Abstract
Accumulating evidence suggests that the regulation of gene expression by histone lysine methylation is crucial for several biological processes. The histone lysine methyltransferase G9a is responsible for the majority of dimethylation of histone H3 at lysine 9 (H3K9me2) and is required for the efficient repression of developmentally regulated genes during embryonic stem cell differentiation. However, whether G9a plays a similar role in adult cells is still unclear. We identify a critical role for G9a in CD4+ T helper (Th) cell differentiation and function. G9a-deficient Th cells are specifically impaired in their induction of Th2 lineage-specific cytokines IL-4, IL-5, and IL-13 and fail to protect against infection with the intestinal helminth Trichuris muris. Furthermore, G9a-deficient Th cells are characterised by the increased expression of IL-17A, which is associated with a loss of H3K9me2 at the Il17a locus. Collectively, our results establish unpredicted and complex roles for G9a in regulating gene expression during lineage commitment in adult CD4+ T cells.
Cellular differentiation relies on the activation of lineage-specific genes and the concomitant repression of lineage-alternative transcription. Contemporary models of transcriptional regulation integrate the activities of classical transcription factors and coactivator proteins with enzymatic activities that influence chromatin function by modifying histone tails or methylating DNA. In particular, histone lysine methylation is considered to be crucially involved in establishing cell type–specific gene expression patterns (Jenuwein and Allis, 2001). A major repressive histone methylation mark in mammalian euchromatin is dimethylation of histone H3 at lysine 9 (H3K9me2). The methyltransferase G9a (in cooperation with its homologue G9a-like protein [GLP]) is responsible for the majority of H3K9me2, and cells that lack G9a (or GLP) display a drastic reduction in this modification (Tachibana et al., 2002). G9a and H3K9me2 play a pivotal role in embryonic development and the epigenetic silencing of developmentally regulated genes (Tachibana et al., 2002, 2005; Wen et al., 2009), as G9a-deficient mice are severely growth retarded and die around embryonic day 8.5 of gestation and G9a-deficient embryonic stem (ES) cells are characterized by a promiscuous pattern of gene expression and fail to faithfully silence pluripotency factors upon differentiation (Feldman et al., 2006; Dong et al., 2008; Epsztejn-Litman et al., 2008; Tachibana et al., 2008). Although recent studies have identified roles for G9a and GLP in postnatal neuronal function and addiction (Schaefer et al., 2009; Maze et al., 2010), the role of G9a in lineage specification of adult cells remains unknown.
Within the CD4+ Th lineage, cells can differentiate into several subsets depending on the anatomical location, cytokine milieu, and other intrinsic and extrinsic factors (Murphy and Reiner, 2002). Naive CD4+ T cells can differentiate into one of several effector lineages, including Th1, Th2, and Th17, that differ in their expression of transcription factors, secretion of cytokines, and biological functions. Th cell lineage commitment is a tightly controlled process that, when dysregulated, can result in a variety of conditions including inflammatory bowel disease, diabetes, asthma, and allergies (Jiang and Chess, 2006). Th cell lineage differentiation provides a powerful model to study the molecular mechanisms of lineage choice in adult cells, as the roles of exogenous cytokines, upstream signaling molecules, and lineage-specific transcription factors in Th cell differentiation are well documented. Recent studies have reported the presence of differential histone methylation marks on lineage-specific effector genes such as cytokines. For example, Ifng in Th1 cells and Il4 in Th2 cells are marked with activating H3K4me3 modifications, whereas silenced lineage-promiscuous genes, such as Il4 in Th1 cells and Ifng in Th2 cells, are decorated with H3K27me3, a mark associated with gene repression (Koyanagi et al., 2005; Barski et al., 2007; Wei et al., 2009). Although the pattern of H3K4me3 and H3K27me3 modifications has been mapped in Th cells, the functional significance of these marks is still unknown. The repressive mark H3K9me2 is also dynamically regulated during development and its distribution pattern has been shown to differ in lymphocytes from normal and diabetic patients (Miao et al., 2008), suggesting a role in regulation of lymphocyte function. To date, a functional role for H3K9me2 has not been described in Th cells.
In this paper, we identify a critical role for G9a in Th cell differentiation in vitro and in vivo. In the absence of G9a, CD4+ T cells failed to optimally differentiate into Th2 cells in vitro, and mice with a T cell–specific deletion of G9a failed to develop a protective Th2 cell response after infection with the helminth parasite Trichuris muris. Furthermore, expression of IL-17A was dysregulated in Th cells in the absence of G9a or in the presence of a specific G9a methyltransferase inhibitor. These results identify G9a as a critical component in the promotion of Th2 cell differentiation and the inhibition of IL-17A expression.
