we cloned and sequenced switch region junctions from B cells stimulated with LPS and IL-4. Microhomology at junctions was mildly increased in the absence of RNF8 (Fig. 2 C). More strikingly, 24% of the total RNF8−/− CSR junctions analyzed showed insertions >2 nt compared with only 7% in WT mice (Fig. 2 C). Thus, RNF8, like 53BP1, prevents insertions at switch junctions.
The inability to complete CSR is frequently associated with accumulation of immunoglobulin heavy chain (IgH)–associated breaks (Franco et al., 2006, 2008; Ramiro et al., 2006). To examine IgH instability, we used an IgH locus–specific/telomere fluorescence in situ hybridization (FISH) assay in which the loss of telomeric signals near the IgH locus on chromosome 12 is indicative of unresolved DSBs (Fig. 2 D; Ramiro et al., 2006). Surprisingly, distinct from 53BP1, H2AX, or ataxia telangiectasia mutated deficiency (Celeste et al., 2003; Ramiro et al., 2006), we failed to detect IgH breaks in RNF8−/− B cells (Fig. 2 E). We considered the possibility that RNF8-deficient cells with aberrations might be eliminated by a p53-dependent pathway before they reach metaphase, as previously observed in LPS-activated DNA-Pkcs−/− cells (Franco et al., 2008). To test this we performed small hairpin RNA (shRNA)–mediated knockdown of p53 in LPS-activated B cells. In two independent experiments, we observed an increase in the accumulation of IgH-associated breaks in p53-depleted RNF8−/− cells relative to controls (Fig. 2 E). These results suggest that RNF8 is necessary for joining a subset of breaks during CSR, but that RNF8−/− cells with unresolved DSBs are normally eliminated in a p53-dependent manner.
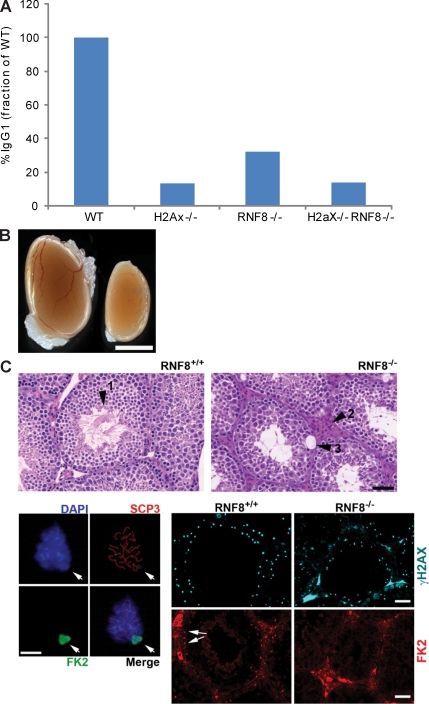
53BP1 and RNF8 localization to DSBs is dependent on H2AX phosphorylation (Celeste et al., 2002; Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007), and H2AX/H2A are ubiquitylated by RNF8 (Huen et al., 2007; Mailand et al., 2007). Nevertheless, RNF8 and H2AX deficiency leads to a milder CSR defect than 53BP1−/− (Fig. 1 B; Ward et al., 2004) and no detectable defect in thymocyte TCRβ expression (Fig. 1 A; Difilippantonio et al., 2008), suggestive of H2AX-independent functions for 53BP1. To determine whether H2AX and RNF8 function epistatically, we produced mice deficient for both H2AX (Celeste et al., 2002) and RNF8 by breeding. RNF8−/−H2AX−/− mice showed impairment of CSR at levels similar to those found in H2AX−/− B cells (Fig. 3 A and Fig. S3), which is still less severe than 53BP1 deficiency (Fig. 1 B). These results indicate that H2AX and RNF8 function in the same pathway during CSR, and that 53BP1 has a unique role that is not entirely regulated by H2AX and RNF8.
Figure 3.
