Abstract
Signaling and repair of DNA double-strand breaks (DSBs) are critical for preventing immunodeficiency and cancer. These DNA breaks result from exogenous and endogenous DNA insults but are also programmed to occur during physiological processes such as meiosis and immunoglobulin heavy chain (IgH) class switch recombination (CSR). Recent studies reported that the E3 ligase RNF8 plays important roles in propagating DNA DSB signals and thereby facilitating the recruitment of various DNA damage response proteins, such as 53BP1 and BRCA1, to sites of damage. Using mouse models for Rnf8 mutation, we report that Rnf8 deficiency leads to impaired spermatogenesis and increased sensitivity to ionizing radiation both in vitro and in vivo. We also demonstrate the existence of alternative Rnf8-independent mechanisms that respond to irradiation and accounts for the partial recruitment of 53bp1 to sites of DNA damage in activated Rnf8−/− B cells. Remarkably, IgH CSR is impaired in a gene dose-dependent manner in Rnf8 mutant mice, revealing that these mice are immunodeficient. In addition, Rnf8−/− mice exhibit increased genomic instability and elevated risks for tumorigenesis indicating that Rnf8 is a novel tumor suppressor. These data unravel the in vivo pleiotropic effects of Rnf8.
Mammalian cells have evolved sophisticated DNA damage signaling and repair mechanisms to prevent the accumulation or transmission of damaged DNA during cell divisions (O’Driscoll and Jeggo, 2006; Bartek and Lukas, 2007; Harper and Elledge, 2007; Hoeijmakers, 2009). Among the various types of DNA damage, DNA double-strand breaks (DSBs) are the most detrimental to our cells. The importance of DSB signaling and repair mechanisms is demonstrated by the association of their defects with various human syndromes characterized by developmental defects, neurodegenerative disorders, immunodeficiency, and increased cancer predisposition (O’Driscoll and Jeggo, 2006; Hakem, 2008; Hoeijmakers, 2009). In addition to DSBs generated by endogenous and exogenous DNA insults, DSBs are also programmed to occur in vivo during normal physiological processes, such as meiosis and during VDJ recombination in T and B cell development, in which the recombination process is essential to amplify the diversity for T and B cell receptor repertoires (Soulas-Sprauel et al., 2007). Furthermore, Ig heavy chain (IgH) class switch recombination (CSR), which is one of the most critical mechanisms for antibody diversification in mammals, also involves programmed generation of DSBs initiated by activation-induced cytidine deaminase (Chaudhuri et al., 2007; Soulas-Sprauel et al., 2007; Stavnezer et al., 2008). The subsequent signaling and repair of these DSBs is required for peripheral B cells to successfully synapse and join these breaks and switch from expressing low-affinity IgM to various high-affinity Ig isotypes, such as IgG1, IgE, and IgA, during an immune response. The joining of DSBs generated during the CSR process involves both classical and alternative nonhomologous end-joining pathways (Yan et al., 2007; Kotnis et al., 2009; Robert et al., 2009). Interestingly, defects in the signaling or the repair of the CSR-associated DSBs inevitably result in immunodeficiency (Durandy et al., 2007; Kotnis et al., 2009).
The signaling of DSBs employs various DNA damage response (DDR) proteins and elaborate posttranslational modifications (PTM) including ubiquitylation, phosphorylation, methylation, and acetylation of chromatin and DDR proteins (Harper and Elledge, 2007; Panier and Durocher, 2009). Within a few minutes after the generation of DSBs, subnuclear foci known as ionizing radiation (IR)–induced foci (IRIF) are assembled at the break sites (Wood and Chen, 2008). These IRIF arise from chromatin remodeling and orchestrated recruitment of various DDR proteins, which are important for mediating the signaling and repair of the damaged DNA as well as cell cycle checkpoint activation or apoptosis. Phosphorylation of the histone variant H2AX at Ser139 (γH2AX) is among the earliest PTMs required for the signaling of DSBs (Su, 2006). These early recruitment and PTM events at the damage sites provide important docking platforms to further enlist downstream DDR proteins. In addition to H2AX, several other DDR proteins, including NBS1, MDC1, 53BP1, and BRCA1, are also phosphorylated by kinases such as ATM, ATR, DNA-PK, Chk2, and Chk1. These phosphorylations provide important mechanisms for these DDR proteins to interact with each other at damage sites and to mediate the signaling and repair of DSBs. The recent demonstration of the roles of the E3 ligases RNF8 and RNF168 in DSB signaling has highlighted the importance of regulatory ubiquitylation in the DNA damage signaling and repair processes (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007; Doil et al., 2009; Panier and Durocher, 2009; Pinato et al., 2009; Stewart et al., 2009). Both E3 ligases are required for the recruitment of DDR proteins such as 53BP1, BRCA1, Rap80, Abraxas, and Brcc36 to DSBs. After the initial recognition of γH2AX by MDC1 at the site of DSBs, RNF8 is recruited to ATM-phosphorylated MDC1 through its FHA domain, and it functions in a complex with the E2 ubiquitin-conjugating enzyme Ubc13 to mediate mono-ubiquitylation of the histones H2AX and H2A at the chromatin-flanking DSBs (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). These RNF8-dependent histone ubiquitylation events serve to recruit the RNF168–Ubc13 complex to DSBs, leading to further K63 ubiquitylation of H2AX and H2A and thereby amplifying the DSBs signals (Doil et al., 2009; Pinato et al., 2009; Stewart et al., 2009). Interestingly, knockdown cell lines for RNF8 or RNF168 and mouse embryonic fibroblasts (MEFs) deficient for Rnf8 are unable to form or retain IRIF for 53BP1, BRCA1, and other downstream DDR proteins (Huen et al., 2007, 2008; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007; Doil et al., 2009; Stewart et al., 2009). These cells also exhibit increased radiosensitivity and impaired cell cycle checkpoints. More importantly, mouse models for mutations of Atm, H2ax, Mdc1, 53bp1, and Brca1 have demonstrated the roles of these signaling proteins in various processes including development, meiosis, CSR, genomic integrity, and cancer. Interestingly, germline mutations of the RNF168 gene have been associated with the human RIDDLE syndrome, which is characterized by defective 53BP1 foci formation, radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties (Stewart et al., 2007, 2009).
In this study, we demonstrate that Rnf8-deficient mice are viable but show a growth retardation phenotype, increased sensitivity to IR both in vitro and in vivo, and impaired spermatogenesis. Rnf8−/− MEFs show a growth defect that is partially rescued under hypoxic conditions. In contrast to Rnf8−/− MEFs, activated Rnf8−/− B cells show a partial recruitment and/or retention of 53bp1 to form IRIF, suggesting the existence of alternative signaling pathways that compensate for the absence of Rnf8 in activated B cells. We also show that CSR is significantly impaired in Rnf8 mutant mice, revealing that these mice are immunodeficient. Remarkably, loss of Rnf8 leads to increased genomic instability and predisposes the Rnf8 mutant mice for tumorigenesis, indicating that Rnf8 is a novel tumor suppressor. Thus, Rnf8 emerges as an important player in DSBs through its essential roles in spermatogenesis, CSR, maintaining genomic integrity, and suppressing cancer.
RESULTS
To examine the in vivo roles of Rnf8, two strains of Rnf8 mutant mice have been generated using independent gene trap embryonic stem cell (ES) clones (Fig. S1 A). Southern blot analysis confirmed the germline transmission of the two Rnf8 mutations (Fig. S1, B and C). Western blot analysis of serial twofold dilutions of cell lysates indicated about a twofold decrease of expression level of Rnf8 protein in heterozygote compared with WT background (Fig. S1 D). Further Western blot analyses demonstrated loss of the full-length Rnf8 protein in Rnf8−/− thymocytes derived from the two different strains (Fig. S1 E). Rnf8 mutants derived from the two clones will be referred to as Rnf8 mutants.
Hypoxic conditions partially rescue impaired in vitro proliferation of Rnf8-deficient MEFs
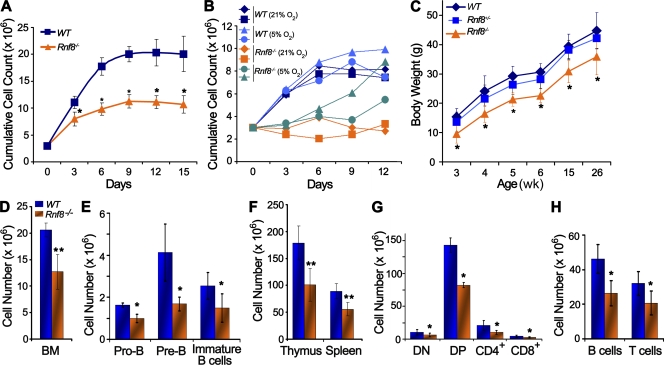
To examine the effect of Rnf8 deficiency on in vitro cellular growth, we derived MEFs from day-14.5 embryos generated from intercrossing of heterozygous mutant mice. Rnf8−/− MEFs of passage 2–5 proliferated at slower rates than WT littermate controls (Fig. 1 A). Furthermore, Rnf8−/− MEFs of passage 6 and 7 displayed lower saturation densities than WT littermate controls.