RESULTS AND DISCUSSION
The proposed role of G9a in repression of gene expression during embryonic development prompted us to investigate the immunological function of G9a in the lymphoid system, as lineage differentiation is a critical process in immune system function. To this end, a conditional G9a knockout strain in which exons 4–20 were flanked with loxP sites was generated (“floxed” G9a; G9afl/fl; Fig. S1 A). We crossed this conditional allele into a transgenic background harboring hematopoietic-specific Cre expression driven by the Vav1 promoter (Stadtfeld and Graf, 2005) to generate mice in which G9a is specifically deleted in the blood including T cells (termed G9a−/− mice; Fig. S1 B). Hematopoietic-specific G9a−/− mice were born at normal Mendelian ratios and displayed no overt phenotypic alteration under homeostatic conditions (unpublished data). In agreement with a previous study (Thomas et al., 2008), G9a is not required for T cell development or homeostasis, as the frequencies and numbers of CD4+ and CD8+ T cells were equivalent in the thymus and spleen of both G9afl/fl and G9a−/− mice (Fig. S2 A and not depicted). Moreover, we found that bone marrow–derived dendritic cells from both G9afl/fl and G9a−/− mice were equally able to promote antigen-specific proliferation of Th cells (Fig. S2 B) and displayed normal inflammatory responses to a variety of Toll-like receptor agonists (Fig. S2 C). Collectively, these findings demonstrate that G9a expression is not required for T cell immune homeostasis or antigen-presenting cell function.
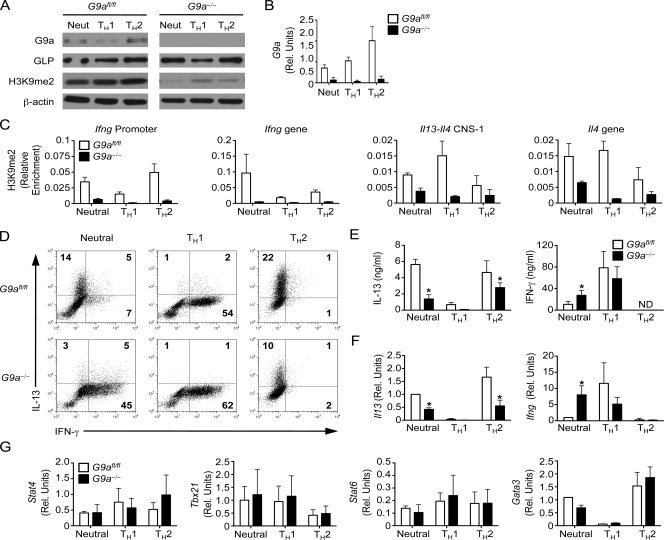
To investigate a potential role for G9a in Th cell differentiation, CD4+ T cells were purified from the spleen and LNs of control G9afl/fl and G9a−/− mice and stimulated in vitro with monoclonal antibodies against CD3 and CD28 under neutral, Th1-promoting, and Th2-promoting conditions. Although G9a was detectable in Th cells isolated from G9afl/fl mice under all conditions tested (Fig. 1 A), its expression level was increased in Th2 cells at the protein (Fig. 1 A) and mRNA level (Fig. 1 B), suggesting a more pronounced requirement for G9a in this lineage. Consistent with previous studies demonstrating that GLP does not compensate for G9a deficiency (Tachibana et al., 2005), Th cells from G9a−/− mice displayed normal levels of GLP but a global decrease in H3K9me2 levels (Fig. 1 A). We next directly examined the levels of H3K9me2 external to and within canonical Th1 and Th2 genetic loci by chromatin immunoprecipitation (ChIP). Consistent with the global reduction in H3K9me2 (Fig. 1 A), G9a deficiency results in decreased levels of H3K9me2 at the Ifng and Il13-Il4 loci (Fig. 1 C). Although we did not observe a high level of enrichment of H3K9me2 at the Ifng and Il13-Il4 loci, H3K9me2 levels were dynamically regulated during Th cell differentiation, with decreased H3K9me2 at lineage-specific loci (i.e., Ifng in Th1 cells and Il13-Il4 in Th2 cells) and increased H3K9me2 at lineage-promiscuous loci, which is consistent with a role for G9a and H3K9me2 in gene repression (Fig. 1 C). Thus, G9a-dependent H3K9me2 modifications are associated with lineage-promiscuous gene silencing and may regulate Th cell differentiation.
Figure 1.
G9a is a critical component of Th cell differentiation. CD4+ Th cells isolated from G9afl/fl and G9a−/− mice were stimulated for 6 d under neutral, Th1, and Th2 conditions and analyzed by immunoblotting for G9a, GLP, H3K9me2, and β-actin (A), qPCR for mRNA levels of G9a (B), ChIP for H3K9me2 (C), intracellular cytokine staining for IL-13 and IFN-γ production by CD4+ T cells by flow cytometry (D), ELISA for IL-13 and IFN-γ (E), qPCR for mRNA levels of Il13 and Ifng (F), and qPCR for expression of Stat4, Tbx21, Stat6, and Gata3 (G). Results are representative of six independent experiments. Rel. units, relative units. ND, not detected. Asterisks indicate P < 0.05. Error bars indicate SEM.
