Functional diversity of histone H1 variants may be caused by differences in phosphorylation during interphase.
Abstract
Histone H1 phosphorylation affects chromatin condensation and function, but little is known about how specific phosphorylations impact the function of H1 variants in higher eukaryotes. In this study, we show that specific sites in H1.2 and H1.4 of human cells are phosphorylated only during mitosis or during both mitosis and interphase. Antisera generated to individual H1.2/H1.4 interphase phosphorylations reveal that they are distributed throughout nuclei and enriched in nucleoli. Moreover, interphase phosphorylated H1.4 is enriched at active 45S preribosomal RNA gene promoters and is rapidly induced at steroid hormone response elements by hormone treatment. Our results imply that site-specific interphase H1 phosphorylation facilitates transcription by RNA polymerases I and II and has an unanticipated function in ribosome biogenesis and control of cell growth. Differences in the numbers, structure, and locations of interphase phosphorylation sites may contribute to the functional diversity of H1 variants.
Introduction
Nonallelic variants of histone H1 in metazoans share a common tripartite structure, with a conserved globular domain flanked by a short N-terminal domain and a longer C-terminal domain (CTD). FRAP analyses of cells expressing H1-GFP fusions have revealed that H1 variants bind chromatin dynamically in vivo and that both the globular domain and CTD contribute to chromatin binding (Lever et al., 2000; Misteli et al., 2000; Hendzel et al., 2004; Brown et al., 2006). H1-binding dynamics affect the chromatin access of high mobility group proteins, MeCP2 (methyl-CpG–binding protein), upstream-binding factor (UBF), the glucocorticoid receptor, and other regulators by modulating H1-mediated chromatin folding and by enabling factors to compete with H1 for chromatin-binding sites (Zlatanova et al., 2000; Phair et al., 2004; Bustin et al., 2005).
CTD interactions with linker DNA are important for higher order folding of chromatin (Allan et al., 1980, 1986; Bednar et al., 1998; Carruthers et al., 1998; Lu et al., 2009). S/TPXK/R Cdk substrate motifs that are repeated in the CTD contribute to its DNA binding (Suzuki, 1989; Vila et al., 2001; Roque et al., 2005), and phosphorylation at these motifs affects CTD–DNA interactions (Roque et al., 2008). These motifs are phosphorylated to varying degrees in H1 prepared from asynchronous or mitosis-arrested mammalian cells (Garcia et al., 2004; Sarg et al., 2006; Wisniewski et al., 2007), but how this affects chromatin processes is unclear. Analyses of synchronized cells suggest that H1 phosphorylation increases progressively during interphase before peaking transiently during mitosis (Gurley et al., 1975; Ajiro et al., 1981a,b), but few details are known about the site specificity of phosphorylation during interphase and mitosis because phosphorylation sites were not identified in these early analyses. Site-specific phosphorylation of an H1 variant during interphase has recently been described, but direct evidence of its significance is lacking (Talasz et al., 2009).
Human somatic cells express six H1 variants with distinct chromatin-binding dynamics that possess CTDs differing in length, net charge, number, and relative positions of S/TPXK/R motifs (Hendzel et al., 2004; Th’ng et al., 2005). FRAP analyses of H1 mutated to mimic dephosphorylation or phosphorylation (Contreras et al., 2003; Hendzel et al., 2004) imply that phosphorylation is likely to have variant-specific and site-specific effects on H1 function, but the paucity of data on how H1 variant phosphorylation is regulated in vivo has hindered investigating this further. To address this problem, we characterized the phosphorylation of the major H1 variants of HeLa S3 cells during interphase and mitosis and generated phosphorylation site–specific antisera to investigate the function of interphase H1 phosphorylation.
Results and discussion
The limited heterogeneity of H1 in HeLa cells
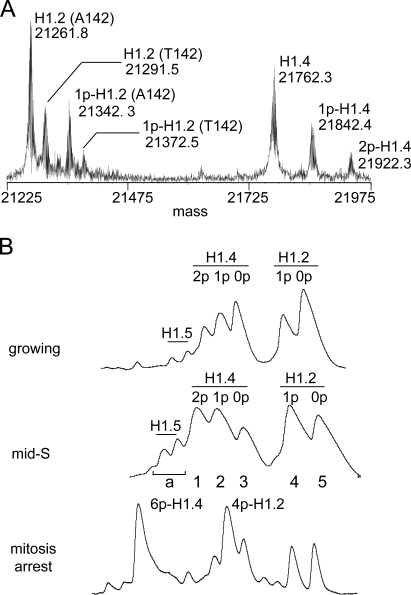
We used top-down mass spectrometry (MS [TDMS]) to analyze H1 phosphorylation because this approach facilitates characterization of multisite histone modification (Pesavento et al., 2008). The mass spectrum of crude H1 from asynchronous HeLa S3 cells was remarkably simple, containing just seven distinct H1 species (Fig. 1 A). A combination of analyses identified four of these to be unmodified and monophosphorylated allelic variants of H1.2 that are polymorphic for an Ala > Thr substitution at residue 142 (H1.2 [A142], H1.2 [T142], 1p-H1.2 [A142], and 1p-H1.2 [T142]; Fig. S1 B and Fig. S2 F). The three remaining forms correspond to unmodified, monophosphorylated, and diphosphorylated forms of H1.4 (H1.4, 1p-H1.4, and 2p-H1.4). Relative quantitation of the mass spectrum revealed that nearly a third of H1.2 and H1.4 is monophosphorylated, and roughly a sixth of H1.4 is diphosphorylated under these conditions. Small amounts of H1.5 were detected in crude H1 by TDMS, but we did not analyze these further. Additional H1 variants have been detected in HeLa cells previously (Garcia et al., 2004), but this is the first work demonstrating that HeLa S3 cells express predominantly H1.2 and H1.4 and that H1.2 is polymorphic in these cells.
Figure 1.
The limited heterogeneity of H1 in HeLa cells. (A) The mass spectrum of intact crude H1 from asynchronous growing HeLa S3 cells. Proteins were identified directly by gas phase fragmentation and MS/MS analysis. Phosphorylation levels were inferred from predicted molecular masses. The A142T polymorphism in H1.2 was confirmed by genotyping (Fig. S1 B). (B) The HIC profiles of crude H1 from asynchronous growing, mid–S phase, and mitosis-arrested HeLa S3 cells. Eluate absorbance (214 nm) is plotted relative to time for equivalent portions of each separation. H1.2, H1.4, and H1.5 represent ∼47%, 48%, and 5% of total H1 in asynchronous cells, respectively, based on chromatographic integration.