Figure 1.
Rnf8−/− MEFs exhibit proliferative defects and Rnf8−/− mice are growth retarded. (A) Rnf8−/− MEFs exhibited reduced ability to proliferate in vitro. Passage 2 WT and Rnf8−/− MEFs were seeded in 6-cm dishes at a density of 3 × 105. After every 3 d, the MEFs were trypsinized, counted, and reseeded at the same density. The cumulative growth curves are shown. Three independent experiments were performed, and a Student’s t test was used for statistical analysis. *, P < 0.05. Error bars represent SD. (B) Passage 4 WT and Rnf8−/− MEFs were cultured as in A, either under normoxic (21% O2) or hypoxic (5% O2) conditions. Data were from two independent experiments. (C) Body weights of age-matched WT (n = 10), Rnf8+/− (n = 14), and Rnf8−/− (n = 9) males. Statistical significance was established at each time point using Student’s t test. *, P < 0.05. Error bars represent SD. (D) Total number of cells in BM (one femur) of WT and Rnf8−/− mice. (E) Absolute numbers of pro–B cells (IgM−CD43+B220+), pre–B cells (IgM−CD43−B220+), and immature B cells (B220+IgM+IgD−) in the BM (one femur) of WT and Rnf8−/− mice. (F) Absolute number of cells in thymus and spleen of WT and Rnf8−/− mice. (G) Absolute numbers of double-negative (CD4−CD8−), double-positive (CD4+CD8+), CD4+, and CD8+ cells in the thymus of WT and Rnf8−/− mice. (H) Absolute numbers of B and T cells in the spleen of WT and Rnf8−/− mice. Data in D–H were generated from seven pairs of WT and Rnf8−/− littermates at the age of 6–10 wk. Student’s t test was used for statistical analysis. *, P < 0.05; **, P < 0.0007. Error bars represent SD.
Because it has been well documented that reactive oxygen species (ROS) play a role in generating endogenous DNA lesions (Bristow and Hill, 2008), hypoxic environment would offer a reduction in ROS-mediated assaults to chromatin. Therefore, we hypothesize that under hypoxic conditions, the proliferation defect of Rnf8−/− MEFs would be alleviated owing to lesser DNA damage. As shown in Fig. 1 B, culturing of passage 4 Rnf8−/− MEFs for 12 d at 5% O2 concentration resulted in a greater proliferation rate than when they were cultured under normoxic conditions. Interestingly, this increase in proliferation became increasingly evident from days 6 to 12 of in vitro culture. At day 12 of culture under hypoxic conditions, one of the independently derived lines of Rnf8−/− MEFs even showed a saturation density close to those of the WT littermate MEFs.
These data indicate that Rnf8 deficiency in MEFs leads to defective proliferation. Furthermore, hypoxic conditions significantly rescue the impaired proliferation of Rnf8−/− MEFs, suggesting that ROS-induced DNA damage in Rnf8−/− MEFs contributes to their in vitro proliferative defects.
Rnf8 deficiency leads to growth retardation and reduced hematopoeitic populations
Rnf8−/− mice generated by intercrossing of heterozygous mutant mice were born at a Mendelian ratio, indicating that Rnf8 is dispensable for embryonic survival. However, Rnf8−/− mice showed a runted phenotype and were underdeveloped compared with WT littermates as determined by their body weight measurements at various time points throughout development (Fig. 1 C and Fig. S2).
Programmed DSBs play essential roles in development and differentiation of hematopoietic cells, and failure to signal or repair these DSBs significantly impairs hematopoietic lineages and can lead to immunodeficiency (Soulas-Sprauel et al., 2007). To assess the effect of Rnf8 deficiency on the immune system development, we examined BM, thymus, and spleen from 6–10-wk-old Rnf8−/− mice. FACS analysis and determination of absolute cell numbers of hematopoietic cells indicated significant hypocellularity of the immune organs of Rnf8−/− mice, and the Rnf8−/− hematopoietic cell subpopulations seemed to suffer this defect to a similar extent (Fig. 1, D–H; and Fig. S3). The total number of BM cells (12.7 ± 3.3 × 106) in a femur from Rnf8−/− mice was significantly reduced compared with WT littermates (20.6 ± 1.3 × 106; P < 0.0003; Fig. 1 D). More importantly, the numbers of Pro-B (IgM−B220+CD43+) and Pre-B (IgM−B220+CD43−) progenitors, as well as the number of immature B cells (IgM+B220+IgD−), were significantly reduced in BM from Rnf8−/− mice compared with WT littermates (P < 0.01; Fig. 1 E).
The absolute number of thymocytes was also significantly reduced in Rnf8−/− mice (101.3 ± 30.4 × 106) compared with WT littermates (178.9 ± 31.5 × 106; P < 0.0006; Fig. 1 F). Further characterization of these thymocytes indicated significant reductions in the absolute numbers of CD4−CD8− (double negative), CD4+CD8+ (double positive), and CD4+ and CD8+ (single positive) thymocyte populations (P < 0.05; Fig. 1 G). As 53bp1 is required for distal V–DJ joining and TCR-β expression (Difilippantonio et al., 2008), we examined the effect of Rnf8 deficiency on TCR-β expression. Our data indicated a mild decrease of the level of TCR-β expression in Rnf8−/− thymocytes compared with WT controls (Fig. S3B), which contrasts with the drastic reduction of TCR-β expression in 53bp1−/− thymocytes (Difilippantonio et al., 2008).
Next, we examined the effect of Rnf8 deficiency on splenocytes and observed a significant reduction in total number of splenocytes from Rnf8−/− mice (55.8 ± 12.7 × 106) compared with WT littermates (89.1 ± 14.2 × 106; P < 0.0006; Fig. 1 F). FACS analysis was further used to determine the absolute numbers of B and T cell populations in the spleen of Rnf8−/− mice and WT littermates. Both Rnf8−/− splenic B and T cell populations were found to be significantly reduced compared with WT controls (P < 0.01; Fig. 1 H). Collectively, these data indicate that Rnf8 plays important roles in maintenance of normal mammalian growth and development of various subpopulations in both primary and secondary immune organs.
Impaired spermatogenesis in Rnf8−/− mice
Programmed DSBs initiated by the meiosis-specific protein Spo11 are essential for meiotic recombination (Longhese et al., 2009). The repair of these breaks is mediated by homologous recombination. Defects in signaling or repair of these programmed DSBs during meiosis can potentially lead to infertility.
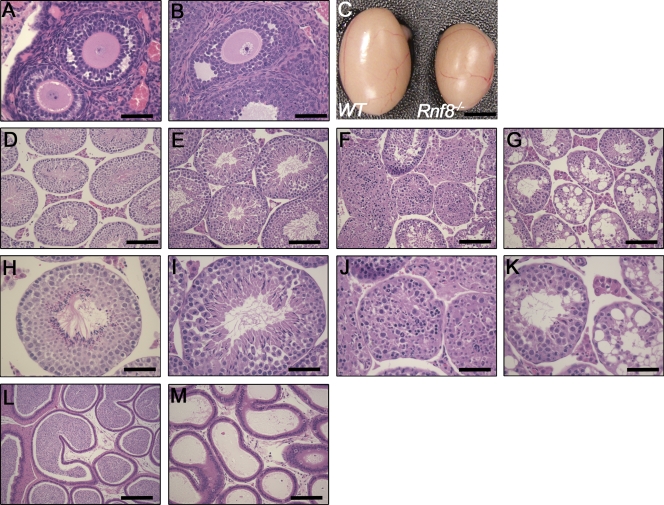
Because Rnf8 plays an essential role in the signaling of DSBs, we examined the effect of its loss on meiosis and fertility. Ovaries from 9-wk-old Rnf8−/− mice and their WT littermates were harvested, fixed, and stained with hematoxylin and eosin (H&E). No overt difference in the size of ovaries from Rnf8−/− females was observed compared with WT littermates. In addition, H&E staining also indicated that Rnf8−/− ovaries are similar to WT ovaries (Fig. 2, A and B). However, young (9 wk of age) and old (1 yr of age) Rnf8−/− males showed a reduction in testicular size compared with WT controls (Fig. 2 C). Examination of H&E-stained testis sections from 1.5–3.5-mo-old Rnf8−/− males showed a spectrum of alterations in spermatogenesis compared with their WT male littermates (Fig. 2, D–M). Within the same Rnf8−/− testis, we observed seminiferous tubules with well-populated layers of spermatogonia, spermatocytes, and mature spermatozoa, whereas other seminiferous tubules showed only few or absent mature spermatozoa in the lumen, as well as an unusually high number of cells in mitosis and immature spermatids (Fig. 2, E–I), suggesting an arrest in the spermatogenesis process. We also observed Rnf8−/− testes with seminiferous tubules that had disorganized epithelial architecture but still showed few spermatogonia and spermatocytes interspersed with apoptotic bodies and the absence of lumen and mature sperm cells (Fig. 2, F–J). Furthermore, more severe phenotypes were also observed, which were characterized by focal vacuolar degeneration of most seminiferous tubules of Rnf8−/− testes, without inflammation and with a normal appearance of interstitial Leydig cells (Fig. 2, G–K). The epididymis of Rnf8−/− mice, although histologically normal, was either poorly populated with or devoid of spermatozoa compared with WT controls (Fig. 2, L and M).
