Apo2 proteins interact with chaperone Unc45b (but not Hsp90) and are required for correct zebrafish skeletal musculature and heart function.
Abstract
The chaperones Unc45b and Hsp90a are essential for folding of myosin in organisms ranging from worms to humans. We show here that zebrafish Unc45b, but not Hsp90a, binds to the putative cytidine deaminase Apobec2 (Apo2) in an interaction that requires the Unc45/Cro1p/She4p-related (UCS) and central domains of Unc45b. Morpholino oligonucleotide-mediated knockdown of the two related proteins Apo2a and Apo2b causes a dystrophic phenotype in the zebrafish skeletal musculature and impairs heart function. These phenotypic traits are shared with mutants of unc45b, but not with hsp90a mutants. Apo2a and -2b act nonredundantly and bind to each other in vitro, which suggests a heteromeric functional complex. Our results demonstrate that Unc45b and Apo2 proteins act in a Hsp90a-independent pathway that is required for integrity of the myosepta and myofiber attachment. Because the only known function of Unc45b is that of a chaperone, Apo2 proteins may be clients of Unc45b but other yet unidentified processes cannot be excluded.
Introduction
Congenital myopathies are a heterogeneous group of muscle disorders that usually become evident at birth or early infancy. Molecular analysis and studies of animal models have broadened our knowledge of the causes and pathophysiology of these diseases immensely (D’Amico and Bertini, 2008; Sewry, 2008). The zebrafish embryo has proven particularly useful for identifying genes that are required to build and maintain the muscle apparatus, offering insights into the underlying mechanisms and providing gene candidates and models for human myopathies (Bassett and Currie, 2004; Behra et al., 2004; Deniziak et al., 2007; Guyon et al., 2007; Flinn et al., 2008; Hawkins et al., 2008; Ingham, 2009). In contrast to humans and mice, zebrafish embryos deficient in proteins of the dystrophin–glycoprotein complex (DGC), such as dystrophin, dystroglycan, delta-sarcoglycan, and the fukutin-related protein (FKRP), already show phenotypes during embryonic stages. These mutants have impaired motility and exhibit a curved body, U-shaped somites, and myofibrils that detach from the vertical myosepta, a connective tissue that separates somites and is functionally equivalent to the mammalian tendon (Parsons et al., 2002; Bassett et al., 2003; Guyon et al., 2003a,b, 2005, 2009; Kudo et al., 2004; Hall et al., 2007; Thornhill et al., 2008). In addition, other proteins, such as obscurin or the TGF-β–related factor Mstn2, have been shown in the zebrafish to influence the expression or organization of the DGC, resulting in dystrophic phenotypes (Raeker et al., 2006; Amali et al., 2008).
The chaperones Unc45 and Hsp90 are necessary for proper myosin folding (Barral et al., 2002; Srikakulam and Winkelmann, 2004; Etard et al., 2007; Hawkins et al., 2008). Mutations in zebrafish unc45b or hsp90a result in paralyzed animals that fail to form myofibrils in skeletal and cardiac muscles (Etard et al., 2007; Hawkins et al., 2008). Unc45 and Hsp90 form complexes with myosin that are required for proper folding and assembly of myosin into thick filaments (Barral et al., 1998, 2002; Srikakulam and Winkelmann, 2004; Etard et al., 2007,2008; Hawkins et al., 2008). The subcellular distribution of the two chaperones is highly dynamic: Unc45b and Hsp90a associate transiently with nascent myosin and locate to the Z line once the fibril has formed in zebrafish muscle (Etard et al., 2008). Damage to the myofiber causes a shift of the two chaperones to the A band, which suggests that the Z line serves as a reservoir for myosin chaperones (Etard et al., 2008). The stability of UNC-45 in Caenorhabditis elegans is tightly regulated via complex formation with the ubiquitin ligase C terminus of Hsp70-interacting protein (CHIP; Hoppe et al., 2004; Nyamsuren et al., 2007), an interaction that has been linked with late-onset hereditary inclusion-body myopathies in humans (Janiesch et al., 2007).
To systematically explore the role of Unc45b in zebrafish and to identify additional candidates for human disease genes, we have performed a two-hybrid screen with Unc45b as bait. Here, we describe an interaction between Unc45b and Apobec2 (Apo2). Apo2 belongs to a small family of structurally related proteins (Wedekind et al., 2003). Apo1, the founding member of this family, edits apolipoprotein mRNA by deaminating a cytosine to uracil (Teng et al., 1993; Yamanaka et al., 1994; Wedekind et al., 2003). This introduces a stop codon, generating two forms of apolipoprotein B (apoB), transporters of cholesterol and triglyceride (Knott et al., 1986). Members of the Apo3 subfamily play critical roles in retroviral defense (Harris et al., 2003; Mangeat et al., 2003). Apo3 proteins bind HIV-1 RNA and physically impede movement of the reverse transcription on the viral template in a cytidine-deaminase–independent manner. In addition, the reverse-transcribed minus strand was shown to be a target for the deaminase activity of Apo3G (Yu et al., 2004). The related activation-induced (cytidine) deaminase enzyme (AID) is expressed in the germinal centers of lymph nodes that contain activated B cells, and is essential for the somatic hypermutations that diversify the variable regions of immunoglobulin genes (Muramatsu et al., 1999, 2000).
In contrast to these members of the Apo family, little is known about either the specific substrates or the physiological functions of Apo2 (Liao et al., 1999; Anant et al., 2001). The deaminase activity of Apo2 itself is discussed controversially: some have claimed there is a low but intrinsic cytidine deaminase activity (Liao et al., 1999), whereas others regard this deaminase activity as a contamination with deaminases from the bacterial host used to produce the protein (Mikl et al., 2005). Expression of apo2 was originally reported to be restricted to cardiac and skeletal muscles (Liao et al., 1999; Mukhopadhyay et al., 2002), but a recent analysis using a cDNA panel derived from different tissues showed ubiquitous expression at a low level in human and mouse (Matsumoto et al., 2006). Apo2 expression is strongly enhanced in the liver in response to both tumor necrosis factor and interleukin-β via a nuclear factor κβ response element in the promoter. This suggests a role of Apo2 in the physiopathology of hepatic inflammation (Matsumoto et al., 2006). However, a function of Apo2 in the skeletal or cardiac muscles has not been demonstrated (Mikl et al., 2005).
In contrast to the situation in mammals, only two members of the Apo2 subfamily and one homologue of AID are detectable in the zebrafish genome, which suggests that they are the most ancient members of the family and that Apo1 and Apo3 emerged later in the evolution of the vertebrate lineage (Liao et al., 1999; Conticello et al., 2005; Mikl et al., 2005). In this study, we show that the two related zebrafish members of the Apo2 subfamily (called Apo2a and Apo2b) interact with Unc45b. This interaction is specific for Unc45b. Morpholino oligonucleotide–mediated knockdown of translation of apo2a or apo2b mRNA resulted in a dystrophic phenotype with cell-free spaces in the somitic musculature and extra long myofibers spanning two somites. These phenotypes are shared with unc45b mutants. Our data suggest that the Apo2 proteins and Unc45b act in a developmental pathway, which is required for the integrity of the myosepta and for myofiber attachment.
Results
Unc45b interacts with Apo2a
In a two-hybrid screen using the full-length zebrafish Unc45b as the bait, an interacting protein with sequence similarity to the mammalian C-to-U deaminase Apo2 (Fig. S1 A) was identified five times, in addition to five other proteins including Hsp90 proteins (Materials and methods; Etard et al., 2007). We refer to this protein as Apo2a. To determine whether the observed interaction in the yeast assay merited follow-up studies, we performed in vitro pull-down experiments with recombinant proteins produced in Escherichia coli and in situ expression analysis in the zebrafish embryo to assess whether unc45b and apo2a are expressed in the same cells.
Pull-down experiments were performed with in vivo and in vitro translated GST-Apo2a and Unc45b-GFP chimeras, respectively, in which Apo2a and Unc45b were fused in-frame with GST and GFP. The results confirmed the interaction observed in the yeast two-hybrid experiments: GST-Apo2a pulled down Unc45b-GFP as revealed by Western blotting with an anti-GFP antibody (Fig. 1, A–C, lane 1).