RNF8 and H2AX function epistatically in class switching and spermatogenesis. (A) Frequency of IgG1 expression in B cells from WT, H2AX−/−, RNF8−/−, and H2AX−/−RNF8−/− mice stimulated with LPS plus IL-4 for 4 d. One representative out of three independent experiments is shown (Fig. S3). (B) Comparison of testes size in RNF8+/+ and RNF8−/− mice. Bar, 3 mm. (C, top) Sections of testes stained with hematoxylin-eosin. (arrowhead 1) In RNF8+/+ testes, mature sperm is organized in a rosette-like manner and is abundant. In RNF8−/− testes, the amount of sperm is much reduced. RNF8 mutant testes show evidence of degeneration such as the accumulation of Leydig cells in intertubular spaces (arrowhead 2) and areas of vacuolization (arrowhead 3). Bar, 50 µm. (bottom left) Immunofluorescence of isolated WT mouse spermatocytes with antibodies against SCP3 (red) and FK2 (green). The image illustrates that the staining of FK2 is restricted to the XY body. Bar, 5 mm. (bottom right) Immunofluorescence of testes from RNF8+/+ and RNF8−/− animals stained with antibodies against γH2AX (green) and FK2 (red). Note that although a normal γH2AX signal was detected in both tissues, the sex body–associated FK2 signal (arrows) was not detectable in RNF8−/− testes. Bars, 50 mm. Two mice of each genotype were analyzed in B and C.
In addition to the regulation of focus formation, H2AX has an essential role in male spermatogenesis. Specifically, H2AX−/− mice exhibit impaired XY synapsis caused by defective synapsis of the XY chromosomes during meiotic prophase (Celeste et al., 2002; Fernandez-Capetillo et al., 2003b). In contrast, 53BP1-deficient mice are fertile, illustrating that H2AX also displays 53BP1-independent functions in the context of meiosis. Similar to H2AX−/− males, RNF8-deficient males failed to breed, had reduced testes size (Fig. 3 B), and showed a marked reduction in sperm (Fig. 3 C). Furthermore, histological analysis revealed that there was considerable degeneration in seminiferous tubules in RNF8−/− mice, indicated by an increase in the number of vacuoles and Leydig cells (Fig. 3 C, top). Given that H2AX phosphorylation coats the XY chromatin of male meiotic cells, we explored whether a similar phenomenon was occurring with respect to ubiquitylation. To this end, we stained spermatocytes with antibodies against FK2, which marks ubiquitinated proteins. Like phosphorylated H2AX, ubiquitylated conjugates coated the sex body in pachytene-stage cells (Fig. 3 C, bottom left). Strikingly, despite normal H2AX phosphorylation of the sex body, FK2 staining was lost in RNF8 knockout spermatocytes (Fig. 3 C, bottom right). Collectively, these results suggest that RNF8 acts downstream of H2AX but distinct from 53BP1 in the regulation of CSR and male meiosis.
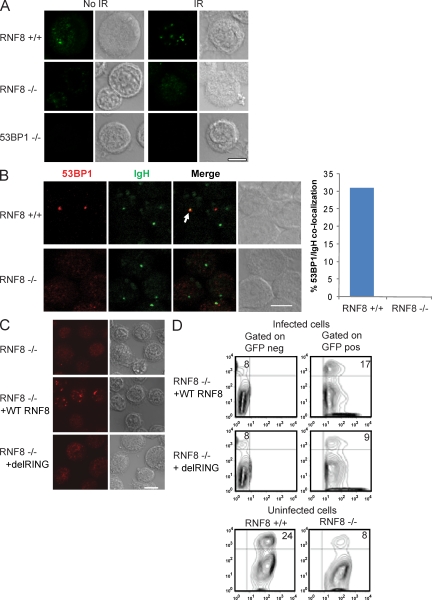
It has been suggested that RNF8-dependent ubiquitylation at DSBs exposes methylated histones, which in turn regulate 53BP1 binding to DNA damage. Indeed, loss of RNF8 impaired the ability of 53BP1 to form IR-induced foci (Fig. 4 A; Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). In B cells stimulated to undergo CSR, 53BP1 accumulates at sites of DNA damage produced by AID on the IgH locus (Reina-San-Martin et al., 2004; Callén et al., 2007), and these foci were absent in RNF8-deficient cells (Fig. 4 B). Therefore, the ability of 53BP1 to form foci at sites of IR- and AID-induced DNA damage is compromised in the absence of RNF8.