Based on the pattern of H3K9me2 in Th cells, we hypothesized that a lack of G9a-dependent H3K9me2 would result in lineage-inappropriate gene expression in differentiating Th cells. Under neutral conditions, Th cells from G9a−/− mice produced significantly higher levels of IFN-γ but decreased amounts of IL-13, IL-4, and IL-5 as determined by intracellular cytokine staining (Fig. 1 D), ELISA (Fig. 1 E and Fig. S3), and quantitative real-time PCR (qPCR, Fig. 1 F). However, although we observed equivalent production of IFN-γ when Th cells from G9afl/fl and G9a−/− mice were polarized toward a Th1 phenotype, we did not observe any inappropriate expression of IL-4, IL-5, or IL-13 (Fig. 1, D–F and Fig. S3). Furthermore, under Th2-promoting conditions, Th cells from G9a−/− mice did not express IFN-γ inappropriately, suggesting that G9a is not required for gene repression in Th cells. In contrast, G9a-deficient Th cells failed to maximally produce type 2 cytokines after stimulation under neutral and Th2-polarizing conditions (Fig. 1, D–F), suggesting that unlike in ES cells, G9a may act as a specific and positive regulator of gene expression in Th cells. Thus, these results demonstrate that in contrast to its role in embryonic development, G9a is not required for lineage-alternative gene silencing in Th1 or Th2 cells but has a direct or indirect role in inducing the Th2-specific cytokines IL-4, IL-5, and IL-13. We reasoned that the impairment in cytokine induction in the absence of G9a could potentially be attributed to altered levels of Th1-associated factors Stat4 and Tbx21 or Th2-associated factors Stat6 and Gata3 (Murphy and Reiner, 2002). We therefore analyzed the expression of these factors by qPCR but observed no difference in expression of any of these genes between Th cells isolated from G9afl/fl or G9a−/− mice under neutral, Th1, or Th2 conditions (Fig. 1 G). Thus, although G9a expression in Th1 and Th2 cells is dispensable for silencing of lineage-promiscuous gene expression and induction of canonical master lineage-specific transcription factors, it is critical for the regulated expression of lineage-specific Th2-associated cytokine genes.
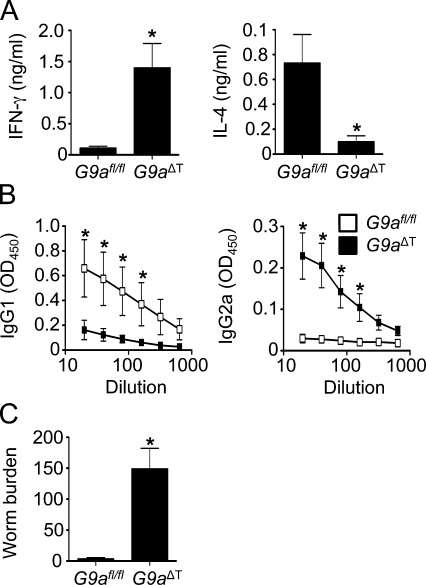
To determine whether G9a also controlled Th2 cell development in vivo, we used the T. muris infection model. T. muris–specific Th2 cells that produce the cytokines IL-4 and IL-13 are required to mediate resistance to infection, whereas IFN-γ–producing Th1 cells are associated with chronic infection (Cliffe and Grencis, 2004). Although we could not detect any difference in antigen-presenting cell function in the G9a−/− mice, and purified CD4+ T cells were defective in Th2 cell differentiation in vitro, we could not rule out a non–cell-autonomous role for G9a in regulating Th2 cell responses in vivo. To directly assess whether T cell–intrinsic expression of G9a was required for the development of protective Th2 cell responses in vivo, G9afl/fl mice were crossed with Cd4-Cre mice (Lee et al., 2001) to generate T cell–specific G9a-deficient mice (G9aΔT mice). Similar to hematopoietic-specific G9a-deficient mice (Fig. S2 A), naive G9aΔT mice also had normal frequencies of CD4+ and CD8+ T cells in the thymus and spleen (unpublished results). After infection, control G9afl/fl mice developed polarized T. muris–specific Th2 responses characterized by increased production of IL-4 after antigen restimulation of draining mesenteric LN (mLN) cells (Fig. 2 A). In contrast, mLN cells isolated from infected G9aΔT mice exhibited reduced expression of protective CD4+ T cell–derived IL-4 and increased IFN-γ expression (Fig. 2, A and B). Other parameters of Th2 cell–dependent immunity were also affected, with decreased levels of T. muris–specific serum IgG1 (Fig. 2 B, left) and increased levels of IgG2a (Fig. 2 B, right). Critically, G9aΔT mice were susceptible to infection, failing to clear parasites from the GI tract (Fig. 2 C). Furthermore, Cre toxicity was not observed, as Cd4-Cre+ G9afl/+ mice had equal numbers of CD4 and CD8 cells in the thymus, spleen, and LNs and were equally as resistant to T. muris infection as Cd4-Cre− G9afl/+ mice (Fig. S4). In summary, these results demonstrate that T cell–intrinsic expression of G9a is a critical regulator of Th2 responses in vivo.
Figure 2.
T cell–intrinsic requirement for G9a in immunity to T. muris. G9afl/fl and G9aΔT mice were infected with T. muris and sacrificed 21 d later. (A) mLN cells were restimulated with T. muris antigen for 72 h and analyzed by ELISA for production of IFN-γ and IL-4. (B) T. muris–specific IgG1 and IgG2a levels in the serum of infected G9afl/fl and G9aΔT mice were determined by ELISA. (C) Worm burdens (number of worms per animal) were determined microscopically. Data are representative of two independent experiments (n = 10). Asterisks indicate P < 0.05. Error bars indicate SEM.