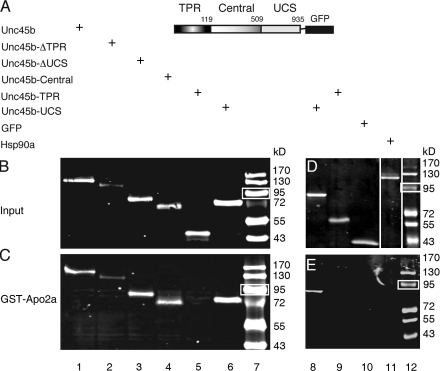
Figure 1.
Apo2a interacts with Unc45b but not Hsp90a in pull-down assays. (A) Schematic representation of the full-length Unc45b protein fused to GFP. Unc45b is composed of three domains: the TPR (amino acids 1–119), the central (amino acids 120–509), and the UCS (amino acid 910–935) domains. The intact Unc45b protein and its subdomains as well as Hsp90a were fused at their C terminus to GFP and used as input protein in the pull-down assays as indicated. (B) In vitro synthesized chimeric proteins visualized by Western blotting with anti-GFP antibody. (C) Full-length Unc45b-GFP fusion protein (lane 1) and deletion variants of Unc45b fused to GFP (TPR-deleted Unc45b [Unc45-ΔTPR] lane 2; UCS-deleted Unc45b [Unc45b-ΔUCS], lane 3; or single Unc45b domains fused to GFP [Unc45b-Central], lane 4; Unc45b-TPR, lane 5; Unc45b-UCS, lane 6) were pulled down with the GST-Apo2a fusion protein. With the exception of Unc45b-TPR (lane 5), all other chimeras were pulled down by GST-Apo2a fusion proteins (lanes 1–4 and 6). (D) Western blot of protein input of Hsp90a-GFP fusion (lane 11). Unc45b-UCS (lane 8), Unc45b-TPR (lane 9), and GFP (lane 10) were used as positive and negative controls, respectively, to test for an interaction of GST-Apo2a with Hsp90a-GFP. Note that relative to protein inputs in binding assays (C and E), 10-fold less protein was loaded onto the control gels shown in B and D. White lines indicate that intervening lanes have been spliced out. (E) Although Unc45b-UCS interacted with Apo2a (lane 8), neither the Unc45b-TPR fusion (lane 9), GFP alone (lane 10), nor Hsp90a-GFP (lane 11) were pulled down by GST-Apo2a. Thus, the interaction of Apo2a with Unc45b is specific. Lane 7 and 12 show protein markers. White boxes indicate 95 kD.
Unc45b protein contains three domains with different functions, namely a tetratricopeptide repeat (TPR) domain necessary for interaction with the cochaperone Hsp90; a Unc45-/Cro1p-/She4p-related (UCS) domain, which binds to myosin motor domain; and a central domain that mediates Z line association (Etard et al., 2008). To determine which domain of Unc45b is required for interaction with Apo2a, Unc45b deletions (Fig. 1, A and B) were tested in pull-down assays. The results show that both the UCS and the central domain have the ability to interact with Apo2a (Fig. 1 C, lanes 2–4 and 6). In contrast, the TPR-GFP fusion protein did not interact with Apo2a (Fig. 1 C, lane 5). GFP alone, without attached Unc45b sequences, was also not pulled down by the GST-Apo2a chimera (Fig. 1 E, lane 10). These results demonstrate that the binding to Apo2a requires the UCS and central domains of Unc45b and is not mediated by interaction with the TPR domain or GFP. In addition, we tested whether the related chaperone Unc45a that is required for blood vessel formation in the zebrafish (Anderson et al., 2008) can also bind Apo2a in the in vitro binding assay. However, we did not detect an interaction between Apo2a and Unc45a (unpublished data).
Unc45b forms complexes with other chaperones such as members of the Hsp90 family (Barral et al., 2002; Etard et al., 2007). We therefore investigated whether Apo2 proteins could also bind to Hsp90a. GST-Apo2a did not pull down Hsp90a-GFP (Fig. 1, D and E, lane 11). The closely related Hsp90a2 and Hsp90b proteins also did not interact with Apo2a in this in vitro pull-down assay (unpublished data). Collectively, these results suggest that Apo2a does not bind Hsp90 chaperones. Furthermore, this underscores the notion that Apo2a interacts with Unc45b in a specific manner.
Apo2a is coexpressed with unc45b in skeletal and cardiac muscle
Unc45b is exclusively expressed in skeletal and cardiac muscles of the zebrafish embryo (Etheridge et al., 2002; Etard et al., 2007). We next assessed whether the apo2a gene is transcribed in the same tissues. Whole mount in situ hybridization showed that apo2a mRNA is expressed strongly in all skeletal muscles of the trunk and the head as well as in the heart (Fig. 2, A–F). Expression was detected as early as the 18-somite stage in the developing somitic musculature (Fig. 2 A). At 24 hours postfertilization (hpf), apo2a transcripts are evident in the heart and in the somites (Fig. 2, B and C). At later stages, expression of apo2a becomes apparent in the musculature of the head, as well as in that of the pectoral fins (Fig. 2, D–F). We did not detect expression in tissues other than the cardiac and skeletal musculature, although we cannot exclude low-level expression in other tissues below the detection limit of the assay used. This pattern of cell-specific mRNA expression is directly comparable to that of unc45b (Etard et al., 2007).
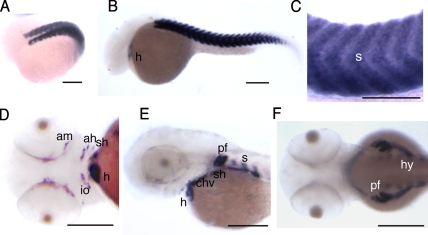
Figure 2.
Apo2a is expressed in the heart and in the skeletal musculature. (A–C) In situ hybridization with an apo2a anti-sense probe to 18 somites (A) and 24-hpf (B and C) embryos showing apo2a mRNA expression in cardiac (h) and skeletal muscles (s). (D–F) At 48 (D and E) and 60 hpf (F), the expression was found in the somitic muscle (s; E and F), hypaxial muscles (hy; F), the pectoral fin muscles (pf; E), and cranial muscles (adductor mandibulae [am], adductor hyomandibulae [ah], sternohyoideus [sh], constrictor hyoideus ventralis [chv], and inferior oblique [io]; D and E). The orientation of embryos in the images is anterior to the left and are as follows: (A) dorso-lateral view; (B, C, and E) lateral view, dorsal up; (D) ventral view; and (F) dorsal view. Bars, 200 µm.
Apo2a is necessary for muscle integrity
To investigate whether Apo2a has a function in the development or maintenance of the muscle apparatus, the translation of apo2a was blocked by injection of a morpholino oligonucleotide (Mo[ATG]-apo2a) directed against a 25-bp mRNA region surrounding the start codon (Fig. 3 A). Morpholino-injected embryos showed impaired motility, in that embryos twitched and contracted upon touch stimulation but failed to escape (70.3% of morpholino-injected embryos [“morphants,” n = 100]). Next, we assessed whether morpholino-mediated knockdown of Apo2a resulted in reduced birefringence of the skeletal musculature, which is an indication of a disturbed arrangement of muscle fibers and/or fibrils. Impaired mobility was correlated with a significant reduction of birefringence on the flanks of the morphants, which indicates disorganized myofibrils (Fig. 3, B–D; 70% of morphants, n = 100). Morphant embryos developed pericardial edema (Fig. 3 O) associated with a slightly decreased heart rate compared with wild-type (Fig. 3 N, n = 20 for both morphant and control embryos). The Mo(ATG)-apo2a–injected embryos also showed brain necrosis, which is a frequent nonspecific effect of morpholino injections (Robu et al., 2007). Necrosis was completely abolished when a morpholino directed against p53 (Mo-p53; Robu et al., 2007) was coinjected. Mo-p53 alone did not affect the muscle birefringence or induce pericardial edema (Fig. 3, K and L; and not depicted), which suggests that the cardiac and skeletal defects are indeed specific for the injected Mo(ATG)-apo2a.