To enhance phosphorylation site identification, we used hydrophobic interaction chromatography (HIC) to enrich phosphorylated forms of H1 before TDMS. HIC resolved five major peaks for crude H1 from asynchronous cells (Fig. 1 B). These correspond to the forms resolved by TDMS except that similarly modified allelic variants of H1.2 were not separated from each other. The same HIC peaks were observed at all times sampled after cells were released from double-thymidine block synchronization (unpublished data), but the relative abundance of phosphorylated forms increased significantly as cells progressed toward mitosis. Phosphorylated H1.2 and H1.4 were more abundant in samples from mid–S phase cells (4 h after release) compared with asynchronous cells, and most H1.2 and H1.4 were tetra- and hexaphosphorylated, respectively, in cells arrested in mitosis with colchicine (Fig. 1 B). These findings are consistent with previous analyses of H1 phosphorylation stoichiometry in synchronized cells (Ajiro et al., 1981a,b; Gurley et al., 1995; Talasz et al., 1996).
H1 is phosphorylated at a specific subset of sites during interphase
We identified interphase phosphorylation sites in HIC fractions from mid–S phase cells because H1 phosphorylation was clearly increasing at this time, but the chance of contamination by hyperphosphorylated H1 from mitotic cells was less than at later time points. SDS gel electrophoresis confirmed that the five major HIC peaks for mid–S phase samples were essentially homogenous fractions of H1.2 or H1.4 (Fig. S1 A). TDMS analyses of these fractions provided striking evidence that interphase H1 phosphorylation is site specific. H1.2 contains four Cdk substrate motifs, but MS/MS analysis of the 1p-H1.2 peak localized phosphorylation exclusively to S173 in both allelic variants. Similarly, although H1.4 has five Cdk substrate motifs, phosphorylation localized exclusively to S187 in the 1p-H1.4 peak and to S172 plus S187 in the 2p-H1.4 peak (Table I, Fig. S1 B, and Fig. S2 F).
Table I.
H1 phosphorylation during interphase and mitosis in HeLa S3 cells
| HIC peak | Molecular mass | Δm | Variant | Modifications | |
| Measured | Predicted | ||||
| D | D | ||||
| Interphase | |||||
| 1 | 21,491.6 | 21,332.8 | +160 | H1.4 | pS172, pS187 |
| 2 | 21,412.3 | 21,332.8 | +80 | H1.4 | pS187 |
| 3 | 21,333.1 | 21,332.8 | 0 | H1.4 | None |
| 4 | 20,912.1 | 20,832.6 | +80 | H1.2 (A142) | pS173 |
| 4 | 2,0941.7 | 20,832.6 | +110 | H1.2 (T142) | pS173 |
| 5 | 20,832.3 | 20,832.6 | 0 | H1.2 (A142) | None |
| 5 | 20,862.2 | 20,832.6 | +30 | H1.2 (T142) | None |
| Mitosis | |||||
| 6p-H1.4 | 21,813.9 | 21,332.8 | +480 | H1.4 | pT18, pS27, pT146, pT154, pS172, pS187 |
| 4p-H1.2 | 21,153.3 | 20,832.6 | +320 | H1.2 (A142) | pT31, pT146, pT154, pS173 |
| 4p-H1.2 | 21,182.9 | 20,832.6 | +350 | H1.2 (T142) | pT31, pT146, pT154, pS173 |
HIC peaks are labeled as in Fig. 1 B. Molecular masses are reported as neutral monoisotopic species. Predicted values were found using NCBI Protein database accession no. NP_005312 (H1.4) and NP_005310 (H1.2), assuming the loss of Met1 during protein maturation in vivo and residues 2–5 during electrospray ionization. Peak identifications are based on MS/MS sequencing of multiple electron capture dissociation fragment ions. The A142 and T142 allelic variants of H1.2 were detected initially by MS and confirmed by genotyping as described in Fig. S1. Phosphorylated residues were identified by MS/MS sequencing of electron capture dissociation fragment ions. Analyses of the H1.2 (T142) forms indicated that phosphorylated residues were identical to those determined for the H1.2 (A142) forms.
Analyses of the 4p-H1.2 and 6p-H1.4 peaks from colchicine-treated cells (Fig. 1 B) revealed uniform phosphorylation at the four and five Cdk substrate motifs present in H1.2 and H1.4, respectively (Table I and Fig. S2 F). A sixth site, S27, in an RKS motif that is unique to H1.4 was phosphorylated concurrently. We refer to T31, T146, and T154 of H1.2 and T18, S27, T146, and T154 of H1.4 as M sites because they appear to be phosphorylated exclusively during mitosis. In contrast, we refer to H1.2-S173, H1.4-S172, and H1.4-S187 as I sites because they can be phosphorylated during mitosis and interphase.
Our results suggest that H1.2-S173 and H1.4-S172 and -S187 are the sole sites of interphase phosphorylation in these proteins in human cells. Notably, monophosphorylation at H1.4-S172 was not detected, implying that interphase H1.4 phosphorylation/dephosphorylation occurs hierarchically or that other mechanisms prevent the accumulation of detectable H1.4-S172 monophosphorylation. Our findings also suggest that interphase H1 kinases preferentially phosphorylate Ser-containing Cdk substrate motifs, which is consistent with evidence that three such sites in H1.5 are phosphorylated during interphase in human cells (Talasz et al., 2009). With the exception of H1.4-S27, all of the M sites identified in this study are Thr-containing Cdk substrate motifs. Identification of H1.4-S27 as an M site is noteworthy because phosphorylation at this residue during mitosis may affect acetylation or methylation at H1.4-K26 or the interaction of factors that recognize these modifications (Kuzmichev et al., 2004; Vaquero et al., 2004; Daujat et al., 2005; Trojer et al., 2007). We did not detect either modification at H1.4-K26 using TDMS, suggesting that they affect <1% of total H1.4 in HeLa S3 cells (Pesavento et al., 2008).
pS173-H1.2 and pS187-H1.4 are enriched in nucleoli
To investigate the roles of specific H1 phosphorylations, we generated antisera against phosphopeptides containing the pS173-H1.2 and pS187-H1.4 I sites and the pS27-H1.4 and pT154-H1.4 M sites (Fig. S2 A). We also generated antisera against recombinant human H1.4 that recognizes H1.4 regardless of its phosphorylation state to use as a control. The specificity of these antisera was validated using Western blotting (Fig. S2, B–E), ELISA assays (not depicted), and immunocytochemistry (Fig. S3).