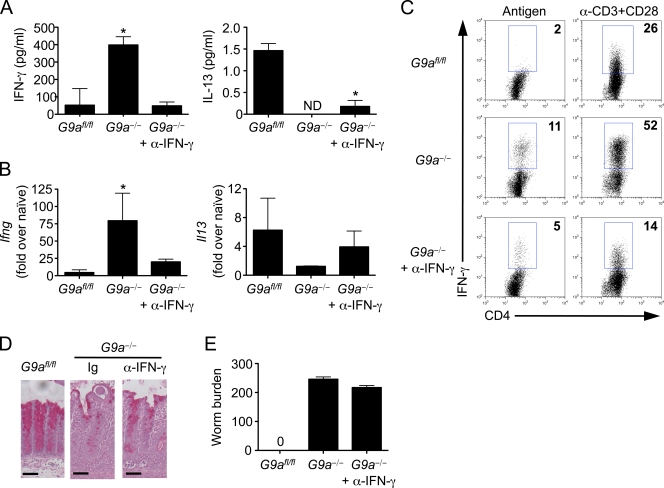
Because elevated levels of IFN-γ after T. muris infection can confer susceptibility to the disease (Else et al., 1994), we administered a monoclonal antibody against IFN-γ to T. muris–infected G9a−/− mice. Antibody treatment resulted in decreased production of IFN-γ after restimulation of cells isolated from mLN (Fig. 3, A and C) and lower expression of Ifng mRNA in the gut (Fig. 3 B). However, levels of IL-13 in the mLN and intestine were not significantly increased after blockade of IFN-γ in G9a−/− mice (Fig. 3, A and B). Accordingly, antibody treatment failed to recover goblet cell hyperplasia (Fig. 3 D) and resistance to infection (Fig. 3 E). Collectively, these findings demonstrate that in G9a−/− mice, dysregulated IFN-γ production is not responsible for susceptibility to infection. In addition, the inability of G9a-deficient Th cells to fully up-regulate type 2 cytokine expression in the absence of IFN-γ suggests a critical cell-autonomous role for G9a in promoting expression of Th2 cell–associated cytokines.
Figure 3.
G9a is required for the development of protective Th2 cell responses in the absence of IFN-γ. G9afl/fl, G9a−/−, and G9a−/− mice treated with α–IFN-γ were infected with T. muris and sacrificed 21 d later. (A) mLN cells were restimulated with T. muris antigen for 72 h and analyzed by ELISA for production of IFN-γ and IL-13. (B) Ifng and Il13 gene expression in large intestinal tissue was analyzed by qPCR. (C) mLN cells were restimulated with T. muris antigen or monoclonal antibodies against CD3 and CD28 (α-CD3/CD28) for 72 h and analyzed by intracellular cytokine staining for production of IFN-γ. (D) Goblet cell hyperplasia and intestinal architecture was analyzed by staining cecal sections with periodic acid–Schiff’s. Bars, 50 µm. (E) Worm burdens were determined microscopically. Data are representative of three independent experiments (n = 12). ND, not detected. Asterisks indicate P < 0.05. Error bars indicate SEM.
We next sought to address how G9a was required to promote expression of Th2 cytokines. One possible mechanism was that G9a was acting to silence expression of a repressive molecule that could bind to the Il13–Il4 locus. Runx3 and Mina are two DNA-binding proteins that inhibit the expression of Th2 cytokines in Th cells (Naoe et al., 2007; Lee et al., 2009; Okamoto et al., 2009). Thus, G9a could be required to silence expression of Runx3 or Mina to allow Th2 cell development. However, we observed no difference in expression of Runx3 or Mina mRNA between G9afl/fl and G9a−/− Th cells after in vitro stimulation (Fig. S5 A). These results demonstrate that G9a is not required for repression of known repressors of the Il13–Il4 locus and suggests an alternative function in promoting Th2 cell development.
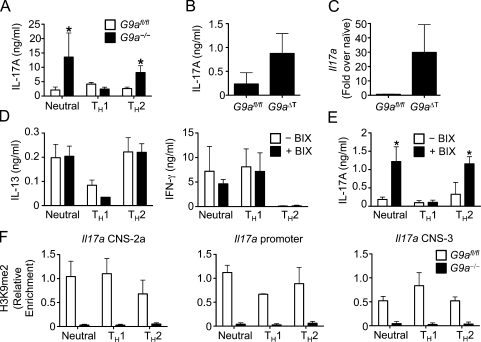
The recruitment of chromatin-modifying enzymes by DNA binding transcription factors aids in determining their target gene specificity. G9a has been shown to interact with several zinc finger proteins to regulate transcription. One of these factors, growth factor independent 1 (Gfi1), is also involved in Th cell differentiation and activation by interacting with Gata3 and preventing its proteasomal degradation (Zhu et al., 2002; Duan et al., 2005; Shinnakasu et al., 2008). However, we could not detect any alterations in the protein levels of either Gfi1 or Gata3 between Th cells from control G9afl/fl and G9a−/− mice (Fig. S5 B), indicating that G9a regulates Th2 cell differentiation independently of Gfi1 and Gata3 levels. A recent study has also identified a role for Gfi1 in repressing the Th17 cell lineage (Zhu et al., 2009), as Gfi1-deficient Th2 cells can be stimulated to produce IL-17A. Strikingly, similar to Gfi1-deficient T cells, loss of G9a had a dramatic effect on lineage-inappropriate expression of IL-17A, with increased expression and secretion of IL-17A by G9a-deficient CD4+ T cells stimulated under neutral and Th2 conditions (Fig. 4 A). This increased IL-17A production in the absence of T cell–intrinsic G9a was also observed in vivo after T. muris infection, as infected G9aΔT mice expressed higher levels of IL-17A in the mLN (Fig. 4 B) and Il17a mRNA in the intestine (Fig. 4 C). Thus, these results suggest that although G9a–Gfi1 interactions are not required for Gata3 stability and Th2 cell development, they may be critical for Il17a gene silencing in vivo.