Figure 3.
Apo2a morphants have defects in the skeletal musculature and the heart. (A) Position of the morpholinos Mo(ATG)-apo2a (ATG) and Mo(UTR)-apo2a (UTR) on the apo2a mRNA. (B–D) Mo(ATG)-apo2a (B and D) and control Mo(ATG)-apo2a-cont (harboring five mismatches relative to the Mo(ATG)-apo2a sequence; C)-injected embryos. The morphants exhibit a curved body axis (B) and show reduced birefringence (D) compared with controls (C). Note that the Mo(ATG)-apo2a–injected embryos (B and D) were coinjected with the Mo-p53 to suppress unspecific effects (see K–M for additional controls for Mo-p53 injection). (E–G) Mo(UTR)-apo2a (E and G)- and control Mo(ATG)-apo2a-cont (F)-injected embryos. The morphants exhibit a curved body axis (E) and show reduced birefringence (G) compared with controls (F). (H–J) Apo2a-GFP expression (H) in embryos injected with an apo2a-gfp encoding plasmid. Co-injection of Mo(ATG)-apo2a with the apo2a-gfp plasmid abolishes Apo2a-GFP expression (I), whereas coinjection of the apo2a-gfp plasmid with the five-mismatch Mo(ATG)-apo2a-cont does not abolish Apo2a-GFP expression (J). (K–M) Uninjected (K), Mo(ATG)-apo2a-cont– and Mo-p53–coinjected (L), and Mo(ATG)-apo2a– and Mo-p53–coinjected embryos (M). Injection of Mo(ATG)-apo2a morpholino reduces birefringence (M), whereas neither the five-mismatch Mo(ATG)-apo2a-cont nor the Mo-p53 had an effect on the birefringence of the musculature (L). (N) Mo(ATG)-apo2a and Mo(UTR)-apo2a morphants have a decreased heart rate (the graph shows heart beats per minute, n = 20 embryos). Black bars represent standard deviation. The datasets were pairwise subjected to the Student’s t test. The difference between the heart rate of Mo(ATG)-apo2a versus Mo(UTR)-apo2a was not significant (P = 0.220), whereas the difference between the heart rate of Mo(ATG)-apo2a versus control or Mo(UTR)-apo2a versus control was highly significant (P = 0.002 and P = 0.003, respectively). (O) Pericardial edema (arrow) in a Mo(ATG)-apo2a– and Mo-p53–coinjected embryo. Embryos shown are 72 hpf, except F–H, which show 24-hpf embryos. Pigmentation of the embryo shown in E was suppressed by 1-phenyl 2-thiourea (PTU) treatment (Karlsson et al., 2001). Bars, 80 µm.
To further prove that the effects of the injected Mo(ATG)-apo2a are specific for the apo2a mRNA and that the observed phenotypes are caused by a lack of Apo2a function, we performed several additional controls. First, we coinjected a plasmid encoding a fusion protein of Apo2a and GFP. Co-injection of the Mo(ATG)-apo2a strongly reduced expression of this Apo2a–GFP fusion protein as measured by reduced GFP fluorescence (Fig. 3, H and I). Second, we designed a mismatch morpholino (Mo(ATG)-apo2a-cont) that contained five mismatches compared with the Mo(ATG)-apo2a. Injection of this morpholino did not show any phenotypes (Fig. 3 L and not depicted) and it did not block Apo2a-GFP expression (Fig. 3 J). Finally, another morpholino (Mo(UTR)-apo2a) directed to sequences in the 5′ untranslated region (UTR) distinct from those covered by Mo(ATG)-apo2a (Fig. 3 A) was designed. Injection of Mo(UTR)-apo2a recapitulated the effects of the Mo(ATG)-apo2a, such as pericardial edema and reduced birefringence of the skeletal musculature (Fig. 3, E–G). Moreover, Mo(UTR)-apo2a did not cause necrosis in the brain, obviating the need to co-inject Mo-p53. Collectively, these results demonstrate that the observed phenotypes result from a specific knockdown of apo2a mRNA translation.
Loss of Apo2a causes disruption of myosepta and myofibers
Examination of apo2a morphants with differential interference contrast (DIC) optics revealed abnormal cell-free spaces (Fig. 4, A and B) in the skeletal muscles at 72 hpf. These myofiber-free spaces were restricted to individual somites in some cases (Fig. 4 A). In other instances, they spanned two somites (Fig. 4 B). Morphant embryos displayed irregular and discontinuous myosepta that were locally interrupted with myofibers extending over two adjacent somites (Fig. 4 C; 100%, n = 20 embryos). None of these defects were seen in control morpholino-injected embryos (100%, n = 60).
Figure 4.
Apo2a morphants have defective myosepta and form intersomitic giant myofibers. (A–C) DIC images of Mo(ATG)-apo2a–injected embryos. Mo(ATG)-apo2a morphants (Mo) exhibit cell-free spaces in the somites (A and B, stars), disrupted myosepta (B and C, arrows), and giant myofibers that span two somites (C, arrowhead). (D–F) Immunohistochemistry with antibodies directed against myoseptal proteins reveals defects in the myosepta of apo2a morphants: α-Dystroglycan (DG) staining of control (cont; D) and Mo(ATG)-apo2a–injected (Mo; E and F) embryos (white arrows, disrupted myosepta; green arrows, split myosepta). (G and H) Laminin staining reveals abnormal myosepta in Mo(ATG)-apo2a morphants (H) compared with wild type (G). Green arrows, split myosepta. (I–K) Double immunohistochemistry with anti–α-dystroglycan (DG) and the anti-slow muscle myosin antibody F59 reveals giant slow muscle myofibers (J, arrowhead) crossing the somite boundary (J, arrow). (I) Control embryos never show these intersomitic myofibers. Detachment of myofibers from the myosepta (K, arrow) generates cell-free spaces (K, star). (L and M) Sarcomeric myosin stained with A4.1025 in Mo(ATG)-apo2a morphants shows detached slow fibers (M, yellow arrows) overlaying fast fibers and generating cell-free areas inside the muscle tissue (star). Most of the fast fibers are still anchored to the myosepta. A few fast fibers have also detached (M, blue arrow). (L) Control. All embryos injected with Mo(ATG)-apo2a were coinjected with Mo-p53. Embryos are 72 h old. The anterior is shown to the left, dorsal side up. Bars, 25 µm.
The disruptions of the myotome boundary as well as the formation of myofiber-free spaces in the somite resembles the phenotype of dystrophic mutants (Bassett et al., 2003; Guyon et al., 2003b, 2005; Cheng et al., 2006). This prompted us to examine in more detail the consequence of Apo2a depletion on the expression of the DGC. β-Dystroglycan is highly enriched in the chevron-shaped myosepta (Fig. 4 D). At 72 hpf, immunolabeling of β-dystroglycan revealed gaps or bifurcations in these structures in the morphants (Fig. 4, E and F). Moreover, the myosepta were not as straight and were more U-shaped (Fig. 4, E and F) in comparison with the chevrons of control embryos (Fig. 4 D). Other proteins of the DGC, such as dystrophin and β-sarcoglycan, showed the same defective patterns in apo2a morphants (unpublished data). Examination of laminin, an extracellular matrix component of the myoseptum, also revealed disrupted and bifurcated somite boundaries in the morphants (Fig. 4, G and H), which suggests that the integrity of the entire myoseptum is disturbed.
To correlate these defects of the myosepta with the cell-free spaces and giant fibers observed under DIC optics, morphants and controls were costained with antibodies directed against β-dystroglycan and slow muscle myosin (Fig. 4, I–K). Cell-free spaces did not always correlate with the disruption of the myoseptum (Fig. 4 K, star), which suggests that they arise independently of the gaps in the myosepta. Some muscle fibers were detached from the myoseptum (Fig. 4 K, arrow). At places where myosepta were disrupted, giant slow myosin fibers were frequently observed that spanned the length of two somites (Fig. 4 J, arrowhead). Formation of these extra-long fibers was not restricted to the superficial slow muscles: immunostaining of fast myosin and sarcomeric actin also confirmed the presence of giant fibers (unpublished data) and of enlarged intercellular spaces (Fig. 4, L and M) in the fast muscles. Instead of being regularly packed, the fast muscle fibers appeared twisted and less well stacked in the morphants (Fig. 4, L and M).