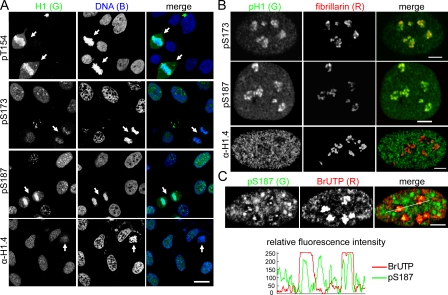
The pT154 antisera stained the chromosomes of mitotic HeLa cells intensely but did not stain interphase nuclei (Fig. 2 A). Similar results were observed for the pS27 and pT146 antisera (unpublished data), confirming that these sites are phosphorylated exclusively in mitosis. In contrast, the pS173 and pS187 antisera stained chromatin in both interphase and mitotic cells. The pS173 antisera stained mitotic chromosomes less intensely than either the pT154 or the pS187 antisera. Because these residues are expected to be phosphorylated to similar degrees during mitosis, this suggests that the pS173 epitope may be less accessible in mitotic chromosomes. Differences were also observed for the staining of interphase cells by the pS173 and pS187 antisera (Fig. 2 A). Most interphase cells displayed stippled nuclear staining and clusters of punctate nucleolar staining for pS173. Many interphase cells showed similar staining for pS187, but others displayed speckled staining similar to that of mouse 10T1/2 cells stained by antiserum to phosphorylated Tetrahymena thermophila H1. The latter antiserum preferentially recognizes phosphorylated mouse H1.5, and it has been suggested that the speckled staining represents the localization of transcriptionally active chromatin near sites of RNA splicing (Chadee et al., 1997). Other evidence suggests that speckled staining with this same antibody occurs primarily during G1 phase in human cells (Lu et al., 1994), but we have not investigated whether this is the case for pS187 staining. In contrast, stippled staining distributed throughout interphase nuclei was observed for the α-H1.4 antisera, with weaker staining associated with nucleoli in some cases. Because this antiserum recognizes H1.4 regardless of its phosphorylation state (Fig. S2 D), the stippled pattern may reflect a nonuniform distribution of H1.4, as suggested previously for two H1 variants (Parseghian et al., 1994) or differences in the accessibility of H1.4 at different loci to the antisera.
Figure 2.
Interphase phosphorylated H1.2/H1.4 are enriched in nucleoli. (A) Confocal images of asynchronous HeLa cells stained with the H1 antisera shown. DNA was stained with TO-PRO-3. Arrows indicate mitotic cells. Bar, 20 µm. (B) Confocal images of asynchronous HeLa cells costained with antisera to fibrillarin and the H1 antisera shown. Bars, 5 µm. (C) Confocal images of asynchronous HeLa cells costained with antisera to pS187-H1.4 and BrdU after pulse labeling with BrUTP to detect nascent transcripts. Bar, 5 µm. Plots of the relative fluorescence intensity of the green (pS187-H1.4) and red (BrUTP) channels along the white line shown in the merged panel demonstrate colocalization of pS187-H1.4 with BrUTP incorporation foci.
Fibrillarin colocalized extensively with clustered punctate pS173 and pS187 staining, but not with α-H1.4 staining, confirming that these H1.2/H1.4 phosphorylations are enriched in nucleoli (Fig. 2 B). Moreover, punctate pS187 staining colocalized with centers of bromo-UTP (BrUTP) incorporation foci when cells were pulse labeled with BrUTP to detect nascent 45S pre–ribosomal RNA (rRNA) transcripts (Fig. 2 C; Koberna et al., 2002; Olson and Dundr, 2005). Similar results were obtained for pS173 staining (unpublished data), suggesting that interphase phosphorylated H1.2/H1.4 are associated with transcribing rDNA (45S preribosomal RNA genes) and may facilitate RNA pol I transcription.
pS187-H1.4 is preferentially associated with active rDNA promoters
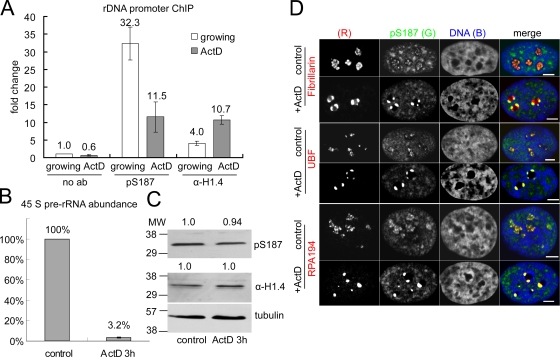
Mammalian cells contain several hundred rDNA repeats. The transcriptional activity of individual repeats is regulated by mechanisms including histone modification to match cellular demand for ribosome biogenesis (Lawrence and Pikaard, 2004; Moss et al., 2007; McStay and Grummt, 2008). To investigate whether interphase H1 phosphorylation contributes to this regulation, we used chromatin immunoprecipitation (ChIP) to compare the association of pS187-H1.4 with rDNA promoters before and after selective inhibition of RNA pol I transcription with actinomycin D (ActD; Jordan et al., 1996; Olson and Dundr, 2005). The levels of 45S pre-rRNA were assessed using RT-PCR to provide a semiquantitative measure of rDNA transcription (Huang et al., 2008; Murayama et al., 2008). A brief ActD treatment that markedly reduced the levels of nascent 45S pre-rRNA significantly reduced the promoter association of pS187-H1.4 compared with untreated cells (Fig. 3, A and B). Although treatments that impair RNA pol II transcription are associated with global H1 dephosphorylation (Chadee et al., 1997), immunoblots revealed that the limited ActD treatment used had little effect on the global levels of pS187-H1.4 and total H1.4 (Fig. 3 C). Moreover, ChIP with the α-H1.4 antisera revealed that ActD treatment actually enhanced the level of total H1.4 at the promoter (Fig. 3 A). Collectively, the data suggest that pS187-H1.4 is enriched at active rDNA promoters and that this association is dynamically regulated.
Figure 3.
pS187-H1.4 is preferentially associated with transcriptionally active rDNA. (A) The levels of pS187-H1.4 and total H1.4 (α-H1.4) at the 45S pre-rRNA promoter in untreated and 50 ng/ml ActD-treated HeLa cells (3 h). The data are expressed as fold change relative to a parallel ChIP without primary antibody from untreated cells. (B) The levels of the 45S pre-rRNA transcript in untreated and ActD-treated cells were assayed by RT-PCR and normalized to β-actin expression. The data in A and B represent the mean of two independent experiments. Error bars indicate the standard error of the mean for triplicate samples. (C) The global levels of pS187-H1.4 and total H1.4 in ActD-treated cells were assayed by immunoblotting and quantified relative to the control samples after normalization to α-tubulin levels. MW, molecular weight. (D) Confocal images of control and ActD-treated HeLa cells costained with antibodies to pS187-H1.4 and fibrillarin, UBF, or the large subunit of RNA polymerase I (RPA 194). DNA was visualized using TO-PRO-3. Bars, 5 µm.