Figure 4.
53BP1 foci formation at sites of CSR is RNF8 dependent. (A) RNF8+/+, RNF8−/−, and 53BP1−/− B cells were stimulated with LPS and IL-4 for 3 d, left untreated (no IR), or exposed to 1 Gy of γ irradiation (IR) and then incubated for 30 min at 37°C before staining for 53BP1 (green). Bar, 5 µm. One representative out of two independent experiments is shown. (B) Distribution of 53BP1 in LPS/IL-4–activated B cells from RNF8+/+ and RNF8−/− mice. B cells were stained with anti-53BP1 antibodies followed by DNA FISH detection of the CH region (left). The arrow indicates colocalization of 53BP1 and IgH. (right) The percentage of cells with colocalization of 53BP1 and IgH. A total of 52 cells positive for both signals were analyzed for each genotype in one experiment. Bar, 7 µm. (C and D) RNF8−/− B cells stimulated with LPS/IL-4 were retrovirally infected with a WT or delRING form of RNF8 and stained for 53BP1 (C) or IgG1 (D) on day 3 (percentages are shown in D). One out of two representative experiments is shown. Bar, 7 µm.
Previous studies have shown that the RING domain of RNF8 is required for DSB-induced ubiquitylation and 53BP1 foci formation (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). To determine whether the RING domain of RNF8 is also necessary for class switching, we retrovirally expressed a WT or RING-deleted (delRING) form of RNF8 in RNF8−/− B cells that were stimulated to undergo CSR. As expected, 53BP1 foci were visible in RNF8-deficient B cells reconstituted with WT RNF8 but not in the delRING RNF8 (Fig. 4 C). In addition, WT RNF8 reconstituted CSR, whereas the delRING failed to do so (Fig. 4 D). Thus, the RING domain of RNF8 is critical for CSR.
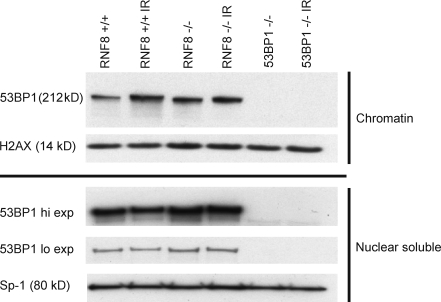
To determine directly whether RNF8 affects the association of 53BP1 with chromatin, we examined the subcellular distribution of 53BP1 in untreated and irradiated cells by cell fractionation. We found that a considerable fraction of cellular 53BP1 was bound to chromatin independently of IR, and that this pool increased only slightly, if at all, after IR (Fig. 5). These results are consistent with photobleaching experiments that indicate that in the absence of DNA damage, 53BP1 interacts with chromatin (Bekker-Jensen et al., 2005). Strikingly, loss of RNF8 did not affect the chromatin binding of 53BP1 (Fig. 5). Thus, although RNF8-dependent ubiquitylation may increase the affinity of 53BP1 for damaged chromatin, the initial loading to chromatin clearly occurs by mechanisms independent of RNF8. As previously proposed in the context of V(D)J recombination, constitutive (DNA damage–independent) 53BP1 chromatin binding might promote long-range CSR via 53BP1 homooligomerization-mediated synapsis of distal switch regions (Difilippantonio et al., 2008). In this model, RNF8 would increase the local concentration of 53BP1 or induce contraction of the switch regions in response to AID DSBs, thereby further promoting the pairing of switch regions.
Figure 5.