Figure 2.
Impaired spermatogenesis in Rnf8−/− mice. (A and B) Histological analysis of ovaries of WT (A) and Rnf8−/− (B) females showed no significant differences in ovarian architecture. Bars, 50 µm. (C) Testes from 12-mo-old Rnf8−/− male exhibited a significant reduction in size compared with those of WT littermates. Bar, 0.25 cm. (D–K) Images of H&E-stained seminiferous tubules from 1.5-mo-old WT (D and H) and Rnf8−/− (E, F, I, and J) mice and a 3.5-mo-old Rnf8−/− male (G and K). H&E staining of testes from Rnf8−/− males showed a spectrum of alterations in spermatogenesis. Rnf8−/− testis showed seminiferous tubules with well-populated layers of spermatogonia, spermatocytes, and mature spermatozoa (E and I) and other seminiferous tubules with only few or absent mature spermatozoa in the lumen and an unusually elevated number of immature spermatids and mitotic cells (E and I). Rnf8−/− testis showed disorganized epithelial architecture of seminiferous tubules, few spermatogonia and spermatocytes mixed together with apoptotic bodies, and the absence of mature sperm cells (F and J). Rnf8−/− testis showed focal vacuolar degeneration of most seminiferous tubules (G and K). In contrast to WT (L), H&E staining of epididymis of Rnf8−/− mice (M, 20X) show no histological abnormalities; however, they have a significant reduction of mature sperms. Bars: (D–G) 100 µm; (H–K) 50 µm; (L and M) 100 µm. A minimum of five mice per genotype have been analyzed.
Interestingly, despite their meiotic defect, and in contrast to the complete infertility of Atm−/−, H2ax−/−, and Mdc1−/− males, Rnf8−/− males derived from AS0574 ES clone successfully produced progenies, although the size of their litters (5.16 ± 0.9) was significantly reduced compared with Rnf8+/− males (9.3 ± 0.6; P = 0.0018). However, attempts to breed Rnf8−/− males from RRR260 ES clone failed to yield any pregnancies, possibly because of the difference of the Rnf8 mutation. We therefore conclude that Rnf8 deficiency impairs spermatogenesis.
53bp1 is partially recruited to DNA DSB sites via Rnf8-independent mechanisms in LPS-activated B cells
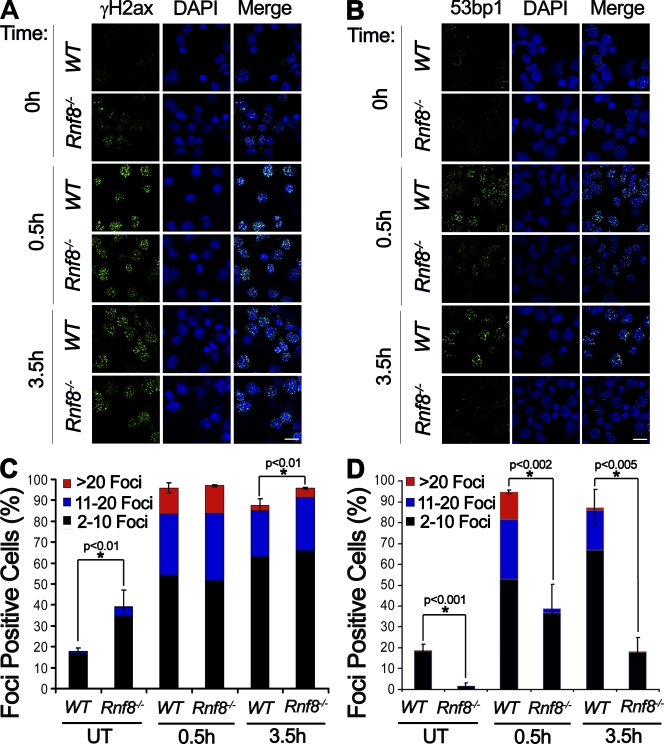
RNF8 plays an essential role in the signaling of DSBs. It mediates ubiquitylation of H2A and H2AX, facilitating the sequential recruitment of downstream DDR proteins to DSBs (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Panier and Durocher, 2009; Wu et al., 2009). Previous studies and our data in Fig. S4 indicate the abolition of recruitment of certain DDR proteins, especially 53bp1, to IR-induced DSBs in Rnf8−/− MEFs and human RNF8 knockdown cell lines (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Panier and Durocher, 2009; Wu et al., 2009). To further study the hierarchy of DDR proteins in immune cells, we examined time course recruitment of γH2ax and 53bp1 to DSBs in Rnf8−/− and WT B cells. B cells from Rnf8−/− and WT mice were activated for 60 h in the presence of LPS. Cells were either left untreated or irradiated with 3 Gy of γ radiation and were then harvested at various time points after IR for immunofluoresence to examine γH2ax and 53bp1 IRIF formation. A significantly higher number of γH2ax foci was observed in untreated Rnf8−/− cells compared with WT control cells, indicating an elevated number of endogenous breaks in the activated Rnf8−/− B cells (Fig. 3, A and C). However, no significant difference in the number or size of γH2ax IRIF was observed between Rnf8−/− and WT cells at 0.5 h after IR, although the number of γH2ax IRIF was slightly greater at 3.5 h after IR. Interestingly, despite the increased number of IRIF for γH2ax in nonirradiated Rnf8−/− activated B cells, a significantly lower number of 53bp1 foci was observed in these cells compared with WT controls (Fig. 3, B and D). About 38% of activated Rnf8−/− B cells was able to form 53bp1 foci at 0.5 h after γ radiation (3 Gy), and these foci appeared to be smaller in size than WT controls. Moreover, in contrast to WT controls, the number of Rnf8−/− B cells showing 53bp1 IRIF dropped by ∼50% at 3.5 h after IR treatment, indicating the unstable nature of 53bp1 foci in the absence of Rnf8. These data indicate the existence in activated B cells of Rnf8-independent mechanisms for the recruitment of 53bp1 to sites of DNA breaks, although Rnf8-dependent mechanisms are the major driving force for the formation and retention of 53bp1 IRIF.
Figure 3.
Partial recruitment of 53bp1 to the sites of DNA damage in the absence of Rnf8. (A and B) LPS-activated B cells either untreated (0 h) or harvested 0.5 and 3.5 h after 3 Gy irradiation were stained with anti-γH2ax (A) and anti-53bp1 (B) antibodies. Images were taken with a confocal microscope. Bars, 10 µm. (C and D) Quantitative analysis of γH2ax and 53bp1 subnuclear foci. Activated B cells harboring two or more than two foci were counted as foci-positive cells. Three independent experiments were performed, and a minimum of 300 cells was counted for each condition and genotype. A Student’s t test was used for statistical analysis. *, P < 0.01. Error bars represent SD.
Rnf8 deficiency leads to increased radiosensitivity
Failure to repair DSBs leads to cell cycle arrest and apoptosis, thus preventing transmission of damaged DNA during cell division. Based on the role of Rnf8 on initiation and maintenance of IRIF for DDR proteins such as 53bp1, we examined the effect of its loss, both in vitro and in vivo, with respect to radiosensitivity. Rnf8−/−, Rnf8+/−, and WT thymocytes were either left untreated or subjected to various dosages of IR (0.5–6 Gy) and were harvested 18 h after IR. The level of cell death was then examined by flow cytometry using 7AAD, as well as Annexin V and propidium iodide assays. A significant increase in radiosensitivity was observed with Rnf8−/− thymocytes, but not Rnf8+/− thymocytes, compared with WT controls (Fig. 4 A and Fig. S5 A).