The ultrastructural elements of nucleoli in higher eukaryotes, the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component, are thought to reflect vectorial organization of major steps in ribosome biogenesis: pre-rRNA transcription, pre-rRNA processing, and ribosome assembly on mature rRNA (Olson and Dundr, 2005; Hernandez-Verdun, 2006). This organization is affected when rDNA transcription is inhibited by ActD (Jordan et al., 1996; Olson and Dundr, 2005), so we investigated how ActD affects pS187-H1.4 colocalization with markers for these nucleolar compartments. Fibrillarin localizes primarily to the DFC in punctate staining that rings FCs in untreated cells but becomes concentrated in large foci at the nucleolar periphery when the FC and DFC dissociate from each other in ActD-treated cells (Olson and Dundr, 2005). Although pS187-H1.4 and fibrillarin colocalized extensively in untreated cells, ActD caused a characteristic change in this relationship (Fig. 3 D). Both proteins formed similar numbers of foci near the nucleolar periphery, but the pS187-H1.4 foci were typically smaller and shifted relative to the fibrillarin foci.
Similarly, pS187-H1.4 colocalized extensively with UBF in untreated cells (Fig. 3 D). ActD caused the characteristic punctate UBF staining to coalesce into a few large granules at the nucleolar periphery, which is in agreement with previous work (Zatsepina et al., 1993). However, in contrast to the fibrillarin results, UBF and pS187-H1.4 foci colocalized completely in ActD-treated cells. ActD had similar effects on the colocalization of pS187-H1.4 and RNA pol I (RPA 194). Like UBF, RPA 194 localizes preferentially to the FC in untreated cells (Matera et al., 1994). Punctate pS187-H1.4 and RPA 194 staining colocalized extensively in the central portions of nucleoli in untreated cells, and they colocalized completely in dense clusters at the edge of nucleoli in ActD-treated cells (Fig. 3 D). The perdurance of pS187-H1.4 staining and its colocalization with UBF and RPA 194 in foci formed upon ActD treatment are consistent with evidence that the latter proteins remain associated with rDNA under these conditions (Jordan et al., 1996). Together, these results suggest that pS187-H1.4 is enriched in active rDNA within the FC or at the FC–DFC interface in untreated cells.
UBF dimers bind rDNA promoters and activate transcription by recruiting SL1/TIF-IB and enhancing preinitiation complex formation (Moss et al., 2007; McStay and Grummt, 2008). UBF binding throughout the transcribed portions of rDNA repeats may also affect other aspects of RNA pol I transcription and help maintain the euchromatic state of rDNA (O’Sullivan et al., 2002; Moss et al., 2007; Sanij et al., 2008; Sanij and Hannan, 2009). Multiple lines of evidence suggest that competition between UBF and H1 determines the proportion of active rDNA repeats (Kermekchiev et al., 1997; Sanij et al., 2008). The dependence of pS187-H1.4 and rDNA promoter association on RNA pol I activity and the colocalization of pS187-H1.4 with UBF and RNA pol I before and after ActD treatment reported in this study (Fig. 3) suggest the possibility that H1 kinases are recruited to transcriptionally active/competent rDNA. S187 phosphorylation could promote rDNA decondensation and transcription by enhancing H1.4 dissociation and facilitating UBF binding.
Induced enrichment of pS187-H1.4 at hormone response elements
Data from approaches that do not account for H1 phosphorylation suggest that H1 represses transcription by RNA pol II (Laybourn and Kadonaga, 1991; Cheung et al., 2002; Lee et al., 2004; Kim et al., 2008). In contrast, ChIP analyses using antisera to phosphorylated T. thermophila H1 suggest that H1 phosphorylation is required for glucocorticoid-dependent transcription from the murine mammary tumor virus (MMTV) promoter in mammalian cells (Lee and Archer, 1998; Bhattacharjee et al., 2001). This led us to investigate whether pS187-H1.4 is involved in transcription by RNA pol II.
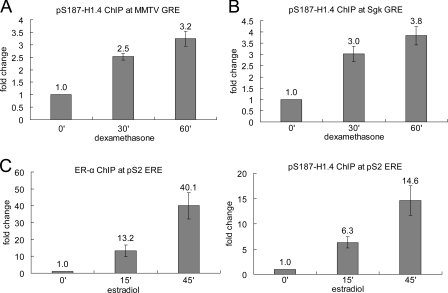
We used ChIP to compare the association of pS187-H1.4 with the multicopy MMTV long terminal repeat glucocorticoid response element (GRE [MMTV-GRE]) in murine 3134 mammary tumor cells before and after hormone stimulation. Dexamethasone rapidly induced pS187-H1.4 association with the MMTV-GRE, increasing the level of pS187-H1.4 at this locus by approximately threefold in 60 min (Fig. 4 A). Rapid, hormone-induced enrichment of pS187-H1.4 was also observed at the single-copy Sgk gene GRE in these same cells (Fig. 4 B). We also compared how the association of pS187-H1.4 and estrogen receptor–α (ER-α) with the estrogen response element (ERE) of the pS2 gene in MCF-7 human breast cancer cells changes after estradiol treatment (Fig. 4 C). Both ER-α and pS187-H1.4 were markedly enriched at the pS2-ERE after just 15 min of estradiol treatment and became further enriched after 45 min of treatment. Collectively, the data in Fig. 4 suggest that H1.4-S187 phosphorylation facilitates gene-specific activation of RNA pol II transcription by nuclear hormone receptors.
Figure 4.
Hormone-induced enrichment of pS187-H1.4 at nuclear hormone receptor response elements. (A) pS187-H1.4 association with the MMTV-GRE in 3134 cells before and after dexamethasone treatment. (B) pS187-H1.4 association with the Sgk gene GRE in 3134 cells before and after dexamethasone treatment. (C) ER-α and pS187-H1.4 association with the pS2 gene ERE in MCF-7 cells before and after estradiol treatment. The data are normalized to the signal obtained for each antibody before hormone treatment and represent the mean of two independent experiments. Error bars indicate the standard error of the mean for triplicate samples.