Constitutive binding of 53BP1 to chromatin is RNF8 independent. RNF8+/+, RNF8−/−, and 53BP1−/− B cells were stimulated with LPS/IL-4 for 3 d and were treated with IR (10 Gy) or left untreated. After 90 min, cells were collected and chromatin and nuclear fractions were isolated (see Materials and methods). Immunoblotting was performed using the indicated antibodies. One out of two independent experiments is shown.
H2AX regulates foci formation of numerous factors that carry out diverse functions in nonhomologous end joining, HR and DNA damage signaling. Nevertheless, many of these factors have essential roles at sites of DSB independent of their ability to form foci (Celeste et al., 2003). For example, Brca1 is essential for viability and HR, whereas H2AX and RNF8 knockout mice are viable and show only a mild defect in HR, despite the fact that BRCA1 foci are impaired in their absence. Although assays using recombination substrates failed to detect impairment in nonhomologous end joining in H2AX knockout mice (Bassing et al., 2002), H2AX functions in CSR and regulates foci formation of numerous factors, including 53BP1 and Nbs1, which are themselves implicated in DNA end joining and CSR (Celeste et al., 2003). RNF8, acting downstream of H2AX, regulates focus formation of 53BP1 but not of Nbs1. We speculate that because RNF8 regulates only a subset of repair proteins that form H2AX-dependent foci, this may explain why the CSR defect in RNF8 knockout mice is slightly milder than that observed in H2AX−/− and H2AX−/−RNF8−/− mice (Fig. 3 A and Fig. S3). The idea that RNF8 acts epistatically with H2AX in vivo is bolstered by the finding that RNF8−/− males are infertile, with a similar meiotic arrest as in H2AX knockouts. In light of the role of H2AX in XY body formation, it is tempting to propose that RNF8-dependent ubiquitylation might play an active role in transcriptional silencing and/or chromatin compaction. Moreover, because RNF168 acts together with RNF8 in promoting efficient histone ubiquitylation, we would predict that patients with RIDDLE syndrome should exhibit male sterility. In conclusion, our results indicate that a significant contribution of the CSR and meiotic defects observed in H2AX knockout mice is likely to be the inability to ubiquitylate proteins at sites of CSR DSBs and throughout the male sex body. Consistent with our findings, a recent report showed that CSR to IgA was impaired in mouse CF12F3-3 cells treated with shRNAs that depleted RNF8 and RNF168 (Ramachandran et al., 2010).
MATERIALS AND METHODS
Mice.
For generation of RNF8−/− mice, the embryonic stem (ES) cell line RRR260 was used (Bay Genomics). The RNF8 gene was disrupted by a neo gene selection cassette inserted between transcribed exons 4 and 5. The exact insertion sites were mapped by genomic PCR and DNA sequencing. These ES cells were injected into C57BL/6 blastocysts to generate chimeric mice that were then backcrossed with C57BL/6 mice to generate RNF8+/− animals. Finally, the heterozygotes were intercrossed to generate RNF8−/− mice. All experiments with mice followed protocols approved by the National Institutes of Health Institutional Animal Care and Use Committee.
Lymphocyte cultures, cell sorting, and flow cytometry.
B cells were isolated from spleens using CD43 microbeads (Miltenyi Biotec), labeled with 5 µM CFDA-SE for 10 min at 37°C (Invitrogen), and cultured (0.5–1 × 106 cells/ml) with 25 µg/ml LPS alone (Sigma-Aldrich) or LPS plus 5 ng/ml IL-4 (Sigma-Aldrich) for 1–4 d. In cell-sorting experiments, propidium iodide was added immediately before laser excitation to exclude dead cells. Cell sorting was performed on a FACSAria (BD). For flow cytometry, the following antibodies were obtained from BD: IgG1-biotin, IgG3-biotin, streptavidin-PE, B220-FITC, and TCRβ-PE. Cell-cycle analysis was performed as previously described (Daniel et al., 2008).
Viral infection and shRNA.