Like the Mo(ATG)-apo2a morphants, embryos injected with Mo(UTR)-apo2a (directed to the 5′ UTR of apo2a mRNA; see above, Fig. 3 A) showed gaps between myofibers, detached fibers, and giant fibers extending across two somites (not depicted). Collectively, we conclude that Apo2a is required for muscle integrity. Importantly, knockdown embryos displayed phenotypes that are identical to those observed in zebrafish models of human muscular dystrophies (Parsons et al., 2002; Bassett et al., 2003; Bassett and Currie, 2004; Thornhill et al., 2008; Guyon et al., 2009).
Apo2b is necessary for muscle integrity
A search of the zebrafish genome uncovered another related apo gene that we named apo2b. The two proteins share 57.8% amino acid identity. Phylogenetic analysis suggests that both genes belong to the apo2 subfamily (Fig. S1 B). With the exceptions of apo2b and a gene related to AID, no further members of the apo gene family were identified in the zebrafish genome.
We first examined the expression pattern of apo2b mRNA by in situ hybridization of whole embryos. Expression of apo2b was detected in the skeletal muscles of the head, in somitic muscles, and in the heart (unpublished data), a pattern identical to that of apo2a. Because apo2a and apo2b share sequence similarity and both are expressed in the muscle of the zebrafish embryo, we next investigated if Apo2b also interacts with Unc45b. Pull-down assays performed with GST-Apo2b and Unc45b-GFP showed that Apo2b can indeed also interact with Unc45b (Fig. 5, B and C, lane 1). Furthermore, this interaction was mediated by the central and UCS domains of Unc45b (Fig. 5 C, lanes 2–4 and 6). As in the case of Apo2a, neither the TPR-GFP chimera nor GFP alone interacted with GST-Apo2b (Fig. 5 C, lane 5; and not depicted).
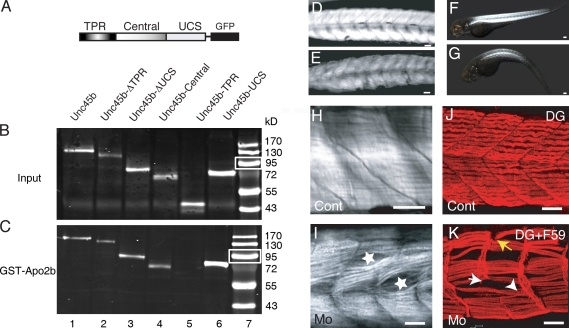
Figure 5.
Apo2b interacts with Unc45b. (A) Schematic diagram of the Unc45b-GFP full-length and deletion proteins used in pull-down assays. (B and C) Western blot of in vitro translated input (B) and Unc45b-GFP proteins pulled down with GST-Apo2b (C). Fusion proteins were detected with an anti-GFP antibody. With the exception of Unc45b-TPR (lane 5), all the other fusions proteins were pulled down by Apo2b-GST (lanes 1–4 and 6), which indicates that both the central as well as the UCS domain can mediate the interaction with Apo2b. Lane 7 shows size markers. Note that relative to protein inputs in binding assays (B), 10-fold less protein was loaded onto the control gels shown in C. White boxes indicate 95 kD. (D and E) Control (D) and Mo(ATG)-apo2b (E)-injected embryos. Injection of the morpholino complementary to sequences surrounding the ATG of apo2b mRNA reduces the birefringence of the skeletal muscle. (F and G) Control (F) and Mo(UTR)-apo2b (G)-injected embryos. The morpholino complementary to the 5′ UTR of the apo2b mRNA reduces the birefringence of the skeletal muscle in a similar manner to the morpholino directed against the ATG of apo2b mRNA. The striking similarity of this phenotype to apo2a morphants suggests that the two Apo2 proteins do not act redundantly. (H and I) Mo(ATG)-apo2b morphants (I) but not controls (H) observed with DIC optics have cell-free spaces in the somites (I, stars). (J and K) Double immunohistochemistry with a slow muscle myosin antibody F59 and/or an α-dystroglycan (DG; J and K, respectively) antibody reveals extra-long intersomitic myofibers spanning two somites (K, arrowhead), detached myofibers, and defective myosepta in the morphants (K; white arrow, disrupted myosepta; yellow arrow, detached fibers). (I) Control. Bars: (D, E, and H–K) 30 µm; (F) 80 µm; (G) 100 µm.
We next tested the function of apo2b by injecting morpholinos designed against the region spanning the start codon or the region further 5′ in the untranslated sequences. Both morphants showed reduced mobility and decreased birefringence (79.8% morphants, n = 200) compared with control embryos (Fig. 5, D–G). In addition, the heart rate was reduced and the blood circulation was impaired (unpublished data). Immunohistological analysis of the muscle structure revealed the same defects as those observed with apo2a morpholinos: detachment of fibers, interrupted myosepta, which are often associated with giant myofibrils, and cell-free cavities in the somites (Fig. 5, H–K). The results of these knockdown experiments suggest that the two apo2 genes cannot compensate for one another.
Although unlikely, as the sequences are very dissimilar, we wished to exclude the possibility that the morpholinos cross-react with the two apo2a and -2b mRNAs. Co-injection of the apo2a-gfp plasmid and Mo(ATG)-apo2b morpholinos did not cause reduction of GFP expression, which indicates that Mo(ATG)-apo2b did not efficiently disrupt translation of Apo2a (unpublished data). When apo2b-gfp plasmids encoding the Apo2b-GFP fusion protein were coinjected with the Mo(ATG)-apo2b, none of the double-injected embryos showed fluorescence (data not shown), which indicates that the morpholinos do efficiently block translation of Apo2b. In parallel, Mo(ATG)-apo2a injected with apo2b-gfp plasmid did not block the expression of the Apo2b-GFP fusion protein (unpublished data). This strongly suggests that Mo(ATG)-apo2a and Mo(ATG)-apo2b morpholinos are specific to apo2a and apo2b mRNA, respectively.
Apo2a and Apo2b form heteromers
One possible explanation for the nonredundant function of Apo2a and Apo2b is that heteromeric complexes between the two proteins are required for function. To test whether Apo2a and Apo2b can form heteromers, we performed pull-down experiments with GST-tagged Apo2a or Apo2b with GFP-labeled Apo2b (Fig. 6 A). No complexes formed when GST-Apo2a was incubated with GFP alone (Fig. 6 A, lane 2). However, when GST-Apo2a was incubated with Apo2b-GFP, a band of the expected size for the chimeric protein was pulled down (Fig. 6 A, lane 3). Thus, Apo2a and Apo2b can form heteromeric complexes. GST-Apo2b was also able to interact with Apo2b-GFP (Fig. 6 A, lane 6), whereas an interaction with GFP alone was not noticeable. This suggests that Apo2b can also form homomeric complexes. The formation of both homomeric as well as heteromeric complexes could also be demonstrated for GST-Apo2a or GST-Apo2b and MYC-Apo2a chimeras (Fig. 6 B). In summary, these data support the notion that Apo2a and Apo2b can form heteromeric complexes in addition to the homomeric complexes suggested previously (Prochnow et al., 2007).
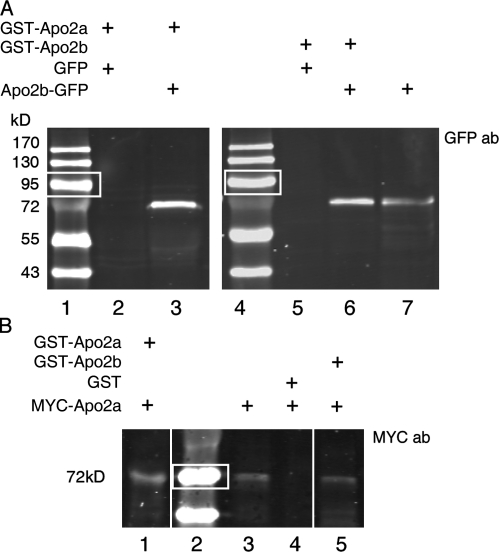
Figure 6.