Analyses of glucocorticoid receptor–regulated transcription at the MMTV promoter in different systems suggest that H1 is depleted immediately after hormone stimulation (Bresnick et al., 1992; Belikov et al., 2007) but reassociates with refractory promoters after prolonged hormone treatment (Lee and Archer, 1998), which is consistent with the notion that H1 generally acts as a repressor. In contrast, overexpression of H1c or H1° enhanced basal and hormone-stimulated transcription of stably integrated MMTV-LTR reporter genes in murine 3T3 cells and prevented their repression during prolonged hormone stimulation (Gunjan and Brown, 1999). Our data are consistent with the proposal that H1 affects MMTV promoter chromatin architecture to facilitate the binding of liganded nuclear hormone receptors, their synergism with transcription factors such as NF1 and AP-1, and the recruitment or activation of kinases that phosphorylate and facilitate H1 displacement after hormone stimulation (Vicent et al., 2002). Together with recent evidence that estradiol stimulates the exchange of HMGB for H1 at the pS2-ERE and sites recognized by AP-1 or by other nuclear hormone receptors (Ju et al., 2006), our data suggest that H1.4-S187 phosphorylation promotes transcriptional activation by nuclear hormone receptors by enhancing chromatin access for other regulatory factors.
Considered together with evidence from FRAP analyses that mutations mimicking phosphorylation or dephosphorylation of Cdk sites enhance or diminish H1-GFP dissociation from chromatin, respectively, our findings imply that phosphorylation at a limited number of specific sites enables H1 to affect chromatin accessibility to factors that regulate transcription and other processes. Our evidence that the association of pS187-H1.4 with specific loci is dynamic implies that H1 kinases and phosphatases are recruited to these loci in a targeted fashion, although little is known about the mechanisms involved. Kinases that mediate interphase H1 phosphorylation in vivo have not been directly identified, but several lines of evidence implicate Cdk2 (Herrera et al., 1996; Bhattacharjee et al., 2001; Contreras et al., 2003). Less is known about whether specific phosphatases regulate interphase H1 phosphorylation. Although our data support the general model that interphase phosphorylation enhances H1 dissociation from chromatin, variation in the numbers, locations, and structures of I sites among H1 variants suggests that interphase phosphorylation may affect H1 chromatin binding in a variant-specific fashion. Such differences may underlie recent evidence for H1 variant–specific effects on transcription and replication (Sancho et al., 2008; Talasz et al., 2009).
Materials and methods
Cell culture
HeLa S3 cells were grown in suspension in Joklik’s modified minimal essential medium supplemented with 10% newborn calf serum (NCS) and synchronized using the double-thymidine block procedure as described previously (Pesavento et al., 2008). 1 µM colchicine was added to growing asynchronous cells for 18 h to enrich for mitotic cells. For the experiments shown in Fig. 3, adherent HeLa cells were grown in DME supplemented with 10% FBS. Cells were treated with 0.05 µg/ml ActD for 3 h to selectively inhibit RNA pol I transcription. 3134 cells were maintained in DME supplemented with 10% FBS, sodium pyruvate, nonessential amino acids, and 2 mM glutamine as described previously (John et al., 2009). Cells were transferred to DME supplemented with 10% charcoal dextran–treated FBS for 48 h before treatment with 100 nM dexamethasone. MCF-7 cells were maintained in DME supplemented with 5% NCS as described previously (Schultz-Norton et al., 2007). Cells were transferred to phenol red–free DME containing 5% charcoal dextran–treated NCS for 72–96 h before treatment with 10 nM estradiol.
Histone preparation, chromatography, and MS
Crude H1 was prepared by 5% perchloric acid fractionation of crude histones (Pesavento et al., 2008). Recombinant human H1.4 was expressed in Escherichia coli BL21 cells from a pET-3d vector using standard procedures.
Reverse phase HPLC used a column (4.6 mm ID × 250 mm; Vydac C18) with a multistep linear gradient from buffer A (0.1% vol/vol TFA in 5% vol/vol CH3CN) to buffer B (0.094% TFA in 90% CH3CN). HIC used a column (4.6 mm ID × 100 mm; PolyPROPYL A; PolyLC Inc.) with a multistep linear gradient from buffer A (2.5 M (NH)2SO4 and 50 mM ethylenediamine, pH 7.0) to buffer B (1.0 M (NH)2SO4 and 50 mM ethylenediamine, pH 7.0). Hydrophilic interaction chromatography used a column (4.6 mm ID × 200 mm; PolyCAT A; PolyLC Inc.) with a multistep linear gradient from buffer A (70% CH3CN and 15 mM triethylamine/H3PO4, pH 3.0) to buffer B (70% CH3CN, 0.68 M NaClO4, and 15 mM triethylamine/H3PO4, pH 3.0).
MS data were acquired on a custom 8.5-T quadrupole Fourier transform ion cyclotron resonance mass spectrometer with an electrospray ionization source operated in positive-ion mode as described previously (Pesavento et al., 2008). Desalted HIC/hydrophilic interaction chromatography fractions or crude H1 were dissolved in 50% methanol + 1% formic acid for infusion into the mass spectrometer. Masses are reported as neutral, monoisotopic species. Figs. 1 and S1 show the spectrum of the most abundant charge state.
Immunochemical methods
Phosphorylated and nonphosphorylated peptides were custom synthesized. Rabbits were immunized with phosphopeptides coupled to keyhole limpet hemocyanin (Thermo Fisher Scientific) using standard procedures. Recombinant human H1.4 was complexed with yeast tRNA before rabbit immunization as described previously (Stollar and Ward, 1970). Antibodies to pT146-H1.4 and fibrillarin were obtained from Abcam. Antibodies to UBF and RPA 194 were obtained from Santa Cruz Biotechnology, Inc. Antibodies to α-tubulin (clone DM1A) and bromodeoxyuridine (clone BRD.3) were obtained from Sigma-Aldrich and Neomarkers, respectively.
Microscopy and BrUTP labeling of cells
HeLa cells grown on glass coverslips were fixed with 4% (wt/vol) paraformaldehyde in PBS for 10 min at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 15 min, and immunostained with primary antisera using standard procedures. Staining was visualized with FITC-conjugated donkey anti–rabbit antibody (1:200; Jackson ImmunoResearch Laboratories, Inc.) and Cy3-conjugated donkey anti–mouse antibody (1:800; Jackson ImmunoResearch Laboratories, Inc.). Nuclei were counterstained with TO-PRO-3 (Invitrogen), and coverslips were mounted with Vectashield media (Vector Laboratories). Images were captured at room temperature with a confocal microscope and software (LSM 510; Carl Zeiss, Inc.) using a Plan Apochromat 63× 1.4 NA oil immersion objective lens and processed with ImageJ (National Institutes of Health) and Photoshop (Adobe). For peptide competition, primary antibodies were preincubated with peptides at 2× final concentration for 2 h at room temperature and diluted to a final peptide concentration of 1.0 µg/ml before use.