Figure 4.
The methyltransferase activity of G9a is required for silencing of IL-17A. (A) CD4+ Th cells isolated from G9afl/fl and G9a−/− mice were stimulated for 6 d under neutral, Th1, or Th2 conditions and analyzed for IL-17A production by ELISA. (B and C) Expression of IL-17A was determined by ELISA of supernatants from T. muris antigen-restimulated mLN cultures (B) and qPCR of mRNA isolated from intestinal tissue (C). (D and E) CD4+ Th cells isolated from G9afl/fl and G9a−/− mice were stimulated for 6 d under neutral, Th1, and Th2 conditions in the presence or absence of 500 nM BIX-01294 (BIX) and analyzed by ELISA for expression of IL-13 and IFN-γ (D) or IL-17A (E). (F) CD4+ Th cells isolated from G9afl/fl and G9a−/− mice were stimulated for 6 d under neutral, Th1, and Th2 conditions and analyzed by ChIP for H3K9me2. Data in A–E are representative of three independent experiments and data in F is from two independent experiments. Asterisks indicate P < 0.05. Error bars indicate SEM.
Our findings demonstrate that G9a has multiple functions in Th cell differentiation, including transcriptional activation and of its target genes. Indeed, such dual roles for G9a have been proposed for β-globin gene expression in erythroid cells (Chaturvedi et al., 2009) and in the control of androgen receptor transcriptional activity (Lee et al., 2006). These studies also suggested that the ability of G9a to activate transcription is independent of its enzymatic activity. To directly test whether the activating function of G9a on Th2 cytokine expression is independent of its methyltransferase activity, CD4+ T cells isolated from wild-type mice were stimulated under neutral, Th1, or Th2 conditions in the absence or presence of the specific G9a methyltransferase inhibitor BIX-01294 (Kubicek et al., 2007). Importantly, inhibition of G9a activity had no effect on production of IL-13 or IFN-γ under either neutral, Th1, or Th2-promoting conditions (Fig. 4 D). Strikingly however, treatment with BIX-01294 resulted in increased expression of IL-17A under both neutral and Th2-polarizing conditions (Fig. 4 E). Thus, repression of IL-17A production is dependent on the methyltransferase activity of G9a, whereas repression of IFN-γ and induction of IL-4 and IL-13 requires G9a but not its known catalytic activity. Consistent with these results, H3K9me2 levels are highly enriched at the Il17a locus in Th cells stimulated under neutral, Th1, and Th2 conditions and that G9a is required for this modification (Fig. 4 F). The levels of H3K9me2 at the Il17a locus are 10-fold higher than the Ifng locus and 100-fold higher than the Il13-Il4 loci (Fig. 1 C), suggesting that G9a-dependent methylation is critical for repression of lineage-alternative expression of IL-17A but is dispensable for silencing of IFN-γ, IL-4, and IL-13. Collectively, we conclude that G9a exhibits activating and inactivating functions in gene regulation in vivo that can be discriminated by their dependence on its catalytic activity.
In summary, our results indicate a complex and unexpected role for G9a in the transcriptional orchestration of T helper cell fates and functions. Although canonical models of G9a-mediated chromatin regulation have almost exclusively focused on its repressive mode of action exerted by its H3K9 methylating activity, we provide the first in vivo evidence for both activating and inactivating roles of G9a in the specification of developmentally regulated gene expression programs. G9a-mediated H3K9me2 is one of the most abundant histone modifications in mammalian euchromatin. Genome-wide analysis of H3K9me2 modifications in pluripotent and differentiated cells has identified G9a-dependent large organized chromatin K9 modifications (termed LOCKs) that increase in size and number as cells become more specialized (Wen et al., 2009). These results support classical models of lineage differentiation that hypothesize the stepwise inactivation of the genome to prevent lineage-promiscuous gene expression. However, a striking observation from our results is that silencing of many lineage-promiscuous genes in Th cells is independent of G9a and H3K9me2, suggesting that in adult cells, G9a-dependent H3K9me2 modifications are not a general mechanism of gene repression but display a high degree of specificity. Although the molecular mechanisms controlling this specificity are unclear, genome-wide analysis of both H3K9me2 modifications and gene expression by ChIP sequencing and microarray will potentially identify genes—in addition to Il17a which is identified here—that are specifically regulated by G9a and H3K9me2.