Apo2a and Apo2b can form homo- and heteromers. (A) GST-Apo2a and GST-Apo2b fusion proteins can pull down Apo2b-GFP proteins. Pulled-down proteins were detected by Western blotting with an anti-GFP antibody. Lanes 1 and 4: protein size markers (white boxes indicate 95 kD); lanes 2 and 5: GFP-only controls; lane 3: pull-down of Apo2b-GFP by GST-Apo2a; lane 6: pull-down of Apo2b-GFP by GST-Apo2b; lane 7: Apo2b-GFP protein input. (B) GST-Apo2a and GST-Apo2b can pull down MYC-Apo2a fusion proteins as revealed by Western blotting with the anti-MYC epitope 9E10 antibody. Lane 1: pull-down of Apo2a-MYC with GST-Apo2a; lane 2: protein size markers (white boxes indicate 72 kD); lane 3: input of MYC-Apo2a protein; lane 4: GST-only control; lane 5: pull-down of MYC-Apo2a by GST-Apo2b.
apo2 morphants share phenotypic traits with unc45b mutants
The most striking phenotype of unc45b mutants is the severe disruption of the myofibrils, presumably caused by a failure of the myosin heavy chains to fold properly (Etard et al., 2007). We did not observe such a phenotype in the apo2a morphants, which suggests that apo2a does not have a function in the genesis of myofibrils. However, if the interaction of Unc45b and Apo2a has any functional relevance in the muscle, one would predict that the more subtle defects seen in apo2a morphants would also be evident in unc45b mutants. Close examination of α-actin-GFP transgenic unc45b mutants indeed revealed the same myofiber-free areas within the muscle structure (Fig. 7, A and B; 100%, n = 10 embryos), a result confirmed by the use of DIC optics (Fig. 7, D and E; n = 10 embryos). Moreover, fibers were also detached from the myosepta in unc45b mutants (Fig. 7 G, arrows). Immunohistochemistry using the anti–β-dystroglycan antibody revealed broader and bifurcated myosepta in unc45b mutants compared with wild-type embryos (Fig. 7, I and H), which indicates that the myosepta are also disrupted.
Figure 7.
unc45b mutants also exhibit cell-free spaces in the somites and disruption of the myosepta like the apo2a morphants. (A–C) α-Actin–GFP transgenic wild-type (A), unc45b (B), and hsp90a (C) mutant embryos. unc45b mutant embryos exhibit cell-free spaces in the somitic musculature (B, stars) as seen in apo2a morphants. In contrast, wild-type siblings (A) and hsp90a (C) mutants do not show these gaps. (D–F) Cell-free spaces are also visible in unlabeled unc45b mutants using DIC optics (E, white stars). In contrast, wild-type siblings (D) and hsp90a mutants (F) do not exhibit these cell-free spaces in the somites. Broken lines indicate myoseptal boundaries. (G) Merge of DIC and α-actin–GFP fluorescence frames shows detachment of myofibrils (arrows) and cell-free spaces (stars) in unc45b mutants, which is seen in apo2a morphants but never seen in wild-type or morphant control embryos. (H and I) Immunohistochemistry with a β-dystroglycan (DG) antibody shows disruption of myosepta in unc45b mutants (I), whereas wild-type siblings (H) have normal chevron-shaped myosepta. (J–L) Unc45b-GFP is localized at the Z line (J, white arrow) as well as at the myosepta (blue arrow), whereas embryos expressing CapZa1-GFP show localization at the Z line exclusively (L, arrow). Immunohistochemistry with an antibody against endogenous Unc45b reveals that endogenous Unc45b is localized at the myosepta (K and K′, blue arrows) in addition to the previously shown Z line staining (K′, white arrow). Bars: (A–I and K) 40 µm; (J and L) 2 µm; (K′): 4 µm.
We also examined hsp90a mutants, which, like unc45b mutants, fail to form myofibrils (Etard et al., 2007; Hawkins et al., 2008). Cell-free spaces were not detectable in hsp90a mutants, and the myosepta were structured as in wild-type embryos (Fig. 7, C and F; n = 20 embryos). In contrast to unc45b mutants, hsp90a mutants have a beating heart. To exclude the possibility that the cell-free spaces and disrupted myosepta in apo2a morphants and unc45b mutants result from impaired blood circulation, we examined the myosin light chain gene 2 (mlc2) mutant, whose heart does not beat (Rottbauer et al., 2006). The homozygous mlc2 mutants did not exhibit cell-free spaces in the somitic muscle, showing that the absence of blood flow cannot account for the observed phenotype (unpublished data, n = 10 embryos).
We have previously reported that a chimeric Unc45b-GFP protein is localized at the Z line in undamaged muscle (Etard et al., 2008). Given the unexpected myoseptal phenotype of unc45b mutants, we reexamined the location of Unc45b-GFP protein after injection of an unc45b-gfp plasmid in wild-type embryos. This protocol results in a strong mosaicism of expressing cells, permitting the study of individual labeled cells in a background-free environment in the intact embryo. As previously noted, the fusion protein localized to the Z line. In addition, we also found accumulation at the myoseptal boundary of expressing cells (Fig. 7 J, blue arrow). We also raised an antibody against zebrafish Unc45b and examined the location of the endogenous protein. As in the case of the fusion protein, endogenous Unc45b accumulated at the Z line. In addition, we also noted an enrichment of Unc45b immunoreactivity at the myoseptal boundary (Fig. 7 K, blue arrow). We also studied the localization of CapZa1-GFP fusion protein (Schafer et al., 1993; Papa et al., 1999) to assess whether Z line proteins in general are enriched at the myoseptal boundary. The CapZa1-GFP fusion protein was enriched at the Z line, but it showed only very low levels at the myoseptal boundary (Fig. 7 L). Collectively, the location of the Unc45b protein at the myoseptal boundary supports a role for Unc45b in myofibril attachment, as suggested by the phenotype of unc45b mutants. To assess whether the localization of Unc45b is affected by a lack of Apo2a proteins, Mo(ATG)-apo2a and Mo(ATG)-apo2b were injected together with a plasmid encoding unc45b-gfp. Unc45b-GFP localization appeared unaffected in the morphants (unpublished data).
Apo2 fusion proteins are localized to the myoseptal boundary
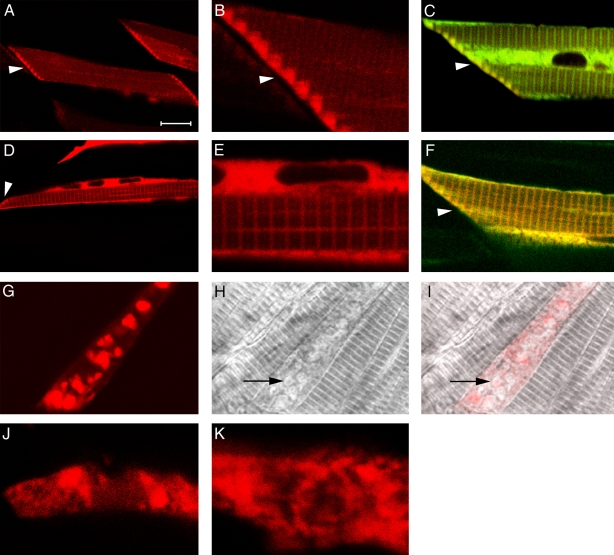
To compare the intracellular localization of Apo2 proteins with that of Unc45b, we generated fusion proteins by linking mOrange1 fluorescent protein (Shaner et al., 2004) to the C terminus of the cDNA of Apo2a and Apo2b. The fusion proteins were expressed under the control of the muscle-specific unc45b promoter in a mosaic fashion. The localization of the fusion proteins was analyzed in 4–5-d-old larvae.