Figure 4.
Rnf8−/− thymocytes and BM cells display increased radiosensitivity. (A) Thymocytes from WT and Rnf8−/− littermates were either left untreated or treated with various dosages of γ radiation. Cells were then stained with 7-AAD 18 h after treatment and analyzed by flow cytometry. Data from all treated samples were normalized to their respective untreated samples, and percentages of viable cells were plotted. Three independent experiments were performed. Student’s t test was used for statistical analysis. *, P < 0.05. Error bars represent SD. (B) Thymocytes from Rnf8−/− mice and WT littermates were either left untreated (0) or irradiated with 2 Gy of γ radiation and harvested at 2 and 5 h after treatment. Western blot analysis was performed to examine protein levels of Chk2, Ser15 phosphorylated p53 (S15-p53), and total p53. Actin was used as a loading control. (C) Representative pictures of dishes showing colonies derived from WT and Rnf8−/− BM cells either left untreated or irradiated with the indicated doses (0.5–6 Gy). All the pictures were taken at day 10 of culture. (D) Quantitative analysis showing the percentages of surviving colonies. Three independent experiments were performed. Data from all treated samples were normalized to their respective untreated samples, and Student’s t test was used for statistical analysis. *, P < 0.05 (statistically significant difference between WT and Rnf8+/− colony survivals); **, P < 0.02 (statistically significant difference of Rnf8−/− compared with WT and Rnf8+/− colony survivals). Error bars represent SD.
The Atm–Chk2–p53 signaling pathway plays essential roles in response to DSBs (Meek, 2009). Therefore, we examined the effect of Rnf8 loss on the activation of this pathway. Rnf8−/− and WT thymocytes were either left untreated or irradiated with 2 Gy of γ radiation, followed by harvesting at 2 and 5 h after treatment. Western blotting analyses indicated a significantly higher level of IR-induced phosphorylation of Chk2 and p53 (Ser15), as well as an increase in total p53 level in Rnf8−/− thymocytes compared with WT controls (Fig. 4 B).
To examine whether Rnf8 deficiency affects radiosensitivity of other cell types, we performed colony-forming assays using Rnf8−/−, Rnf8+/−, and WT BM cells either untreated or subjected to various dosages of radiation (0.5–6 Gy). Colonies were counted at day 10 and pictures of the dishes were taken (Fig. 4, C and D). After 0.5 Gy of irradiation, the total number of colonies for WT controls was, on average, 1.5-fold greater than that of Rnf8−/−. Remarkably, the fold differences between WT and Rnf8−/−colony numbers displayed an increasing trend with increased IR dosages (2 Gy, 3.3-fold difference; 4 Gy, 4.3-fold difference; 6 Gy, 13.2-fold difference). In addition, Rnf8+/− BM cells only displayed a very modest sensitivity to 4 Gy and 6 Gy of γ radiation compared with WT controls (P < 0.05), whereas they were significantly less sensitive to irradiation compared with Rnf8−/− BM cells at all doses (P < 0.02; Fig. 4 D).
To determine the effect of Rnf8 inactivation regarding radiosensitivity in vivo, cohorts of age-matched Rnf8−/− and WT littermates (n = 10 per genotype) were subjected to 8 Gy of γ radiation, followed by daily monitoring for 51 d (Fig. S5 B). Although 60% of irradiated WT mice survived and recovered from the IR treatment, only 10% of irradiated Rnf8−/− mice remained viable. Therefore, Rnf8−/− mice were significantly more sensitive to γ radiation compared with WT littermates (Kaplan Meier analysis, Log-rank test, P < 0.003). Collectively, these data indicate the important role Rnf8 plays in the response to DNA damage and that its loss increases radiosensitivity.
Rnf8 deficiency impairs CSR
IgH CSR is one of the three important mechanisms for antibody diversification in peripheral B lymphocytes (Soulas-Sprauel et al., 2007; Stavnezer et al., 2008). It is initiated by activation-induced cytidine deaminase, which induces DSBs into the repetitive switch (S) regions upstream of the heavy chain constant (C) region exons of the Ig locus. The subsequent repair is mainly accomplished by the nonhomologous end-joining repair pathway. Interestingly, CSR is reduced in B cells deficient for several DDR proteins that act upstream of Rnf8 including Atm, H2ax, and Mdc1 (Kotnis et al., 2009). Furthermore, defective CSR has also been observed with 53bp1−/− mice (Manis et al., 2004; Difilippantonio et al., 2008) and with the RIDDLE syndrome, which is associated with RNF168 mutations (Stewart et al., 2007, 2009).
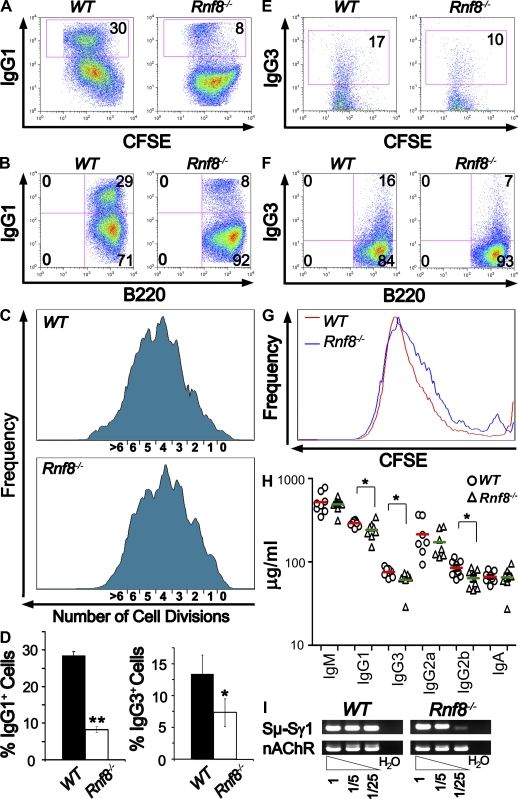
To determine whether Rnf8 plays a role in CSR, purified Rnf8−/− and WT B cells labeled with CFSE were stimulated with anti-CD40 plus IL4 for 4.5 d, and their ability to switch into IgG1 isotype was examined by FACS analysis (Fig. 5, A and B). Interestingly, the percentage of IgG1+ cells in activated Rnf8−/− B cells (8.2 ± 0.8%) was significantly reduced compared with WT cells (28.4 ± 1.1%; P < 0.0001; Fig. 5 D). These activated IgG1+ cells were B220+ and had similar cell division profiles, as shown by CFSE tracking (Fig. 5, A–C), thus ruling out a role for impaired proliferation in the reduced CSR of Rnf8−/− B cells. Similar analysis of the expression level of IgG3 on B cells stimulated with LPS for 4.5 d indicated a mild decrease in the percentage of IgG3+ B cells from Rnf8−/− mice (7.3 ± 2.2%) compared with WT controls (13.3 ± 3.0%; P = 0.048; Fig. 5, D–G). Interestingly, Rnf8+/− B cells also indicated a significantly impaired CSR to IgG1 compared with WT controls (Fig. S6, A and B; P < 0.03); however, the defect of IgG1 CSR observed with Rnf8+/− B cells was not as marked as with Rnf8−/− B cells (P < 0.02), suggesting a gene dosage effect.
Figure 5.
Impaired Igh CSR in Rnf8−/− B cells. (A and B) Flow cytometric analysis of IgG1 expression was performed on CFSE-labeled B cells 4.5 d after anti-CD40 and IL4 stimulation. Populations of IgG1+ switched cells were outlined by the boxes with percentages indicated. Results are representative of four independent experiments. (C) Cell divisions tracked by CFSE, after 4.5 d of stimulation with anti-CD40 plus IL4, were analyzed by flow cytometry. Each peak indicates a cell division. (D) Percentages of switched cells under anti-CD40 and IL4 stimulation for 4.5 d from four independent experiments (left) and percentages of switched cells under LPS stimulation for 4.5 d from three independent experiments (right). Statistical significance was established by Student’s t test. *, P < 0.05; **, P < 0.0002. Error bars represent SD. (E and F) Flow cytometric analysis of IgG3 expression was performed on CFSE-labeled B cells 4.5 d after LPS stimulation. Populations of IgG3+ switched cells were outlined by the boxes with percentages indicated. Three independent experiments were performed. (G) Cell division profiles of WT and Rnf8−/− B cells tracked by CFSE at day 4.5 after LPS stimulation. (H) Isotypes of serum Ig’s quantified in 14-mo-old Rnf8−/− (n = 8) and WT (n = 8) mice. Red horizontal bars represent the mean value of serum Ig levels of WT mice, whereas the green bars represent the mean value of serum Ig levels of Rnf8−/− mice. Student’s t test was used for statistical analysis. *, P < 0.05. (I) Sμ-Sγ1 recombination products were quantified by digestion-circularization PCR analysis. Genomic DNA was obtained from B cells stimulated for 4 d with anti-CD40 and IL4. Fivefold dilutions of the genomic DNA were used as templates in the PCR reactions. nAChR was used as the control DNA. H2O indicates no input DNA. Results are representative of three independent experiments.