FuGENE 6 was diluted 1:10 in Hepes-buffered saline (25 mM Hepes, 0.75 mM Na2HPO4, and 140 mM NaCl, pH 7.05) and incubated at room temperature for 5 min. BrUTP was added to achieve 5 mM BrUTP (final) and incubated for 15 min at room temperature. Cells grown on coverslips were pulse labeled by placing coverslips over BrUTP transfection mixture drop on parafilm for 15 min at room temperature followed by a 15-min chase in DME containing 10% NCS at 37°C.
ChIP
Cells were cross-linked by adding formaldehyde directly to cultures (1% final) and incubating for 10 min at room temperature. 125 mM final glycine was added, and cultures were incubated for 10 min on ice. Cells were washed twice with cold PBS, scraped, and resuspended in ChIP lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris, pH 8.0) containing 0.5 mM AEBSF and 5 nM microcystin. Chromatin was sheared to ∼500-bp mean length by repeated cycles of sonication alternated with cooling in ice water.
After clarification by centrifugation (18,000 g for 10 min), supernatants were diluted 10-fold with dilution buffer (1.11% Triton X-100, 1.11 mM EDTA, 16.7 mM Tris, and 167 mM NaCl, pH 8.0). Aliquots representing 1–2 × 106 cells in 1.0 ml final volume were used for each pull-down. Samples were first precleared by incubation with 15 µl protein G beads (Lake Placid Bio) for 30 min at 4°C, and the resulting supernatants were incubated with 15 µl pS187-H1.4 overnight at 4°C. Samples were incubated with 12.5 µl protein G beads for 1 h at 4°C. Beads were pelleted by centrifugation and washed sequentially with 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.0, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl, pH 8.0, 0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0, and twice with 1 mM EDTA and 10 mM Tris-HCl, pH 8.0. Beads were eluted twice with 200 µl 1% SDS in 0.1 M NaHCO3 at 65°C for 10 min. The combined eluates were made 200 mM NaCl (final), incubated at 65°C overnight to reverse cross-links, digested with 50 µg/ml RNase A at 37°C for 30 min, and then digested with 50 µg/ml proteinase K at 50°C for 1 h. The DNA fragments were then purified by phenol/chloroform extraction, recovered by ethanol precipitation using 20 µg glycogen as a carrier, and dissolved in 50 µl of deionized water.
ChIP products were quantitated by real-time PCR using SYBR Green master mix (Applied Biosystems) and the following primers: rRNA promoter (forward), 5′-GTGGCTGCGATGGTGGCGTTTT-3′ and (reverse) 5′-TGCCGACTCGGAGCGAAAGA-3′; MMTV-GRE (forward), 5′-TTTCCATACCAAGGAGGGGACAGTG-3′ and (reverse) 5′-CTTACTTAAGCCTTGGGAACCGCAA-3′; Sgk-GRE (forward), 5′-CTTCCCTTATCCAGCATGTCTTGTG-3′ and (reverse) 5′-TGCATCGTGCAATCTGTGGC-3′; and pS2-ERE (forward), 5′-CCCGTGAGCCACTGTTGTC-3′ and (reverse) 5′-CCTCCCGCCAGGGTAAATAC-3′.
ChIP to monitor the association of ER-α and pS187-H1.4 with the pS2-ERE included minor modifications as described previously (Schultz-Norton et al., 2007). The ER-α antibody (sc-8002) for ChIP was obtained from Santa Cruz Biotechnology, Inc.
Online supplemental material
Fig. S1 shows SDS gel and TDMS analyses of H1 fractions with different levels of phosphorylation prepared from mid–S phase HeLa cells using HIC. Fig. S2 shows the specificity of antisera for individual H1 phosphorylation sites, the antisera to total H1.4 in immunoblots, and the phosphorylation sites identified in this study relative to an alignment of the human H1.1–H1.5 protein sequences. Fig. S3 shows the specificity of the pS173-H1.2 and pS187-H1.4 antisera using peptide competition in immunofluorescence microscopy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001148/DC1.
Supplementary Material
Acknowledgments
We thank Rafael E. Herrera for flag-H1.4 vectors and Michel Bellini for help with microscopy.
This work was funded by grants from the Roy J. Carver Charitable Trust (grant 04-76), the March of Dimes (Basil O’Connor Scholar Award FY05-1232), and the National Science Foundation (MCB–0821893) to C.A. Mizzen, by grants from the Packard Foundation, the Sloan Foundation, and a Cottrell Scholar Award to N.L. Kelleher, and National Institutes of Health grants to N.L. Kelleher (GM 067193) and A.M. Nardulli (R01 DK53884). J.J. Pesavento was a recipient of a National Institutes of Health Institutional National Research Service Award in Molecular Biophysics (5T32 GM 08276).