In addition to a low-level distribution in the cytoplasm, we could detect enrichment of the Apo2a-mOrange1 protein at the myoseptal boundaries (Fig. 8, A and B). In this region, the expression clearly overlapped with that of Unc45b (Fig. 8 C and Fig. 7, J and K). When mOrange1 was expressed by itself without being fused to Apo2a, we did not detect this distribution (unpublished data). Interestingly, the Apo2b–mOrange fusion protein was distributed in a similar but not identical pattern. Although Apo2b-mOrange1 was expressed at lower levels at the myoseptal boundary, it was strongly enriched at the Z line of the myofibril (Fig. 8, D and E), colocalizing with Unc45b at this structure (Fig. 8 F).
Figure 8.
Localization of Apo2a and Apo2b fusion proteins. (A–C) Mosaic expression of Apo2a-mOrange1 fusion proteins in individual cells of the skeletal muscles. Cells expressing Apo2a-mOrange1 at low levels show enrichment of the fusion proteins at the myoseptal boundary (A and B, arrowheads). Low levels and more diffuse staining are present in the cytoplasm and over the myofibril. Enrichment of Apo2a-mOrange1 at the myoseptal boundary overlaps with Unc45b-TFP (C, arrowhead). (D–F) Mosaic expression of Apo2b-mOrange1 fusion proteins in individual cells of the skeletal muscles. Diffuse staining in the cytoplasm and in a striated manner at the Z line of the myofibrils was noted for this fusion protein. Apo2b–mOrange is colocalized with Unc45b over the Z line and also at the myoseptal boundary (D and F, arrowhead) in cells coexpressing Unc45b-TFP fusion protein (C). (G–I) Expression of Apo2a-mOrange1 at high levels results in aggregation of the protein in the cytoplasm (G). This results in loss of striation in highly expressing cells (H and I, arrows; overlay of G and H). (J and K) Lack of Unc45b protein leads to aggregation of Apo2a-mOrange1 (J) and Apo2b-mOrange1 fusion proteins. Bars: (A, C, D, and F) 16 µm; (B, E, J, and K) 4 µm; (G, H, and I) 10 µm.
The injection/expression protocol we used normally results in different levels of expression in individual myofibers even in the same embryo. Cells expressing Apo2a-mOrange1 or Apo2b-mOrange1 fusion proteins at high levels always formed large aggregates of fluorescence in the cytoplasm (Fig. 8 G). These cells also showed defects in striation (Fig. 8, H and I), which suggests that high levels of Apo2 proteins impair the formation of myofibrils. Such protein aggregates were not observed when high levels of mOrange1 alone or other fusion proteins such as Unc45b-GFP or Hsp90a-GFP were expressed in zebrafish embryos using the same expression systems (Etard et al., 2008; unpublished data). This suggests that Apo2 proteins have a tendency to aggregate at high levels. In addition, the Apo2 fusion proteins lose their characteristic subcellular localization when expressed in unc45b−/− embryos (Fig. 8, J and K). These punctuate aggregates in the mutant also form in cells that express only low levels of fusion protein, which suggests that in these cases, it is not caused by overexpression but rather indicates a requirement for Unc45b in the correct localization of Apo2 proteins.
Discussion
We have identified and characterized Apo2 proteins as interaction partners of Unc45b, and we have shown that lack of function of the Apo2 proteins causes defects in myoseptal structure of the developing skeletal muscle of the embryo. This interaction is specific for Unc45b and does not include Hsp90a, which is known to cooperate with Unc45b in myofibrillogenesis. This role of Unc45b and Apo2 proteins in myoseptal formation and integrity represents a novel function of Unc45b that is distinct from Unc45b’s well-established role in myofibrillogenesis.
Embryos lacking apo2 function have a dystrophic phenotype
Embryos in which Apo2a or Apo2b were knocked out showed muscle disorders that resemble those of mutations in DGC genes (Parsons et al., 2002; Bassett et al., 2003; Guyon et al., 2005; Cheng et al., 2006) and in genes that are involved in myoseptal integrity (Kudo et al., 2004; Raeker et al., 2006; Hall et al., 2007). For example, apo2a and apo2b morphants had impaired mobility and displayed cell-free spaces in the somites, just as the dystrophin-deficient sapje (Bassett et al., 2003) and the laminin-α2–deficient candyfloss mutant (Hall et al., 2007). These cell-free spaces are either the result of failed attachment during development or are caused by rupture of the attachment of the myofiber (Hall et al., 2007). The vertical myosepta, which are part of the connective tissue that transmits the force between the contractile units of each somite, were split apart at places. This suggests that not only the attachment of myofibrils to the myoseptum, but also the integrity of the myoseptum itself, is weakened in the morphants. Gaps in the myosepta were frequently correlated with the presence of giant fibers that span two somites. It has previously been shown that in the absence of normal somite boundaries, muscle cell progenitors elongate until they reach the next intact myoseptum (Henry et al., 2005). It is thus conceivable that the myoseptal defects already existed before the onset of contractions of the somitic muscle. In agreement with this idea, unc45b mutants, which are completely paralyzed from early stages of development, also show fiber detachment.
In addition to skeletal muscle, apo2a and apo2b transcripts were detected in the heart. Morphants showed reduced heart rates and developed pericardial edema, which suggests that Apo2a and -2b also have a function in the heart. It remains to be established whether adhesion of myocardiocytes is affected by lack of Apo2a/b function in the same manner as skeletal myocytes.
This is the first report ascribing a function to Apo2-like genes in any organism. Histological examination of heart and calf muscles of Apo2 knockout mice did not reveal any abnormalities (Mikl et al., 2005). The Apo2 subgroup appears to contain the most ancient members of this DNA/RNA-editing enzyme family, as it is present throughout the vertebrate lineage. In contrast, the Apo1 and Apo3 subgroups seem to be restricted to mammals (Conticello et al., 2005). It is possible that Apo1 or -3 act redundantly with Apo2 in the heart and skeletal muscle of mammals. The zebrafish model thus provides a powerful tool to test the function of apo2 in the potential absence of redundancy. Also, given the fact that the mutations in DGC genes generally present pathological defects much later in the ontogeny of mammals than in fish development, the phenotype may be subtle and may have been overlooked in the knockout mice. In light of the fact that so far only half of human congenital myopathies can be assigned to a specific gene defect (North, 2008; Sewry et al., 2008), our work identifies Apo2a and -2b as well as Unc45b as novel disease candidates for human muscular dystrophies.
The myosin chaperone Unc45b interacts with Apo2a and Apo2b
Both Apo2 proteins interact with the chaperone Unc45b. Multiple lines of evidence suggest that this interaction is specific, and that it delineates a novel function for Unc45b, distinct from the myosin folding pathway that it has previously been implicated in (Barral et al., 2002; Srikakulam and Winkelmann, 2004; Etard et al., 2007). As a prerequisite for any physical interaction, unc45b, apo2a, and apo2b are indeed coexpressed in the muscles of the skeleton and the heart. The Apo2a protein interacts with Unc45b in the yeast two-hybrid screen, and both Apo2a and Apo2b pull down Unc45b in in vitro assays. This interaction was not observed for Hsp90a and related chaperone proteins, which suggests that the interactions of Apo2a and -2b are restricted to the chaperone Unc45b. These interactions of Apo2a and 2b with Unc45b require the central and the UCS domains of Unc45b and are not mediated by the TPR domain of Unc45b, which is an Hsp90 interaction domain. Mutants of unc45b showed phenotypic traits that resemble the dystrophic phenotype seen in apo2a and apo2b morphants. Specifically, they exhibited cell-free spaces in the somites and myoseptal defects, and their myofibers were found to be detached from the myosepta. These defects were not observed in hsp90a mutants. Together with the fact that Hsp90a protein did not interact with Apo2 proteins in vitro, this underscores the notion that the Unc45b/Hsp90a and Unc45b/Apo2 interactions represent distinct pathways. This suggests a role for Unc45b that is different from myofibrillogenesis, the only hitherto known biochemical function of Unc45b in zebrafish. During myofibrillogenesis, Unc45b functions as a co-chaperone that in concert with Hsp90 folds the myosin heavy chain into a functional conformation. This is a prerequisite for myofibrillogenesis to proceed to completion (Srikakulam and Winkelmann, 2004). Our findings indicate that Unc45b, probably via its interaction with Apo2, also plays a role in myofibril attachment to the myoseptum.