To examine the effect of Rnf8 deficiency on CSR in vivo, we examined Ig levels in serums of 14-mo-old Rnf8−/− mice (n = 8) and WT littermates (n = 8). ELISA analysis indicated no significant differences in the levels of serum IgM, IgG2a, and IgA in Rnf8−/− mice compared with WT littermates (Fig. 5 H). However, serum levels of IgG1, IgG3, and IgG2b were significantly reduced in Rnf8−/− mice compared with WT littermates (P < 0.002, 0.05, and 0.04, respectively). Therefore, these data indicate that Rnf8 deficiency leads to defective CSR in vivo.
The CSR defect associated with the loss of Rnf8 was further investigated using digestion-circularization PCR assay. Genomic DNA, extracted from Rnf8−/− and WT B cells stimulated for 4.5 d in the presence of anti-CD40 plus IL4, was used in this assay to examine the efficacy of Sμ-Sγ1 recombination. The nicotinic acetylcholine receptor (nAChR) locus was used as a control. Digestion-circularization PCR assay, using serial dilutions of the genomic DNA template, confirmed the impaired CSR in Rnf8−/− B cells (Fig. 5 I) and demonstrated that the CSR defect occurs at the genomic level and is intrinsic to Rnf8−/− B cells. Overall, these data demonstrate a significant role for Rnf8 in CSR, and that its deficiency increases the risk for immunodeficiency.
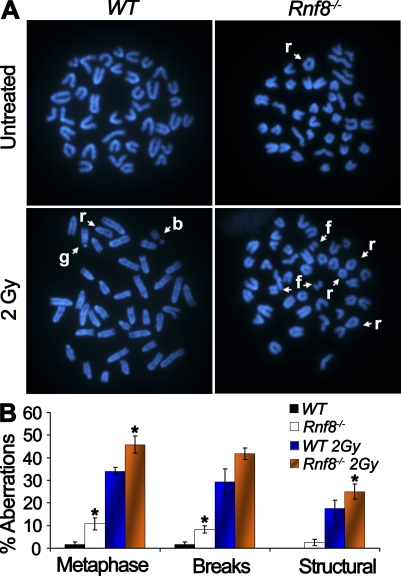
Rnf8 suppresses spontaneous and IR-induced genomic instability
Mutations in the DDR components often result in failure to signal or repair DSBs and, consequently, lead to increased genomic instability, which serves as a driving force for cancer development (Bartek et al., 2007). To examine the potential role of Rnf8 in maintaining genomic integrity, metaphase spreads of LPS-activated Rnf8−/− and WT B cells were analyzed for chromosomal aberrations (Fig. 6, A and B). Under untreated conditions, Rnf8−/− cells displayed an elevated frequency of cells harboring total chromosomal aberrations (10.8 ± 2.7%) compared with WT controls (1.7 ± 1%; P = 0.046). Furthermore, we subcategorized cells harboring chromosomal aberrations into cells harboring chromosomal breaks as well as those that exhibit structural chromosomal aberrations. Notably, the level of breaks (8.3 ± 1.7%) in untreated Rnf8−/− activated B cells was ∼4.9-fold greater than that of WT controls (1.7 ± 1%; P = 0.017). Moreover, structural chromosomal aberrations (2.5 ± 1.4%) were observed in untreated Rnf8−/− activated B cells, whereas such aberrations were completely absent in WT controls. In response to 2 Gy of radiation, the frequency of metaphases with chromosomal aberrations (45.8 ± 3.8%) in Rnf8−/− cells was significantly increased compared with WT controls (34 ± 1.7%; P = 0.048). The level of IR-induced breaks appeared to be higher in Rnf8−/− cells than WT controls, although this difference did not reach statistical significance (P = 0.22). Interestingly, the frequency of IR-induced structural chromosomal aberrations was significantly greater in Rnf8−/− cells (25 ± 3.4%) compared with WT controls (17.5 ± 3.8%; P = 0.037). Therefore, these data demonstrate that Rnf8 is required for maintaining genomic integrity, as its loss results in elevation of both spontaneous and IR-induced genomic instability.
Figure 6.
Rnf8 deficiency leads to increased genomic instability. (A) Representative metaphases of WT and Rnf8−/− LPS-activated B cells. Cells were either left untreated or irradiated with 2 Gy of γ radiation and harvested 12 h after treatment. (B) The graph shows the incidence of total spontaneous and IR-induced chromosomal aberrations. A minimum of 40 metaphase spreads of untreated or irradiated Rnf8−/− and WT cells were analyzed for each genotype and treatment in three independent experiments. Data are presented as mean ± SD. *, P < 0.05 compared with WT. r, ring; b, break; f, fragment; g, gap.
Increased cancer predisposition in Rnf8 mutant mice
The fundamental and most important role of DDR proteins is to safeguard the integrity of the genome, thus minimizing the risk for disorders including cancer. Because Rnf8−/− cells exhibit elevated levels of genomic instability, we examined the potential role of Rnf8 in cancer. Cohorts of Rnf8−/− (n = 27), Rnf8+/− (n = 35), and WT (n = 29) mice were monitored daily for survival for 465 d. Only 56% of Rnf8−/− (15 out of 27) mice were viable at the end of the monitoring period, whereas 77% of Rnf8+/− mice and 96.6% of WT littermates remained alive. Kaplan-Meier survival analysis (Fig. S7A) indicated that the life spans of Rnf8−/− and Rnf8+/− mice were significantly reduced compared with those of WT mice (Log-rank test, P < 0.0005 and 0.04, respectively).
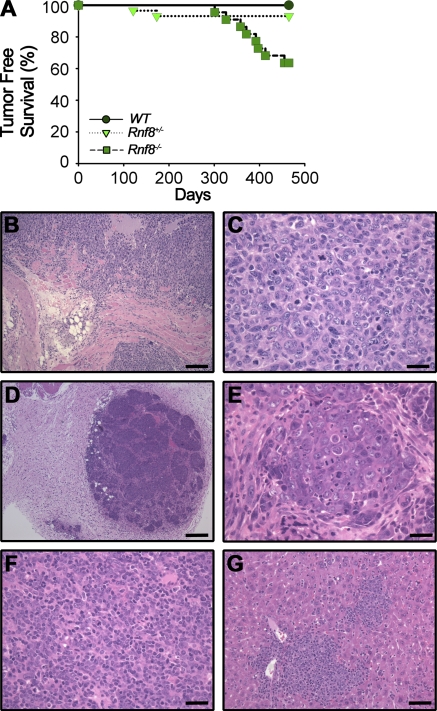
Kaplan-Meier tumor-free survival curves were established (Fig. 7 A), and biopsies of the moribund mice were examined using H&E staining in an effort to determine their cause of death (Fig. 7 B–G). 36.4% of the Rnf8−/− mice cohort developed cancer, and Kaplan-Meier tumor-free survival analysis indicated statistically significant differences of the Rnf8−/− cohort of mice from both the WT and Rnf8+/− cohorts of mice regarding cancer development (Log-rank test, P < 0.001 and 0.02, respectively). Characterization of Rnf8−/− tumors by H&E staining indicated that four were lymphomas (50%), three were thymomas (38%), one was mammary carcinoma (13%), one was a skin tumor (13%), and one was a sarcoma (13%). FACS analysis of the Rnf8−/− thymomas indicated that they were of CD4+CD8+ or CD4+ in origin, whereas the lymphomas were B220+ or B220− (Fig. S7, C and D). In addition, 6.9% of Rnf8+/− mice also developed tumors (one thymoma and one brain tumor; Fig. S7, E and F), although the difference between the Rnf8+/− and WT cohorts of mice did not reach statistical significance based on Kaplan Meier analysis (Log-rank test, P = 0.16). The tumor cells from the Rnf8 mutants were frequently found to infiltrate other organs such as liver (Fig. 7 G).