Our results also suggest a role for G9a in gene activation during lineage commitment that is independent of methyltransferase activity. In Th cells, the physical conformation of the Ifng and Il13-Il4 loci plays a critical role in cytokine gene expression (Spilianakis et al., 2005; Cai et al., 2006). SATB1 (Special AT-rich sequence binding protein) is induced in Th2 cells and organizes the Il13-Il4 locus into loops allowing for the optimal induction of type 2 cytokine expression (Cai et al., 2006). SATB1 interacts with the homeodomain protein CCAAT displacement protein/cut homologue (Liu et al., 1999), which has been shown to interact directly with G9a (Nishio and Walsh, 2004). Thus, G9a may play an activating role in Th cell differentiation by directly influencing chromatin structure. In agreement with this, a recent study has demonstrated that G9a specifically regulates genes at the nuclear periphery (Yokochi et al., 2009), a site of inducible gene expression which links transcription to mRNA export (Rodríguez-Navarro, 2009). Therefore, in addition to established pathways involved in Th cell differentiation, including regulated transcription factor expression, histone acetylation, and DNA methylation, results presented in this paper support a critical role for G9a in the development of Th responses in vitro and in vivo and identify a dual role for G9a in cellular lineage development.
MATERIALS AND METHODS
Mice.
Vav-Cre mice were obtained from T. Graf (Centre for Genomic Regulation, Barcelona, Spain). CD4-Cre mice were obtained from Taconic. OT-II mice were obtained from The Jackson Laboratory. The conditional G9a allele was generated by gene targeting in C57BL/6 C2 ES cells (from A. Nagy, University of Toronto, Toronto, Canada). A loxP site was inserted into a BamHI site upstream of exon 4 and an frt-flanked neomycin selection cassette with a second loxP site was inserted into an NdeI site downstream of exon 20. The resulting Cre-mediated excision of the conditionally targeted locus results in the deletion of exons 4–20, coding for most of the protein, including the central ankyrin repeats, and causes a frameshift within all exons downstream of exon 20. Further breeding to Actb-Flpe transgenic mice (The Jackson Laboratory) eliminated the Neo cassette and generated a functional floxed G9a allele in the subsequent generation. Animals were maintained on a pure C57BL/6 background in a specific pathogen-free environment and tested negative for pathogens in routine screening. All experiments were performed at the University of British Columbia according to institutional and Canadian Council on Animal Care guidelines approved by the UBC Committee on Animal Care.
Parasites and infections.
Isolation of T. muris excretory-secretory antigen and eggs was performed as previously described (Zaph et al., 2007). Mice were infected on day 0 with 150–200 embryonated eggs, and parasite burdens were assessed on day 21 after infection.
Cell culture.
CD4+ T cells were isolated from spleen and LNs by negative selection using RoboSep (STEMCELL Technologies Inc.). Purities were routinely 90–95% CD4+ cells. 0.5–1 × 106 CD4+ cells were cultured for 6 d in DME supplemented with 10% heat-inactivated FBS, 2 mM glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 25 mM Hepes, and 5 × 10−5 M 2-ME with 1 µg/ml each of plate-bound anti-CD3 (145-2C11) and CD28 (37.51; eBioscience) in the presence of 10 ng/ml IL-2 alone (neutral conditions) or under Th1-promoting (10 ng/ml IL-2 and IL-12 [eBioscience] and 5 µg/ml anti-IL-4 [11B11]) or Th2-promoting (10 ng/ml IL-2 and IL-4 and 5 µg/ml of anti–IFN-γ [XMG6]) conditions. For infected animals, the mLN was harvested and cells were plated in medium alone or in the presence 50 µg/ml of T. muris ES antigen or 1 µg/ml of soluble antibodies against CD3 and CD28. Bone marrow–derived dendritic cells were generated as previously described (Zaph et al., 2007) and stimulated with 500 ng/ml LPS (Sigma-Aldrich), 20 µg/ml poly(I:C) (Sigma-Aldrich), and 10 µg/ml peptidoglycan (from Staphylococcus aureus; Sigma-Aldrich) for 24 h. In some experiments, dendritic cells were pulsed with 100 µg/ml ovalbumin (Worthington Biochemical Corporation) in the presence of 10 ng/ml LPS for 24 h, followed by addition of CFSE (Invitrogen)-labeled OT-II CD4+ T cells (10:1 T cell/DC ratio) for 4 d.
Cytokine analysis and flow cytometry.
Cell-free supernatants were harvested and analyzed for cytokine secretion by sandwich ELISA for IL-4, IL-5, IL-13, IFN-γ, and IL-17A (eBioscience). Intracellular cytokine staining for IL-13 and IFN-γ was performed after stimulation for 5 h with 50 ng/ml PMA and 750 ng/ml ionomycin in the presence of 10 µg/ml brefeldin A (all from Sigma-Aldrich). Intestinal tissue was homogenized using a TissueLyser (QIAGEN), RNA was isolated using RNeasy spin columns (QIAGEN) and reverse transcribed, and gene expression was analyzed by real-time PCR using QuantiTect primers (QIAGEN) with SYBR green chemistry (Fermentas) on a real-time PCR system (7900 Fast; Applied Biosystems).
ChIP analysis.