In mature myofibrils, Unc45b was found to be enriched at the Z line, whereas the cytoplasm of the myofibers contained lower levels of the chaperone (Etard et al., 2008). Here, we show that Unc45b is not only enriched at the Z line, but that high levels of Unc45b are also detected in the cytoplasm immediately adjacent to the myosepta. In this region, high levels of Apo2a and to a lesser degree also Apo2b fusion proteins are found. In addition, Apo2b-mOrange1 is enriched over the Z lines. Thus, Apo2 proteins colocalize to subcellular sites with high levels of Unc45b protein, in line with the notion of a direct interaction between the Apo2 and Unc45b proteins.
The sub-myoseptal cytoplasm is also enriched for the mRNAs of DGC proteins (Bolaños-Jimenez et al., 2001), which suggests that this subcellular region is an active site of protein synthesis. A straightforward interpretation of our results is that the Apo2 proteins and possibly other myoseptal proteins are clients of the chaperone Unc45b, requiring Unc45b for correct folding. In agreement, Apo2 fusion proteins tend to form cytoplasmic aggregates when expressed at high levels in muscle cells of wild-type embryos. However, it cannot be excluded that the proteins act in complexes that have different functions; for example, directly maintaining the integrity of the myoseptal junction.
The role of Apo2 proteins
Apo2a and 2b are highly related and coexpressed in the skeletal and cardiac muscles. Both of them interact with Unc45b. The phenotype of the Apo2a and -2b morphants suggests that the two proteins act in the same pathway, but in a nonredundant fashion. The absolute levels of Apo2a and Apo2b proteins may be critical; however, knockdown of both proteins simultaneously does not increase the severity of the phenotype (unpublished data). Analysis of the crystal structure of human Apo2 suggests that the proteins form tetramers (Prochnow et al., 2007). Hence, an alternative explanation for the nonredundant action of the two Apo2 proteins could be that they function in the context of heteromeric complexes composed of Apo2a and Apo2b subunits. In support of this notion, our pull-down experiments suggest that the two Apo2 proteins can indeed form heteromeric complexes.
Apo proteins contain the active site of the cytidine deaminases ([H/C]-[A/V]-E-[X]24-30-P-C-[X]2-C), in common with other members of the family (Navaratnam and Sarwar, 2006). This motif is also present in zebrafish Apo2 proteins and the Apo2 homologues of other species. It is, however, unclear whether Apo2 subfamily members act indeed as deaminases (Liao et al., 1999; Mikl et al., 2005). In contrast to Apo3 proteins, Apo1 has a very high substrate specificity (Blanc and Davidson, 2003). Only a few mRNA substrates in addition to the apolipoprotein mRNA have been described so far (Skuse et al., 1996; Yamanaka et al., 1997). Interestingly, mammalian Apo2 inhibits the deaminase activity of Apo1 on apolipoprotein mRNA (Anant et al., 2001). This suggests that Apo2 protein can interfere with the formation or function of the Apo1 protein–mRNA complex. Unfortunately, the recognition sequence of the Apo1 complex is so degenerate and short that a search of the zebrafish genome with the mammalian Apo1 recognition sequence did not yield significant results (unpublished data). The precise targets of Apo2a and -2b thus remain a mystery. Our work, however, implicates the mRNAs of myoseptal proteins as potential targets. It has recently been demonstrated that splicing of minor-class introns occurs in the cytoplasm, which indicates that RNA processing is not restricted to the nucleus (König et al., 2007). Thus, Apo2a and -2b proteins may have a function in cytoplasmic RNA editing or repair, modifying or maintaining the large mRNAs of the proteins that form the DGC. Alternatively, Apo2a and Apo2b proteins could interact with Unc45b and other myoseptal proteins to establish/maintain the integrity of the myoseptum. Irrespective of the precise targets, our work has uncovered a novel interaction between the myosin chaperone Unc45b and Apo2 proteins, and has implicated apo2 genes as new potential candidates underlying human dystrophic myopathies.
Materials and methods
Fish stocks
Fish were bred and raised at 28.5°C as described previously (Westerfield, 1993). unc45bsb60 (previously called steifsb60) and hsp90au45 (previously called hsp90a.1u45) have been described previously (Etard et al., 2007, 2008; Hawkins et al., 2008). The mlc2 mutant was a gift of S. Just and W. Rottbauer (Otto Meyerhof Zentrum, Heidelberg, Germany; Rottbauer et al., 2006). The AB wild-type line (University of Oregon, Eugene) was used for all the experiments, and the mutations were also kept in this background. Sexually mature fish were crossed in couples, and eggs were collected after being laid. For experiments, fertilized eggs were raised in 1× Instant Ocean salt solution (Aquarium Systems, Inc.) supplemented with 200 µM 1-phenyl 2-thiourea (PTU) to suppress melanogenesis.
Two-hybrid screen
The two-hybrid interaction screen was performed by Hybrigenics. In brief, full-length Unc45b protein was used as the bait, sub-cloned into pB27 as a C-terminal fusion with LexA, and then screened against a prey library cloned from 18–20-hpf zebrafish embryos. The yeast two-hybrid library is based on 10 million primary clones and was screened with 9.8× coverage.
Cloning
The construction of unc45b-gfp and the truncated variants has been described previously (Etard et al., 2008). In summary, all the unc45b-gfp expression constructs were cloned into the pCS2+gfp vector, to the N terminus of GFP. The expression is driven by a cytomegalovirus promoter. Apo2a cDNA (Zebrafish Gene Collection, 110762, seventh assembly [Zv7]) was obtained from Hybrigenics (see Two-hybrid screen). 5′ and 3′ rapid amplification of cDNA ends (RACE) PCRs were performed to clone the full-length coding sequence (FirstChoice RLM-RACE kit; Applied Biosystems). For construction of myc-apo2a and apo2a-gfp, the apo2a cDNA was amplified from cDNA obtained from 72-hpf embryos, and cloned into pCS2+ plasmids providing in-frame 5′ myc tags and 3′ gfp coding sequences, respectively.
The zebrafish genome was searched (Zebrafish Gene Collection release Zv7) for apo2a homologues by blasting the genome with the apo2a sequence. A sequence corresponding to apo2b was found (LOC567047). The apo2b cDNA was amplified with primers matching the apo2b gene. The resulting fragments were cloned into pCS2+gfp. For the generation of gst-apo2a and gst-apo2b, full-length cDNAs were cloned into the pGEX-4T-3 vector (GE Healthcare).
The mOrange1 fluorescent protein was fused in frame with the C terminus of Apo2a and -2b. We used a long flexible linker between the fluorescent protein and the cDNA of the gene of interest, which consisted of Ser + Arg + Ala + 3(4Gly + Ser). The mTFP1 fluorescent protein (Ai et al., 2006) was fused to the C terminus of Unc45b using the linker sequence Glu + Pro + Arg + Ala + 3(4Gly + Ser). In both cases, the start codon of the fluorescent protein was removed. The fusion proteins were expressed under the highly muscle-specific zebrafish Unc45b promoter.
For the synthesis of in situ hybridization probes, we used the full-length apo2 cDNAs cloned into the pGEM-T Easy Vector System (Promega). All constructs were verified by sequencing.
Histology, immunohistochemistry, in situ hybridization, and microscopy
We performed whole-mount in situ hybridization and immunohistochemistry as described previously (Crow and Stockdale, 1986; Oxtoby and Jowett, 1993; Costa et al., 2002). We used monoclonal antibodies directed against slow muscle myosin (F59, (Crow and Stockdale, 1986), sarcomeric myosin (A4.1025, 1:100; Developmental Studies Hybridoma Bank [DSHB], University of Iowa), β-sarcoglycan (1:100 Novocastra; Leica), laminin (1:60; Sigma-Aldrich), and α-dystroglycan (1:60; Sigma-Aldrich), and the anti Myc-epitope antibody 9E10 (1:25; DSHB). Antibodies for Unc45b were produced by injection of rabbits with the following peptide: GVSLLKEMYKKTKN (BioGenes). This peptide is unique in the zebrafish genome (Zebrafish Gene Collection release Zv7). For anti-Unc45b staining, fish were fixed at −20°C in DMSO/methanol (1:5) for 2 h.