To knock down p53 in B cells, we used an shRNA vector against Trp53 that was previously described (Hemann et al., 2003). For expression of WT and delRING RNF8 in B cells, we cloned the previously described constructs (Huen et al., 2007) into the PMX-PIE retroviral vector. Retroviral supernatants were obtained from BOSC23 cells 48 h after transient cotransfection (FuGENE; Roche) with the retroviral and pCLEco plasmids. B cells were infected 24 h after LPS and IL-4 stimulation with retroviral supernatants by centrifugation for 1.5 h at 2,500 rpm at room temperature in the presence of 10 µg/ml polybrene. Supernatants were replaced with LPS and IL-4–containing RPMI 1640 12–16 h after infection.
Quantitative RT-PCR.
RNA was isolated using the RNeasy Mini Kit (QIAGEN). cDNA was synthesized with the SuperScriptII kit (Invitrogen). Real-time quantitative RT-PCR was performed on an ABI 7900 (Applied Biosystems). Transcripts were amplified using SYBR Green PCR Master Mix (Applied Biosystems). The following primers were used: GAPDH (Fwd), 5′-TGAAGCAGGCATCTGAGGG-3′; GAPDH (Rev), 5′-CGAAGGTGGAAGAGTGGGAG-3′; AID (Fwd), 5′-GCCACCTTCGCAACAAGTCT-3′; and AID (Rev), 5′-CCGGGCACAGTCATAGCAC-3′.
Mutation analysis.
Analysis of Sµ mutations and switch junction sequences were performed in B cells stimulated with LPS/IL-4 and CFSE labeled. Cells were sorted for five cell divisions and surface IgM expression, and mutant analysis was performed as previously described (Reina-San-Martin et al., 2003).
FISH analysis, immunofluorescence, and immunohistochemistry.
B cells cultured for 3 d with LPS plus IL-4 were arrested at mitosis with colcemid (Invitrogen) treatment, swollen in prewarmed 0.075 M KCl for 15 min at 37°C, and fixed by methanol/acetic acid treatment, and dropped in microscope slides using a humidity chamber. Slides were denatured at 75°C/70% formamide for 7 min; hybridized with the following probes overnight at 37°C; washed, dehydrated, and stained with DAPI; and mounted with VECTASHIELD (Vector Laboratories). We used bacterial artificial chromosome (BAC) probes containing IgH Cα (from Cγ1 3′ of Cα; Difilippantonio et al., 2002) and telomere repeat–specific peptide nucleic acid probes (Applied Biosystems). For 53BP1 and Rad51 foci analysis, B cells were spun onto coverslips after 72 h of stimulation and immunofluorescence was performed as previously described (Petersen et al., 2001). For coupled immunofluorescence and FISH analysis, day 3 stimulated B cells were spun onto coverslips and processed as described previously (Chen et al., 2000).The IgH-specific BAC probe spanned from Cγ1 to 3′ of Cα. Rabbit polyclonal anti-53BP1 (Novus Biologicals), rabbit polyclonal anti-Rad51 (H-92; Santa Cruz Biotechnology, Inc.), and goat anti–rabbit IgG Alexa Fluor 488 (Invitrogen) were used. Metaphase spread images were captured with a microscope (Axio Imager Metasystems; Carl Zeiss, Inc.) and foci images were captured with a confocal microscope (LSM 510 MetaII; Carl Zeiss, Inc.). Testes were fixed in formalin and embedded in paraffin for subsequent processing. 2.5-mm sections were treated with citrate for antigenic recovery and processed for immunohistochemistry with FK2 (Invitrogen) antibodies. Hematoxiline was used to counterstain. Sections were scanned with a digitalized system (MIRAX; Carl Zeiss, Inc.). For tissue immunofluorescence, sections were previously deparaffinized and γH2AX (Millipore) antibodies were also used. Immunofluorescence of isolated mouse spermatocytes has been previously described (Fernandez-Capetillo et al., 2003a). Combined SCP3 (gift from M. Tarsounas, Gray Institute for Radiation Oncology and Biology, Oxford, England, UK) and FK2 antibody staining was processed with Alexa Fluor 488 and Alexa Fluor 568 (Invitrogen) secondary antibodies.