Figure 7.
Rnf8 is a novel tumor suppressor. (A) Kaplan-Meier tumor-free survival analysis for cohorts of WT (n = 28), Rnf8+/− (n = 29), and Rnf8−/− (n = 22) mice. Mice were monitored for survival for 465 d. A log-rank test was used for statistical analysis. There was a statistically significant difference between WT and Rnf8−/− curves (P < 0.0005) and between Rnf8+/− and Rnf8−/− curves (P < 0.01). (B and C) Sarcoma invading soft tissues, including skeletal muscle, adipose tissue, and a peripheral nerve, is shown (B). High magnification shows that the tumor is poorly differentiated and has a high proliferative index and atypical mitosis (C). (D) Malignant transformation of the mammary gland with a multinodular growth pattern and invasion into adjacent connective tissues. The tumor is well vascularised and shows no necrosis. (E) A higher magnification shows that tumor cells are growing in small groups, with prominent mitosis and invasion. (F) A tumor composed of small cells with round, sometimes eccentric nuclei and small cytoplasm, is compatible with a lymphoma of B cell origin. (G) Despite its well differentiated appearance, this tumor aggressively infiltrates the periportal areas of the liver. Bars: (B) 100 µm; (C, E, and F) 25 µm; (D) 250 µm; (G) 50 µm.
In addition to the Rnf8 mutant mice that developed malignancies, ∼11% of Rnf8−/− and Rnf8+/− cohorts of mice were extremely runted and displayed premature death with no sign of tumorigenesis (Fig. S7, A and B). Although the exact cause of death for these mice remains unknown, based on the impaired CSR in homozygous and heterozygous Rnf8 mutant backgrounds (Fig. 5 and Fig. S6) we speculate that immunodeficiency might account for a subset of these premature deaths. Collectively, these data demonstrate that Rnf8 is a novel tumor suppressor and that its loss also results in non–cancer-related death, likely as a result of immunodeficiency.
DISCUSSION
The fundamental roles of signaling and repair mechanisms of DSBs are best demonstrated by mutations that impair these mechanisms and that associate with several human syndromes characterized by developmental defects, neurodegenerative disorders, immunodeficiency, and increased cancer predisposition (O’Driscoll and Jeggo, 2006; Hakem, 2008; Hoeijmakers, 2009). For example, mutations of ATM lead to the human autosomal recessive disorder ataxia–telangiectasia, which is characterized by cerebellar ataxia, progressive mental retardation, neurological defects, impaired immune functions, and increased cancer risk. Furthermore, germline mutations of the BRCA1 gene, which is involved in the signaling and repair of DSBs, predispose to hereditary breast and ovarian cancer.
The E3 ligases RNF8 and RNF168 have been recently demonstrated to play critical roles in DSBs signaling. Interestingly, mutations of RNF168 have been shown to be associated with the human RIDDLE syndrome, which is characterized by features including radiosensitivity and immunodeficiency (Stewart et al., 2007, 2009). However, the in vivo roles of RNF8 and whether its inactivation results in disease remained unknown.
Our data indicate that, similar to Atm (Barlow et al., 1996), H2ax (Celeste et al., 2002), Mdc1 (Lou et al., 2006), and 53bp1 (Ward et al., 2003) but in contrast to Brca1 (Hakem et al., 1996), Rnf8 is dispensable for embryonic development. However, in line with the effect of null or hypomorphic mutations of genes involved in early DSB signaling, such as Atm, H2ax, Mdc1, 53bp1, and Brca1 (Celeste et al., 2002; Ward et al., 2003; McPherson et al., 2004; Lou et al., 2006), Rnf8−/− mice are growth retarded and display a reduced number of hematopoietic cells. Interestingly, RNF8 mediates the recruitment of RNF168 to the break sites, and the growth retardation of Rnf8−/− mice is reminiscent of the growth defects associated with RNF168 mutation in the human RIDDLE syndrome.
The sequential recruitment of proteins, which is involved in DNA damage signaling and repair as well as cell cycle checkpoint activation, to DNA break sites allows cells to efficiently repair their DSBs through elegantly orchestrated mechanisms. Knockdown experiments of RNF8 in human cell lines and MEFs deficient for Rnf8 have demonstrated the requirement for RNF8 for the recruitment and maintenance of several DDR proteins, including 53BP1, at the site of damage (Huen et al., 2007, 2008; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). Remarkably, loss of Rnf8 in activated Rnf8−/− B cells failed to completely mitigate IR-induced formation of 53bp1 foci. This finding indicates the coexistence of Rnf8-dependent and Rnf8-independent signaling pathways involved in the recruitment and retention of 53bp1, and likely other downstream DDR proteins, to IR-induced DSBs.
The Spo11-mediated generation of DSBs and their repair by homologous recombination are required for meiosis (Longhese et al., 2009). Interestingly, although no meiotic problems were observed in Rnf8−/− females, increased cell death and a reduced number to a complete absence of spermatocytes was observed in seminiferous tubules from Rnf8−/− mice. The meiotic defect of Rnf8−/− males is reminiscent of loss of the upstream DDR proteins including Atm, H2ax, and Mdc1. Interestingly, the meiotic defect observed with Rnf8−/− males contrasts with the lack of any apparent role for 53bp1 in meiosis, suggesting that the role of Rnf8 in spermatogenesis is independent of 53bp1.
Besides meiosis, programmed DSBs are also required for CSR. The generation of these DSBs within the Ig locus results in exchange of IgH constant region exons, providing an effective mechanism for resting B cells expressing low affinity IgM to switch to produce specific Ig isotypes such as IgG1 and IgA (Soulas-Sprauel et al., 2007; Stavnezer et al., 2008). This ultimately enhances the effector function of the Ig’s produced during an immune response to protect against foreign pathogens in a highly effective and specific manner. Although CSR efficiency is defective in Atm−/−, H2ax−/−, Mdc1−/−, and 53bp1−/− mice, it is the loss of 53bp1 that most significantly impairs this process (Kotnis et al., 2009). Interestingly, CSR to IgG1 in Rnf8−/− B cells is reduced by >66%, although this defect did not reach the severity of 53bp1-deficient B cells. This finding indicates that the role of 53bp1 in CSR is only partially affected by the loss of Rnf8, and this is consistent with our observation of the incomplete abolition of 53bp1 recruitment to DSBs in irradiated Rnf8−/− activated B cells, suggesting the coexistence of Rnf8-dependent and Rnf8-independent mechanisms for the recruitment and retention of 53bp1 to DSBs in activated B cells. Exactly how Rnf8 regulates CSR requires further investigation. Interestingly, the CSR defect observed in Rnf8-deficient mice indicates their immunodeficient state and is reminiscent of the immunodeficiency associated with the human RIDDLE syndrome.
A major role of DDR proteins in mammalian cells is to maintain genomic integrity. The impairment of this process increases cancer risk, as it can lead to inactivation of tumor suppressor genes or deregulation of the expression of oncoproteins. Interestingly, the increased spontaneous and IR-induced genomic instability in Rnf8-deficient cells further demonstrates the importance of Rnf8 in the signaling of DSBs and in the maintenance of genomic integrity, and it is in line with the increased radiosensitivity and IR-induced activation of the Chk2–p53 response pathway in the absence of Rnf8. The role of Rnf8 in maintaining genomic integrity is also consistent with the involvement of several other DDR proteins in this process, and it further highlights the importance of the integrity of the DSB signaling network for the maintenance of genomic integrity.
Consistent with their genomic instability, Rnf8−/− mice suffered a statistically significant increase in the incidence of cancer compared with their WT littermates, thus demonstrating that Rnf8 is a novel tumor suppressor. The increased cancer risk is associated, to a various extent, with inactivation of individual components of the DDR pathway that function upstream or downstream from Rnf8. This highlights the need for DDR proteins to function in close collaborations to prevent cancer development.
Interestingly, a gene dose effect for Rnf8 has also been identified in this study. Loss of one allele of Rnf8 reduced the level of Rnf8 protein by approximately twofold, impaired CSR in B cells, and resulted in a mild increase in radiosensitivity of BM cells but not thymocytes. Although the increased cancer predisposition of Rnf8+/− mice did not reach a statistical significance, their increased lethality further supports a gene dose effect for Rnf8. Although, no data are yet available to link RNF8 to human pathologies, the pleiotropic defects associated with Rnf8 inactivation, including increased radiosensitivity, impaired spermatogenesis, and defective CSR together with growth retardation, immunodeficiency, and increased cancer predisposition, all highlight the critical physiological functions of RNF8.