Footnotes
Abbreviations used in this paper:
- ActD
- actinomycin D
- BrUTP
- bromo-UTP
- ChIP
- chromatin immunoprecipitation
- CTD
- C-terminal domain
- DFC
- dense fibrillar component
- ER-α
- estrogen receptor–α
- ERE
- estrogen response element
- FC
- fibrillar center
- GRE
- glucocorticoid response element
- HIC
- hydrophobic interaction chromatography
- MMTV
- murine mammary tumor virus
- MS
- mass spectrometry
- NCS
- newborn calf serum
- rRNA
- ribosomal RNA
- TDMS
- top-down MS
- UBF
- upstream-binding factor
References
- Ajiro K., Borun T.W., Cohen L.H.. 1981a. Phosphorylation states of different histone 1 subtypes and their relationship to chromatin functions during the HeLa S-3 cell cycle. Biochemistry. 20:1445–1454. 10.1021/bi00509a007 [DOI] [PubMed] [Google Scholar]
- Ajiro K., Borun T.W., Shulman S.D., McFadden G.M., Cohen L.H.. 1981b. Comparison of the structures of human histone 1A and 1B and their intramolecular phosphorylation sites during the HeLa S-3 cell cycle. Biochemistry. 20:1454–1464. 10.1021/bi00509a008 [DOI] [PubMed] [Google Scholar]
- Allan J., Hartman P.G., Crane-Robinson C., Aviles F.X.. 1980. The structure of histone H1 and its location in chromatin. Nature. 288:675–679. 10.1038/288675a0 [DOI] [PubMed] [Google Scholar]
- Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C.. 1986. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 187:591–601. 10.1016/0022-2836(86)90337-2 [DOI] [PubMed] [Google Scholar]
- Bednar J., Horowitz R.A., Grigoryev S.A., Carruthers L.M., Hansen J.C., Koster A.J., Woodcock C.L.. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA. 95:14173–14178. 10.1073/pnas.95.24.14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikov S., Astrand C., Wrange O.. 2007. Mechanism of histone H1-stimulated glucocorticoid receptor DNA binding in vivo. Mol. Cell. Biol. 27:2398–2410. 10.1128/MCB.01509-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee R.N., Banks G.C., Trotter K.W., Lee H.L., Archer T.K.. 2001. Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol. Cell. Biol. 21:5417–5425. 10.1128/MCB.21.16.5417-5425.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick E.H., Bustin M., Marsaud V., Richard-Foy H., Hager G.L.. 1992. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 20:273–278. 10.1093/nar/20.2.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.T., Izard T., Misteli T.. 2006. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat. Struct. Mol. Biol. 13:250–255. 10.1038/nsmb1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Catez F., Lim J.H.. 2005. The dynamics of histone H1 function in chromatin. Mol. Cell. 17:617–620. 10.1016/j.molcel.2005.02.019 [DOI] [PubMed] [Google Scholar]
- Carruthers L.M., Bednar J., Woodcock C.L., Hansen J.C.. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 37:14776–14787. 10.1021/bi981684e [DOI] [PubMed] [Google Scholar]
- Chadee D.N., Allis C.D., Wright J.A., Davie J.R.. 1997. Histone H1b phosphorylation is dependent upon ongoing transcription and replication in normal and ras-transformed mouse fibroblasts. J. Biol. Chem. 272:8113–8116. 10.1074/jbc.272.13.8113 [DOI] [PubMed] [Google Scholar]
- Cheung E., Zarifyan A.S., Kraus W.L.. 2002. Histone H1 represses estrogen receptor alpha transcriptional activity by selectively inhibiting receptor-mediated transcription initiation. Mol. Cell. Biol. 22:2463–2471. 10.1128/MCB.22.8.2463-2471.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A., Hale T.K., Stenoien D.L., Rosen J.M., Mancini M.A., Herrera R.E.. 2003. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol. Cell. Biol. 23:8626–8636. 10.1128/MCB.23.23.8626-8636.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S., Zeissler U., Waldmann T., Happel N., Schneider R.. 2005. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 280:38090–38095. 10.1074/jbc.C500229200 [DOI] [PubMed] [Google Scholar]
- Garcia B.A., Busby S.A., Barber C.M., Shabanowitz J., Allis C.D., Hunt D.F.. 2004. Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J. Proteome Res. 3:1219–1227. 10.1021/pr0498887 [DOI] [PubMed] [Google Scholar]
- Gunjan A., Brown D.T.. 1999. Overproduction of histone H1 variants in vivo increases basal and induced activity of the mouse mammary tumor virus promoter. Nucleic Acids Res. 27:3355–3363. 10.1093/nar/27.16.3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley L.R., Walters R.A., Tobey R.A.. 1975. Sequential phsophorylation of histone subfractions in the Chinese hamster cell cycle. J. Biol. Chem. 250:3936–3944. [PubMed] [Google Scholar]
- Gurley L.R., Valdez J.G., Buchanan J.S.. 1995. Characterization of the mitotic specific phosphorylation site of histone H1. Absence of a consensus sequence for the p34cdc2/cyclin B kinase. J. Biol. Chem. 270:27653–27660. 10.1074/jbc.270.46.27653 [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Lever M.A., Crawford E., Th’ng J.P.. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279:20028–20034. 10.1074/jbc.M400070200 [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. 2006. The nucleolus: a model for the organization of nuclear functions. Histochem. Cell Biol. 126:135–148. 10.1007/s00418-006-0212-3 [DOI] [PubMed] [Google Scholar]
- Herrera R.E., Chen F., Weinberg R.A.. 1996. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc. Natl. Acad. Sci. USA. 93:11510–11515. 10.1073/pnas.93.21.11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Ji Y., Itahana K., Zhang Y., Mitchell B.. 2008. Guanine nucleotide depletion inhibits pre-ribosomal RNA synthesis and causes nucleolar disruption. Leuk. Res. 32:131–141. 10.1016/j.leukres.2007.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Johnson T.A., Sung M.H., Biddie S.C., Trump S., Koch-Paiz C.A., Davis S.R., Walker R., Meltzer P.S., Hager G.L.. 2009. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology. 150:1766–1774. 10.1210/en.2008-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P., Mannervik M., Tora L., Carmo-Fonseca M.. 1996. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 133:225–234. 10.1083/jcb.133.2.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B.G., Lunyak V.V., Perissi V., Garcia-Bassets I., Rose D.W., Glass C.K., Rosenfeld M.G.. 2006. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 312:1798–1802. 10.1126/science.1127196 [DOI] [PubMed] [Google Scholar]
- Kermekchiev M., Workman J.L., Pikaard C.S.. 1997. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol. Cell. Biol. 17:5833–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Choi J., Heo K., Kim H., Levens D., Kohno K., Johnson E.M., Brock H.W., An W.. 2008. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J. Biol. Chem. 283:9113–9126. 10.1074/jbc.M708205200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna K., Malínský J., Pliss A., Masata M., Vecerova J., Fialová M., Bednár J., Raska I.. 2002. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J. Cell Biol. 157:743–748. 10.1083/jcb.200202007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Jenuwein T., Tempst P., Reinberg D.. 2004. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell. 14:183–193. 10.1016/S1097-2765(04)00185-6 [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Pikaard C.S.. 2004. Chromatin turn ons and turn offs of ribosomal RNA genes. Cell Cycle. 3:880–883. [PubMed] [Google Scholar]