Chromatin fractionation and Western blotting.
B cells were isolated and cultured with LPS and IL-4 for 3 d as described. A portion of the cultures was subjected to 10 Gy of γ irradiation and allowed to recover at 37°C for 1.5 h. Nuclei from 5 million cells were isolated with the ProteoJET Cytoplasmic and Nuclear Protein Extraction Kit (Fermentas) and lysed; the manufacturer’s nuclei lysis buffer was supplemented with 30 mM EDTA, 2 mM EGTA, and 10 mM dithiothreitol. The nuclear extract was centrifuged at 1,700 g for 20 min at 4°C; the supernatant was saved at −80°C as the “nuclear-soluble fraction,” and the chromatin pellet was washed twice in 250 µl of 0.1× lysis buffer supplement. Chromatin was resuspended in 30 µl of 10 mM Hepes, 10 mM KCl, 1 mM MgCl2, 10% glycerol, 1 mM CaCl2, 1 mM EDTA, 1× protease inhibitors (Roche), and 5 U of micrococcal nuclease (New England Biolabs, Inc.), and then incubated for 45 min at 37°C. The reaction was stopped by the addition of EGTA to 1 mM, and the digested pellet was stored at −80°C as the “chromatin-bound fraction.” Extracts were run on 3–8% Tris-acetate gels and analyzed by Western blotting with antibodies against 53BP1 (Bethyl Laboratories, Inc.) and H2AX (Novus Biologicals). To examine RNF8 expression, total cell extracts derived from RNF8 WT and -deficient mouse embryonic fibroblasts were incubated with anti-RNF8 antibodies and protein A agarose beads for 2 h at 4°C with gentle agitation under denaturing conditions. Beads were washed twice with denaturing buffer and bound proteins were eluted by boiling in sampling buffer. Thereafter, immunoprecipitates were separated by SDS-PAGE and immunoblotted with antibodies generated against full-lengh human RNF8.
Online supplemental material.
Fig. S1 shows the strategy used to disrupt the mouse RNF8 gene. Fig. S2 shows that Rad51 foci formation is impaired in the absence of RNF8. Fig. S3 shows two more independent experiments for the data shown in Fig. 3 A. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092308/DC1.
Acknowledgments
We thank M. Kruhlak, L. Granger, and A. Adams for technical assistance, and members of the Nussenzweig laboratory for helpful discussions.
I.A. Klein was supported by Medical Scientist Training Program grant GM07739 and is a predoctoral fellow of the Cancer Research Institute. This work was supported by the Intramural Research program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- CSR
- class switch recombination
- delRING
- RING deleted
- DSB
- DNA double-strand break
- FISH
- fluorescence in situ hybridization
- HR
- homologous recombination
- IgH
- immunoglobulin heavy chain
- IR
- ionizing radiation
- shRNA
- small hairpin RNA
References
- Alpi A.F., Patel K.J. 2009. Monoubiquitylation in the Fanconi anemia DNA damage response pathway. DNA Repair (Amst.). 8:430–435 10.1016/j.dnarep.2009.01.019 [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Chua K.F., Sekiguchi J., Suh H., Whitlow S.R., Fleming J.C., Monroe B.C., Ciccone D.N., Yan C., Vlasakova K., et al. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA. 99:8173–8178 10.1073/pnas.122228699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Melander F., Bartek J., Lukas J. 2005. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170:201–211 10.1083/jcb.200503043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75 10.1016/j.cell.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M.J., et al. 2002. Genomic instability in mice lacking histone H2AX. Science. 296:922–927 10.1126/science.1069398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., Nussenzweig A. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679 10.1038/ncb1004 [DOI] [PubMed] [Google Scholar]