MATERIALS AND METHODS
Mice.
Two Rnf8 ES gene trap clones (RRR260 and AS0574) were obtained from MMRRC and used to generate Rnf8 chimeras. Germline transmission of the two Rnf8 mutations was successfully obtained, and Rnf8−/− mice were generated by intercrossing Rnf8+/− mice. All mice in this study were in a mixed 129/J × C57BL/6 genetic background, maintained in a specific pathogen-free environment, and genotyped by Southern blotting and PCR (Primer sequences for AS0574 clone: mutant forward 5′-TCAAAGGTTTGCCCTCTGAT-3′, mutant reverse 5′-CGGAGCGGATCTCAAACTCT-3′, WT forward 5′-TGATGACACCTGGGCATGT-3′, and WT reverse 5′-TCTTTGAGACAGCGCCTGG-3′; primer sequences for RRR260 clone: mutant forward 5′-GAGCCTGAAAGGCATCTTGG-3′, mutant reverse 5′-CCGGCTAAAACTTGAGACCTT-3′; WT forward 5′-GAGCCTGAAAGGCATCTTGG-3′, and WT reverse 5′-TGTAAGCCGCTCACTGTGCT-3′). All experiments were performed in compliance with the Ontario Cancer Institute animal care committee guidelines. Animal protocols were approved by the Animal Resource Center of Ontario Cancer Institute.
Flow cytometry.
Single-cell suspensions from BM (femurs), thymus, spleen, and lymph node of 6–10-wk-old mice were stained with monoclonal antibodies (eBioscience) against B220, IgM, IgD, Thy-1.2, TCR-β, CD4, CD8α, and CD43. FACS analyses were performed using a FACSCalibur (BD) and data were subsequently analyzed by FlowJo software (Tree Star, Inc.).
In vitro MEF proliferation assay under oxic and hypoxia conditions.
Rnf8−/− and WT MEFs were generated according to standard procedures. For the in vitro proliferation assay, 3 × 105 passage 2 MEFs were seeded onto each 6-cm dish containing culture media and then incubated at 37°C under 5% CO2. After 3 d, the cells were trypsinized, counted, and reseeded at the same density of 3 × 105 cells/6-cm dish. This process was repeated until day 15. For the hypoxia experiments, the same procedure was performed using passage 4 MEFs. Hypoxia (0.2 or 5% O2 with 5% CO2 and balanced N2) was achieved using Whitley H35 hypoxystations (Don Whitley Scientific).
In vitro activation of B cells and lymphocyte analysis.
Single-cell suspensions from spleens of 6–10-wk-old mice were prepared according to standard procedures. Splenic B cells were subsequently enriched using the Mouse B Cell Negative Isolation kit (Invitrogen). Purified B cells were labeled with 5 µM CFSE for 10 min at 37°C (Invitrogen) and then cultured at a density of 1 × 106 cells/ml in 6-well flat-bottom plates in RPMI medium supplemented with 10% FCS, 5 × 10−5 M 2-ME, and 500 ng/ml anti-CD40 (HM40-3; BD) plus 1,000 U/ml IL4 (eBioscience) or 25 µg/ml LPS alone (Sigma-Aldrich). Anti-CD40– and IL4-stimulated cells were harvested from cultures at day 4.5 to assess surface expression of IgG1, whereas B cells activated by LPS to class switch to IgG3 isotype were harvested from cultures at day 4.5. Cells were then stained, respectively, with PE-conjugated anti–mouse IgG1 (BD) or PE-conjugated anti–mouse IgG3 (SouthernBiotech) and APC-conjugated B220 (eBioscience), followed by analysis on a FACSCalibur. Data were subsequently interpreted using FlowJo software.
Immunofluorescence.
Passage 3 MEFs derived from WT and Rnf8−/− littermates were seeded onto coverslips and either left untreated or subjected to 3 Gy of γ radiation. At various time points after IR, the MEFs were fixed with 2% paraformaldehyde, permeabilized in 0.5% NP-40, and blocked in 2% BSA/1% donkey serum, followed by staining with rabbit anti-53bp1 (Bethyl Laboratories, Inc.) and mouse monoclonal anti-γH2ax (Ser139; Millipore). Donkey anti–rabbit Alexa Fluor 568 and donkey anti–mouse Alexa Fluor 488 (Invitrogen) were used, respectively for the secondary staining, followed by DAPI (Invitrogen) counterstaining. Images were taken on a microscope (IX81; Olympus) under 60× magnification. Image quantification was performed on maximum intensity projections using Image Pro Plus software (Media Cybernetics), and ImageJ software (National Institutes of Health) was used for processing the raw data. Furthermore, splenocytes from WT and Rnf8−/− mice were activated with 10 ng/µl LPS for 60 h. Cells were either left untreated or subjected to 3 Gy of γ radiation. At various time points after IR, the cells were cytospinned onto glass slides (Thermo Fisher Scientific), fixed with 2% paraformaldehyde, blocked with antibody dilution buffer (10% normal goat serum, 3% BSA, and 0.05% Triton X-100 in PBS), and then stained with rabbit anti-53bp1 and rabbit anti-γH2ax (Ser139; Millipore). Goat anti–rabbit Alexa Fluor 488 (Invitrogen) was used for the secondary staining. Subsequently, slides were counterstained with DAPI (Invitrogen) and mounted with Mowiol (Sigma-Aldrich). Images were taken on a laser confocal microscope (LSM510; Carl Zeiss, Inc.) under 63× magnification. ImageJ software was used for processing the raw data, and foci-positive cells were quantified by blind manual counting.
Digestion-circularization PCR.
1 µg of genomic DNA from cultured B cells under anti-CD40 and IL4 stimulation for 4 d was digested with EcoRI overnight, and 100 ng of the digested product was ligated with T4 DNA ligase (New England Biolabs, Inc.). Two rounds of PCR were then performed on the ligated DNA using nested primer pairs for Sµ-Sγ and nAChR. Primer sequences for the first round of PCR are as follows: Sµ-Sγ1, 5′-GAGCAGCTACCAAGGATCAGGGA-3′ and 5′-CTTCACGCCACTGACTGACTGAG-3′; and nAChR, 5′-GCAAACAGGGCTGGATGAGGCTG-3′ and 5′-GTCCCATACTTAGAACCCCAGCG-3′. Primer sequences for the second round are as follows: Sµ-Sγ1, 5′-GGAGACCAATAATCAGAGGGAAG-3′ and 5′-GAGAGCAGGGTCTCCTGGGTAGG-3′; and nAChR, 5′-GGACTGCTGTGGGTTTCACCCAG-3′ and 5′-GCCTTGCTTGCTTAAGACCCTGG-3′.
Antibody detection by ELISA.
Total IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM levels were determined in serum from eight pairs of 14-mo-old WT and Rnf8−/− littermates using Mouse Immunoglobulin Isotype Panel kits (SouthernBiotech). 96-well plates (MaxiSorp; Thermo Fisher Scientific) were precoated with 9 µg/ml of goat anti–mouse Ig capture antibody overnight and were then blocked with 1% BSA/PBS for 1 h. The various isotypes of serum Ig were diluted and captured onto the plates overnight at 4°C. Detections were made, respectively, through diluted HRP-labeled goat anti–mouse IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM, and ABTS substrate. Standard curves were generated using purified mouse IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM isotype controls (SouthernBiotech), and absorbance was measured at 405 nm.
Protein analysis and antibodies.
Western blot analysis was performed using rabbit anti-p53 (DO1; Santa Cruz Biotechnology, Inc.) and anti-phospho-p53 (S15; Cell Signaling Technology) antibodies. Rabbit anti-Chk2 antibody was raised against aa 81–95 of mouse Chk2. Rabbit polyclonal anti-RNF8 antibody was a gift from J. Chen (University of Texas, Houston, TX).
BM colony-forming assay.
BM cells from femurs of 6–10-wk-old mice were seeded on 35-mm culture dishes (1 × 105 cells/ml in MethoCult GF M3434 media; STEMCELL Technologies Inc.). Cultures were either left untreated or subjected to various doses of γ radiation and incubated at 37°C under 5% CO2. Pictures of the dishes were taken, and numbers of colonies were quantified at day 10.
In vitro and in vivo sensitivity to IR.