ChIP for H3K9me2 was conducted as previously described (Dong et al., 2008) using 2.5 µg of antibody specific for H3K9me2 (ab1220; Abcam). After washing, elution, and reversion of cross-links, the DNA was isolated and used in qPCR reactions on a real-time PCR system (ABI 7900 Fast). Data are presented as the ratio of immunoprecipitated to input CT values. The primers used for PCR analysis were the following: Ifng promoter forward, 5′-CATAGCCACTGTGTAATGTTAATG-3′ and reverse, 5′-TTCTCTGTCTTTGGTAACTTATCT-3′; Ifng gene forward, 5′-TCAGCAACAACATAAGCGTC-3′ and reverse, 5′-ACCTCAAACTTGGCAATACTCA-3′; Il13-Il4 CNS-1 forward, 5′-TGATTTCTCGGCAGCCAGG-3′ and reverse, 5′-TGCGTCACCTCTGACCACAC-3′; Il4 gene, forward, 5′-GGGTGTGAATAAGCCATATTG-3′ and reverse, 5′-CCCAGCGTTTACATGAGC-3′; Il17a CNS-2a forward, 5′-CAGCGTGTGGTTTGGTTTAC-3′ and reverse, 5′-CTAGGTGGGTTCCTCACTGG-3′; Il17a promoter forward, 5′-GCAGCAGCTTCAGATATGTCC-3′ and reverse, 5′-TGAGGTCAGCACAGAACCAC-3′; and Il17a CNS-3 forward, 5′-TTAGTGAGGTCGGGGAAGTG-3′ and reverse, 5′-TTTGATGGCAGCACATTCAT-3′.
In vivo antibody treatment.
Monoclonal antibody against IFN-γ (XMG-6) was purchased from Bio X Cell. Mice were treated intraperitoneally with 1 mg of antibody every 4 d between day 4 and 20 after infection.
Goblet cell responses.
Segments of cecum were removed and fixed in 4% paraformaldehyde. For detection of intestinal goblet cells, 5-µm paraffin-embedded sections were cut and stained with periodic acid Schiff’s reagent.
Statistics.
Results represent the mean ± SEM. Statistical significance was determined by Student’s t test (between two groups or conditions) or analysis of variance with a post-hoc test (three or more groups or conditions) using Prism 4.0 (GraphPad Software, Inc.).
Online supplemental material.
The knockout construct and deletion of G9a is shown in Fig. S1. Fig. S2 depicts normal T cell development and antigen-presenting cell function in G9a−/− mice. Fig. S3 shows decreased IL-4 and IL-5 production by G9a−/− Th cells. Fig. S4 shows normal expression of Runx3, Mina, Gfi1, and Gata3 in G9afl/fl and G9a−/− Th cells. Fig. S5 shows that G9a is not required for repression of Runx3 and Mina or Gfi1-dependent Gata3 stability. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100363/DC1.
Acknowledgments
The authors thank C.A. Hunter and M.C. Lorincz for critical reading of the manuscript.
This work was supported by the Canadian Institutes of Health Research (MOP-183792 to C.Z. and MOP-53332 to F.M.V. Rossi) and Canada Foundation for Innovation (to C. Zaph). B. Lehnertz is funded by a doctoral fellowship of the German Academic Exchange Service (DAAD). C. Zaph is a CIHR New Investigator. F.M.V. Rossi is the Canada Research Chair in Regenerative Medicine.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- ES
- embryonic stem
- GLP
- G9a-like protein
- mLN
- mesenteric LN
- qPCR
- quantitative real-time PCR
References
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell. 129:823–837 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Cai S., Lee C.C., Kohwi-Shigematsu T. 2006. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38:1278–1288 10.1038/ng1913 [DOI] [PubMed] [Google Scholar]
- Chaturvedi C.P., Hosey A.M., Palii C., Perez-Iratxeta C., Nakatani Y., Ranish J.A., Dilworth F.J., Brand M. 2009. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc. Natl. Acad. Sci. USA. 106:18303–18308 10.1073/pnas.0906769106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe L.J., Grencis R.K. 2004. The Trichuris muris system: a paradigm of resistance and susceptibility to intestinal nematode infection. Adv. Parasitol. 57:255–307 10.1016/S0065-308X(04)57004-5 [DOI] [PubMed] [Google Scholar]
- Dong K.B., Maksakova I.A., Mohn F., Leung D., Appanah R., Lee S., Yang H.W., Lam L.L., Mager D.L., Schübeler D., et al. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 27:2691–2701 10.1038/emboj.2008.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Zarebski A., Montoya-Durango D., Grimes H.L., Horwitz M. 2005. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 25:10338–10351 10.1128/MCB.25.23.10338-10351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else K.J., Finkelman F.D., Maliszewski C.R., Grencis R.K. 1994. Cytokine-mediated regulation of chronic intestinal helminth infection. J. Exp. Med. 179:347–351 10.1084/jem.179.1.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S., Feldman N., Abu-Remaileh M., Shufaro Y., Gerson A., Ueda J., Deplus R., Fuks F., Shinkai Y., Cedar H., Bergman Y. 2008. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 15:1176–1183 10.1038/nsmb.1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8:188–194 10.1038/ncb1353 [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. 2001. Translating the histone code. Science. 293:1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Jiang H., Chess L. 2006. Regulation of immune responses by T cells. N. Engl. J. Med. 354:1166–1176 10.1056/NEJMra055446 [DOI] [PubMed] [Google Scholar]
- Koyanagi M., Baguet A., Martens J., Margueron R., Jenuwein T., Bix M. 2005. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 280:31470–31477 10.1074/jbc.M504766200 [DOI] [PubMed] [Google Scholar]