- Laybourn P.J., Kadonaga J.T.. 1991. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 254:238–245. 10.1126/science.1718039 [DOI] [PubMed] [Google Scholar]
- Lee H.L., Archer T.K.. 1998. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 17:1454–1466. 10.1093/emboj/17.5.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Habas R., Abate-Shen C.. 2004. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 304:1675–1678. 10.1126/science.1098096 [DOI] [PubMed] [Google Scholar]
- Lever M.A., Th’ng J.P., Sun X., Hendzel M.J.. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 408:873–876. 10.1038/35048603 [DOI] [PubMed] [Google Scholar]
- Lu M.J., Dadd C.A., Mizzen C.A., Perry C.A., McLachlan D.R., Annunziato A.T., Allis C.D.. 1994. Generation and characterization of novel antibodies highly selective for phosphorylated linker histone H1 in Tetrahymena and HeLa cells. Chromosoma. 103:111–121. [DOI] [PubMed] [Google Scholar]
- Lu X., Hamkalo B., Parseghian M.H., Hansen J.C.. 2009. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry. 48:164–172. 10.1021/bi801636y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Tycowski K.T., Steitz J.A., Ward D.C.. 1994. Organization of small nucleolar ribonucleoproteins (snoRNPs) by fluorescence in situ hybridization and immunocytochemistry. Mol. Biol. Cell. 5:1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Grummt I.. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24:131–157. 10.1146/annurev.cellbio.24.110707.175259 [DOI] [PubMed] [Google Scholar]
- Misteli T., Gunjan A., Hock R., Bustin M., Brown D.T.. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881. 10.1038/35048610 [DOI] [PubMed] [Google Scholar]
- Moss T., Langlois F., Gagnon-Kugler T., Stefanovsky V.. 2007. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 64:29–49. 10.1007/s00018-006-6278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K., et al. 2008. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 133:627–639. 10.1016/j.cell.2008.03.030 [DOI] [PubMed] [Google Scholar]
- O’Sullivan A.C., Sullivan G.J., McStay B.. 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22:657–668. 10.1128/MCB.22.2.657-668.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.O., Dundr M.. 2005. The moving parts of the nucleolus. Histochem. Cell Biol. 123:203–216. 10.1007/s00418-005-0754-9 [DOI] [PubMed] [Google Scholar]
- Parseghian M.H., Harris D.A., Rishwain D.R., Hamkalo B.A.. 1994. Characterization of a set of antibodies specific for three human histone H1 subtypes. Chromosoma. 103:198–208. 10.1007/BF00368013 [DOI] [PubMed] [Google Scholar]
- Pesavento J.J., Yang H., Kelleher N.L., Mizzen C.A.. 2008. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell. Biol. 28:468–486. 10.1128/MCB.01517-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R.D., Scaffidi P., Elbi C., Vecerová J., Dey A., Ozato K., Brown D.T., Hager G., Bustin M., Misteli T.. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402. 10.1128/MCB.24.14.6393-6402.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque A., Iloro I., Ponte I., Arrondo J.L., Suau P.. 2005. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J. Biol. Chem. 280:32141–32147. 10.1074/jbc.M505636200 [DOI] [PubMed] [Google Scholar]
- Roque A., Ponte I., Arrondo J.L., Suau P.. 2008. Phosphorylation of the carboxy-terminal domain of histone H1: effects on secondary structure and DNA condensation. Nucleic Acids Res. 36:4719–4726. 10.1093/nar/gkn440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho M., Diani E., Beato M., Jordan A.. 2008. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 4:e1000227. 10.1371/journal.pgen.1000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E., Hannan R.D.. 2009. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics. 4:374–382. [DOI] [PubMed] [Google Scholar]
- Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T.P., Quin J., Robb E., Wong L.H., Thomas W.G., Stefanovsky V., et al. 2008. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 183:1259–1274. 10.1083/jcb.200805146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarg B., Helliger W., Talasz H., Förg B., Lindner H.H.. 2006. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. J. Biol. Chem. 281:6573–6580. 10.1074/jbc.M508957200 [DOI] [PubMed] [Google Scholar]
- Schultz-Norton J.R., Walt K.A., Ziegler Y.S., McLeod I.X., Yates J.R., Raetzman L.T., Nardulli A.M.. 2007. The deoxyribonucleic acid repair protein flap endonuclease-1 modulates estrogen-responsive gene expression. Mol. Endocrinol. 21:1569–1580. 10.1210/me.2006-0519 [DOI] [PubMed] [Google Scholar]
- Stollar B.D., Ward M.. 1970. Rabbit antibodies to histone fractions as specific reagents for preparative and comparative studies. J. Biol. Chem. 245:1261–1266. [PubMed] [Google Scholar]
- Suzuki M. 1989. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 8:797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talasz H., Helliger W., Puschendorf B., Lindner H.. 1996. In vivo phosphorylation of histone H1 variants during the cell cycle. Biochemistry. 35:1761–1767. 10.1021/bi951914e [DOI] [PubMed] [Google Scholar]
- Talasz H., Sarg B., Lindner H.H.. 2009. Site-specifically phosphorylated forms of H1.5 and H1.2 localized at distinct regions of the nucleus are related to different processes during the cell cycle. Chromosoma. 118:693–709. 10.1007/s00412-009-0228-2 [DOI] [PubMed] [Google Scholar]
- Th’ng J.P., Sung R., Ye M., Hendzel M.J.. 2005. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J. Biol. Chem. 280:27809–27814. 10.1074/jbc.M501627200 [DOI] [PubMed] [Google Scholar]
- Trojer P., Li G., Sims R.J., III, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S.D., et al. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 129:915–928. 10.1016/j.cell.2007.03.048 [DOI] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D.. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 16:93–105. 10.1016/j.molcel.2004.08.031 [DOI] [PubMed] [Google Scholar]
- Vicent G.P., Koop R., Beato M.. 2002. Complex role of histone H1 in transactivation of MMTV promoter chromatin by progesterone receptor. J. Steroid Biochem. Mol. Biol. 83:15–23. 10.1016/S0960-0760(02)00253-4 [DOI] [PubMed] [Google Scholar]
- Vila R., Ponte I., Collado M., Arrondo J.L., Suau P.. 2001. Induction of secondary structure in a COOH-terminal peptide of histone H1 by interaction with the DNA: an infrared spectroscopy study. J. Biol. Chem. 276:30898–30903. 10.1074/jbc.M104189200 [DOI] [PubMed] [Google Scholar]
- Wisniewski J.R., Zougman A., Krüger S., Mann M.. 2007. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol. Cell. Proteomics. 6:72–87. [DOI] [PubMed] [Google Scholar]
- Zatsepina O.V., Voit R., Grummt I., Spring H., Semenov M.V., Trendelenburg M.F.. 1993. The RNA polymerase I-specific transcription initiation factor UBF is associated with transcriptionally active and inactive ribosomal genes. Chromosoma. 102:599–611. 10.1007/BF00352307 [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Caiafa P., Van Holde K.. 2000. Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J. 14:1697–1704. 10.1096/fj.99-0869rev [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.