- Chen H.T., Bhandoola A., Difilippantonio M.J., Zhu J., Brown M.J., Tai X., Rogakou E.P., Brotz T.M., Bonner W.M., Ried T., Nussenzweig A. 2000. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 290:1962–1965 10.1126/science.290.5498.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.A., Pellegrini M., Lee J.H., Paull T.T., Feigenbaum L., Nussenzweig A. 2008. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J. Cell Biol. 183:777–783 10.1083/jcb.200805154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio M.J., Petersen S., Chen H.T., Johnson R., Jasin M., Kanaar R., Ried T., Nussenzweig A. 2002. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196:469–480 10.1084/jem.20020851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S., Gapud E., Wong N., Huang C.Y., Mahowald G., Chen H.T., Kruhlak M.J., Callen E., Livak F., Nussenzweig M.C., et al. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 456:529–533 10.1038/nature07476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Liebe B., Scherthan H., Nussenzweig A. 2003a. H2AX regulates meiotic telomere clustering. J. Cell Biol. 163:15–20 10.1083/jcb.200305124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Mahadevaiah S.K., Celeste A., Romanienko P.J., Camerini-Otero R.D., Bonner W.M., Manova K., Burgoyne P., Nussenzweig A. 2003b. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell. 4:497–508 10.1016/S1534-5807(03)00093-5 [DOI] [PubMed] [Google Scholar]
- Franco S., Gostissa M., Zha S., Lombard D.B., Murphy M.M., Zarrin A.A., Yan C., Tepsuporn S., Morales J.C., Adams M.M., et al. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21:201–214 10.1016/j.molcel.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Franco S., Murphy M.M., Li G., Borjeson T., Boboila C., Alt F.W. 2008. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J. Exp. Med. 205:557–564 10.1084/jem.20080044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Fridman J.S., Zilfou J.T., Hernando E., Paddison P.J., Cordon-Cardo C., Hannon G.J., Lowe S.W. 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 33:396–400 10.1038/ng1091 [DOI] [PubMed] [Google Scholar]
- Huang J., Huen M.S., Kim H., Leung C.C., Glover J.N., Yu X., Chen J. 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11:592–603 10.1038/ncb1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Marteijn J.A., Bekker-Jensen S., Mailand N., Lans H., Schwertman P., Gourdin A.M., Dantuma N.P., Lukas J., Vermeulen W. 2009. Nucleotide excision repair–induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 186:835–847 10.1083/jcb.200902150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S., Durocher D. 2009. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst.). 8:436–443 10.1016/j.dnarep.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Petersen S., Casellas R., Reina-San-Martin B., Chen H.T., Difilippantonio M.J., Wilson P.C., Hanitsch L., Celeste A., Muramatsu M., Pilch D.R., et al. 2001. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 414:660–665 10.1038/414660a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Chahwan R., Nepal R.M., Frieder D., Panier S., Roa S., Zaheen A., Durocher D., Scharff M.D., Martin A. 2010. The RNF8/RNF168 ubiquitin ligase cascade facilitates class switch recombination. Proc. Natl. Acad. Sci. USA. 107:809–814 10.1073/pnas.0913790107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109 10.1038/nature04495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778 10.1084/jem.20030569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen H.T., Nussenzweig A., Nussenzweig M.C. 2004. ATM is required for efficient recombination between immunoglobulin switch regions. J. Exp. Med. 200:1103–1110 10.1084/jem.20041162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen J., Nussenzweig A., Nussenzweig M.C. 2007. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur. J. Immunol. 37:235–239 10.1002/eji.200636789 [DOI] [PubMed] [Google Scholar]
- Stewart G.S., Stankovic T., Byrd P.J., Wechsler T., Miller E.S., Huissoon A., Drayson M.T., West S.C., Elledge S.J., Taylor A.M. 2007. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc. Natl. Acad. Sci. USA. 104:16910–16915 10.1073/pnas.0708408104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136:420–434 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Wang B., Elledge S.J. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA. 104:20759–20763 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J.S., Cascalho M., Chen L., Nussenzweig A., Livak F., et al. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459–464 10.1083/jcb.200403021 [DOI] [PMC free article] [PubMed] [Google Scholar]