- Kubicek S., O’Sullivan R.J., August E.M., Hickey E.R., Zhang Q., Teodoro M.L., Rea S., Mechtler K., Kowalski J.A., Homon C.A., et al. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell. 25:473–481 10.1016/j.molcel.2007.01.017 [DOI] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Lee D.Y., Northrop J.P., Kuo M.H., Stallcup M.R. 2006. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 281:8476–8485 10.1074/jbc.M511093200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Jeong H.M., Choi J.M., Cho Y.C., Kim T.S., Lee K.Y., Kang B.Y. 2009. Runx3 inhibits IL-4 production in T cells via physical interaction with NFAT. Biochem. Biophys. Res. Commun. 381:214–217 10.1016/j.bbrc.2009.02.026 [DOI] [PubMed] [Google Scholar]
- Liu J., Barnett A., Neufeld E.J., Dudley J.P. 1999. Homeoproteins CDP and SATB1 interact: potential for tissue-specific regulation. Mol. Cell. Biol. 19:4918–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I., Covington H.E., III, Dietz D.M., LaPlant Q., Renthal W., Russo S.J., Mechanic M., Mouzon E., Neve R.L., Haggarty S.J., et al. 2010. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 327:213–216 10.1126/science.1179438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao F., Smith D.D., Zhang L., Min A., Feng W., Natarajan R. 2008. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes. 57:3189–3198 10.2337/db08-0645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.M., Reiner S.L. 2002. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2:933–944 10.1038/nri954 [DOI] [PubMed] [Google Scholar]
- Naoe Y., Setoguchi R., Akiyama K., Muroi S., Kuroda M., Hatam F., Littman D.R., Taniuchi I. 2007. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf β binding to the Il4 silencer. J. Exp. Med. 204:1749–1755 10.1084/jem.20062456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio H., Walsh M.J. 2004. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. USA. 101:11257–11262 10.1073/pnas.0401343101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Van Stry M., Chung L., Koyanagi M., Sun X., Suzuki Y., Ohara O., Kitamura H., Hijikata A., Kubo M., Bix M. 2009. Mina, an Il4 repressor, controls T helper type 2 bias. Nat. Immunol. 10:872–879 10.1038/ni.1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro S. 2009. Insights into SAGA function during gene expression. EMBO Rep. 10:843–850 10.1038/embor.2009.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A., Sampath S.C., Intrator A., Min A., Gertler T.S., Surmeier D.J., Tarakhovsky A., Greengard P. 2009. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 64:678–691 10.1016/j.neuron.2009.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnakasu R., Yamashita M., Kuwahara M., Hosokawa H., Hasegawa A., Motohashi S., Nakayama T. 2008. Gfi1-mediated stabilization of GATA3 protein is required for Th2 cell differentiation. J. Biol. Chem. 283:28216–28225 10.1074/jbc.M804174200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., Flavell R.A. 2005. Interchromosomal associations between alternatively expressed loci. Nature. 435:637–645 10.1038/nature03574 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Graf T. 2005. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 132:203–213 10.1242/dev.01558 [DOI] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Shinkai Y. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16:1779–1791 10.1101/gad.989402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. 2005. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19:815–826 10.1101/gad.1284005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Matsumura Y., Fukuda M., Kimura H., Shinkai Y. 2008. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 27:2681–2690 10.1038/emboj.2008.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L.R., Miyashita H., Cobb R.M., Pierce S., Tachibana M., Hobeika E., Reth M., Shinkai Y., Oltz E.M. 2008. Functional analysis of histone methyltransferase g9a in B and T lymphocytes. J. Immunol. 181:485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.Y., Watford W.T., et al. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 30:155–167 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B., Wu H., Shinkai Y., Irizarry R.A., Feinberg A.P. 2009. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41:246–250 10.1038/ng.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T., Poduch K., Ryba T., Lu J., Hiratani I., Tachibana M., Shinkai Y., Gilbert D.M. 2009. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc. Natl. Acad. Sci. USA. 106:19363–19368 10.1073/pnas.0906142106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaph C., Troy A.E., Taylor B.C., Berman-Booty L.D., Guild K.J., Du Y., Yost E.A., Gruber A.D., May M.J., Greten F.R., et al. 2007. Epithelial-cell-intrinsic IKK-β expression regulates intestinal immune homeostasis. Nature. 446:552–556 10.1038/nature05590 [DOI] [PubMed] [Google Scholar]
- Zhu J., Guo L., Min B., Watson C.J., Hu-Li J., Young H.A., Tsichlis P.N., Paul W.E. 2002. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity. 16:733–744 10.1016/S1074-7613(02)00317-5 [DOI] [PubMed] [Google Scholar]
- Zhu J., Davidson T.S., Wei G., Jankovic D., Cui K., Schones D.E., Guo L., Zhao K., Shevach E.M., Paul W.E. 2009. Down-regulation of Gfi-1 expression by TGF-β is important for differentiation of Th17 and CD103+ inducible regulatory T cells. J. Exp. Med. 206:329–341 10.1084/jem.20081666 [DOI] [PMC free article] [PubMed] [Google Scholar]