Thymocytes (1 × 106/ml of RPMI medium supplemented with 10% FCS and 5 × 10−5 M 2-ME) were either left untreated or subjected to various doses of γ radiation, and then incubated at 37°C under 5% CO2. After 18 h, the cells were stained with 7AAD or Annexin V and PI and then analyzed on a FACSCalibur. To assess radiosensitivity in vivo, 10 pairs of age-matched WT and Rnf8−/− littermates were subjected to IR (8 Gy). Mice were monitored daily for survival and Kaplan-Meier survival curves were established. A log-rank test was used for statistical analysis.
Chromosomal aberration analysis.
Splenocytes from mice of 6–10 wk of age were cultured in the presence of 10 µg/ml LPS for 48 h. Cells were either left untreated or subjected to 2 Gy IR. 0.1 µg/ml colcemid was added to each sample 24 h after irradiation. Cells were then harvested 3 h after colcemid treatment, followed by hypotonic lysis (0.075 M KCl, 37°C, 15 min) and fixation (methanol 3:1 acetic acid, −20°C, overnight). Fixed cells were dropped onto glass slides (Thermo Fisher Scientific) and were then stained with 0.5 mg/ml DAPI for 10 min (Sigma-Aldrich), followed by mounting with Mowiol. Chromosome number and chromosomal aberration types were examined under an epifluorescence microscope (DMIRB; Leica). Images were acquired and processed using a digital camera (DC 300RF; Leica) and Image Manager Software (Leica).
Histology.
Paraffin sections of tumor and normal tissue were stained with H&E for histological analysis as described in Pamidi et al. (2007).
Online supplemental material.
Fig. S1 describes the generation of Rnf8-deficient mice. Fig. S2 describes body weight of female Rnf8-deficient mice. Fig. S3 describes FACS analysis of hematopoietic cell populations. Fig. S4 describes the transient recruitment of 53bp1 in Rnf8−/− MEFs. Fig. S5 describes post-IR survival of Rnf8−/− mice. Fig. S6 describes a gene dose-dependent CSR defect. Fig. S7 describes Kaplan-Meier survival analysis of Rnf8-deficient mice and characterization of tumors developed by Rnf8 mutants. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092437/DC1.
Acknowledgments
The authors would like to thank P. Cheung, B. Lemmers, M. Bohgaki and members of R. Hakem’s laboratory for reviewing the manuscript and for valuable comments and suggestions. The authors also thank J. Chen for sharing his polyclonal anti-RNF8 antibody.
R. Hakem is supported by a salary award from the Canadian Institute of Health Research. This work was supported by grants from the Canadian Cancer Society Research Institute and Canadian Institutes of Health Research to R. Hakem and by the Ontario Ministry of Health and Long-Term Care.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- CSR
- class switch recombination
- DDR
- DNA damage response
- DSB
- double-strand break
- ES
- embryonic stem cell
- IgH
- Ig heavy chain
- IR
- ionizing radiation
- IRIF
- IR-induced foci
- MEF
- mouse embryonic fibroblast
- nAChR
- nicotinic acetylcholine receptor
- PTM
- posttranslational modification
- ROS
- reactive oxygen species
References
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., Wynshaw-Boris A. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 86:159–171 10.1016/S0092-8674(00)80086-0 [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas J. 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19:238–245 10.1016/j.ceb.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Bartek J., Bartkova J., Lukas J. 2007. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 26:7773–7779 10.1038/sj.onc.1210881 [DOI] [PubMed] [Google Scholar]
- Bristow R.G., Hill R.P. 2008. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer. 8:180–192 10.1038/nrc2344 [DOI] [PubMed] [Google Scholar]
- Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M.J., et al. 2002. Genomic instability in mice lacking histone H2AX. Science. 296:922–927 10.1126/science.1069398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Basu U., Zarrin A., Yan C., Franco S., Perlot T., Vuong B., Wang J., Phan R.T., Datta A., et al. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94:157–214 10.1016/S0065-2776(06)94006-1 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Gapud E., Wong N., Huang C.Y., Mahowald G., Chen H.T., Kruhlak M.J., Callen E., Livak F., Nussenzweig M.C., et al. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 456:529–533 10.1038/nature07476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Durandy A., Taubenheim N., Peron S., Fischer A. 2007. Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies. Adv. Immunol. 94:275–306 10.1016/S0065-2776(06)94009-7 [DOI] [PubMed] [Google Scholar]
- Hakem R. 2008. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 27:589–605 10.1038/emboj.2008.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R., de la Pompa J.L., Sirard C., Mo R., Woo M., Hakem A., Wakeham A., Potter J., Reitmair A., Billia F., et al. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 85:1009–1023 10.1016/S0092-8674(00)81302-1 [DOI] [PubMed] [Google Scholar]
- Harper J.W., Elledge S.J. 2007. The DNA damage response: ten years after. Mol. Cell. 28:739–745 10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2009. DNA damage, aging, and cancer. N. Engl. J. Med. 361:1475–1485 10.1056/NEJMra0804615 [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M.S., Huang J., Yuan J., Yamamoto M., Akira S., Ashley C., Xiao W., Chen J. 2008. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol. Cell. Biol. 28:6104–6112 10.1128/MCB.00987-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotnis A., Du L., Liu C., Popov S.W., Pan-Hammarström Q. 2009. Non-homologous end joining in class switch recombination: the beginning of the end. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:653–665 10.1098/rstb.2008.0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese M.P., Bonetti D., Guerini I., Manfrini N., Clerici M. 2009. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst.). 8:1127–1138 10.1016/j.dnarep.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M.A., Celeste A., Manis J.P., van Deursen J., Nussenzweig A., Paull T.T., et al. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 21:187–200 10.1016/j.molcel.2005.11.025 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Manis J.P., Morales J.C., Xia Z., Kutok J.L., Alt F.W., Carpenter P.B. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481–487 10.1038/ni1067 [DOI] [PubMed] [Google Scholar]
- McPherson J.P., Lemmers B., Hirao A., Hakem A., Abraham J., Migon E., Matysiak-Zablocki E., Tamblyn L., Sanchez-Sweatman O., Khokha R., et al. 2004. Collaboration of Brca1 and Chk2 in tumorigenesis. Genes Dev. 18:1144–1153 10.1101/gad.1192704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D.W. 2009. Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer. 9:714–723 [DOI] [PubMed] [Google Scholar]
- O’Driscoll M., Jeggo P.A. 2006. The role of double-strand break repair - insights from human genetics. Nat. Rev. Genet. 7:45–54 10.1038/nrg1746 [DOI] [PubMed] [Google Scholar]
- Pamidi A., Cardoso R., Hakem A., Matysiak-Zablocki E., Poonepalli A., Tamblyn L., Perez-Ordonez B., Hande M.P., Sanchez O., Hakem R. 2007. Functional interplay of p53 and Mus81 in DNA damage responses and cancer. Cancer Res. 67:8527–8535 10.1158/0008-5472.CAN-07-1161 [DOI] [PubMed] [Google Scholar]
- Panier S., Durocher D. 2009. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst.). 8:436–443 10.1016/j.dnarep.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Pinato S., Scandiuzzi C., Arnaudo N., Citterio E., Gaudino G., Penengo L. 2009. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol. 10:55 10.1186/1471-2199-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert I., Dantzer F., Reina-San-Martin B. 2009. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J. Exp. Med. 206:1047–1056 10.1084/jem.20082468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulas-Sprauel P., Rivera-Munoz P., Malivert L., Le Guyader G., Abramowski V., Revy P., de Villartay J.P. 2007. V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene. 26:7780–7791 10.1038/sj.onc.1210875 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., Schrader C.E. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Stankovic T., Byrd P.J., Wechsler T., Miller E.S., Huissoon A., Drayson M.T., West S.C., Elledge S.J., Taylor A.M. 2007. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc. Natl. Acad. Sci. USA. 104:16910–16915 10.1073/pnas.0708408104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136:420–434 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Su T.T. 2006. Cellular responses to DNA dazmage: one signal, multiple choices. Annu. Rev. Genet. 40:187–208 10.1146/annurev.genet.40.110405.090428 [DOI] [PubMed] [Google Scholar]
- Wang B., Elledge S.J. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA. 104:20759–20763 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Minn K., van Deursen J., Chen J. 2003. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556–2563 10.1128/MCB.23.7.2556-2563.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.L., Chen J. 2008. DNA-damage checkpoints: location, location, location. Trends Cell Biol. 18:451–455 10.1016/j.tcb.2008.07.006 [DOI] [PubMed] [Google Scholar]
- Wu J., Huen M.S., Lu L.Y., Ye L., Dou Y., Ljungman M., Chen J., Yu X. 2009. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol. Cell. Biol. 29:849–860 10.1128/MCB.01302-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482 10.1038/nature06020 [DOI] [PubMed] [Google Scholar]