Anti–mouse or –rabbit IgG Cy3-conjugated secondary antibodies were used for fluorescent detection (1:1,000; Sigma-Aldrich).
A confocal microscope (TCS SP2) and the accompanying software (LCS; both from Leica) were used to analyze antibody staining and fusion protein localization. For visualization of fusion proteins, living zebrafish larvae were immobilized with 0.02% 3-aminobenzoic acid methylester (MESAB; Invitrogen), immersed in a water droplet on a microscopy slide, and imaged with a dip-in 63× objective lens (0.90 NA, HCX Apochromat water; Leica). 458-nm and 476-nm laser lines were used for mTFP1 excitation; 488 nm was used for GFP excitation and 514 nm was used for mOrange1 excitation. The observations were performed at 22°C. Birefringence was analyzed with a microscope (MZ16F; Leica) as described previously (Behra et al., 2002). Image editing was limited to minimal sharpening and exposure adjustment.
Protein pull-down experiments
Expression of GST-Apo2a and GST-Apo2b recombinant proteins was induced in BL21D3 cells by adding 1 mM IPTG at OD600 = 0.8. After incubation for 4 h at 30°C, proteins were extracted by sonication in lysis buffer (1× PBS, 0.1% Triton X-100, 1 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride). For pull-down experiments, the bacterial protein lysates were incubated for 1 h at 4°C with beads coated with the tripeptide glutathione (GE Healthcare). The beads were pelleted for 1 min at 550 g and washed three times with TEN buffer (10 mM tris/HCl, pH 7.4, 150 mM NaCl, 0.5% nonidet P40, and 1 mM EDTA). Unc45b-GFP and all the deletion variants as well as MYC-Apo2a and Apo2b-GFP proteins were synthesized in rabbit reticulocyte lysate (TNT kit; Promega). 15 µl of the TNT lysate was incubated at room temperature for 1 h with glutathione-sepharose–coupled proteins. After three successive washes, bound proteins were detached from the beads by boiling for 10 min at 95°C with 20 µl of 5× SDS-PAGE sample buffer (0.5M Tris/HCl, pH 6.8, 10% [wt/vol] SDS, 20% glycerol, 0.0025% pyronin Y, and 25% β-mercaptoethanol). Proteins were separated by 10% SDS-PAGE, transferred onto a nitrocellulose filter, and incubated with different primary antibodies (1:5,000 GFP [Torrey Pines Biolabs, Inc.] or 1:500 MYC [9E10; DSHB]) for one night at 4°C. After washing three times, a secondary antibody (goat anti–rabbit Alexa Fluor 680 for GFP and goat anti–mouse Alexa Fluor 680 for MYC; Invitrogen) was applied for 1 h at room temperature. Blots were visualized using an infrared imaging system (Odyssey; LI-COR Biosciences).
Microinjections
Injections were performed as described previously (Müller et al., 1999). In brief, zebrafish eggs were collected shortly after being laid. Cleaned eggs were transferred to a Petri dish with a minimal amount of water. Embryos were injected (FemtoJet; Eppendorf) through the chorion into the yolk compartment at the one-cell stage. Injection needles were pulled from borosilicate glass capillary tubes with filament (Warner Instruments) using a micropipette puller (Sutter Instrument Co.). Plasmid DNA was purified with the Plasmid Maxi kit (QIAGEN) and diluted in water to 20–50 ng/µl for injection. Phenol red was added to the DNA and RNA samples before injection (0.1% final concentration). Morpholinos (Gene Tools, LLC) were injected at the following concentrations: 0.2 mM Mo(ATG)-apo2 (5′-TGCTGCCCTTTCTATCGGCCATCTC-3′), 0.2 mM Mo(ATG)-apo2a-contr (5′-TGCTCCCGTTTGTATCCGCGATCTC-3′), 1.6 mM Mo(UTR)apo2a (5′-ACACACCTCAGTATTTATAACAGC-3′), 0.5 mM Mo-p53 (Robu et al., 2007), 0.6 mM Mo(ATG)-Apo2b (5′-CTTGCTGTCCTTTTTGTCTGCCATG-3′), and 1.8 mM Mo(UTR)-apo2b (5′-TTAGCCGTGCTGTAAAATGAGTCTC-3′). All dilutions were made in distilled water.
Online supplemental material
Fig. S1 shows a sequence comparison of different Apo2 orthologues and a corresponding phylogram. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912125/DC1.
Acknowledgments
We thank N. Borel and her fish house team for fish care, M. Rastegar for help with microscopy, S. Just and W. Rottbauer for the gift of mlc2 mutant embryos, and T. Dickmeis, Nicholas S. Foulkes, and J. Sleeman for critical reading of the manuscript. The yeast two-hybrid screens were carried out by Hybrigenics, Paris.
This work was supported by the Forschungszentrum Karlsruhe in the Helmholtz Association, the European commission IP ZF-MODELS and the Association Française contre les Myopathies (AFM). U. Roostalu was supported by PhD Scholarship from the Boehringer Ingelheim Fonds.
Footnotes
Abbreviations used in this paper:
- AID
- activation-induced (cytidine) deaminase enzyme
- Apo
- Apobec
- DGC
- dystrophin–glycoprotein complex
- DIC
- differential interference contrast
- hpf
- hours postfertilization
- TPR
- tetratricopeptide repeat
- UTR
- untranslated region
References
- Ai H.W., Henderson J.N., Remington S.J., Campbell R.E. 2006. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem. J. 400:531–540 10.1042/BJ20060874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amali A.A., Lin C.J., Chen Y.H., Wang W.L., Gong H.Y., Rekha R.D., Lu J.K., Chen T.T., Wu J.L. 2008. Overexpression of Myostatin2 in zebrafish reduces the expression of dystrophin associated protein complex (DAPC) which leads to muscle dystrophy. J. Biomed. Sci. 15:595–604 10.1007/s11373-008-9250-2 [DOI] [PubMed] [Google Scholar]
- Anant S., Mukhopadhyay D., Sankaranand V., Kennedy S., Henderson J.O., Davidson N.O. 2001. ARCD-1, an apobec-1-related cytidine deaminase, exerts a dominant negative effect on C to U RNA editing. Am. J. Physiol. Cell Physiol. 281:C1904–C1916 [DOI] [PubMed] [Google Scholar]
- Anderson M.J., Pham V.N., Vogel A.M., Weinstein B.M., Roman B.L. 2008. Loss of unc45a precipitates arteriovenous shunting in the aortic arches. Dev. Biol. 318:258–267 10.1016/j.ydbio.2008.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral J.M., Bauer C.C., Ortiz I., Epstein H.F. 1998. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J. Cell Biol. 143:1215–1225 10.1083/jcb.143.5.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral J.M., Hutagalung A.H., Brinker A., Hartl F.U., Epstein H.F. 2002. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 295:669–671 10.1126/science.1066648 [DOI] [PubMed] [Google Scholar]
- Bassett D., Currie P.D. 2004. Identification of a zebrafish model of muscular dystrophy. Clin. Exp. Pharmacol. Physiol. 31:537–540 10.1111/j.1440-1681.2004.04030.x [DOI] [PubMed] [Google Scholar]
- Bassett D.I., Bryson-Richardson R.J., Daggett D.F., Gautier P., Keenan D.G., Currie P.D. 2003. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 130:5851–5860 10.1242/dev.00799 [DOI] [PubMed] [Google Scholar]
- Behra M., Cousin X., Bertrand C., Vonesch J.L., Biellmann D., Chatonnet A., Strähle U. 2002. Acetylcholinesterase is required for neuronal and muscular development in the zebrafish embryo. Nat. Neurosci. 5:111–118 10.1038/nn788 [DOI] [PubMed] [Google Scholar]
- Behra M., Etard C., Cousin X., Strähle U. 2004. The use of zebrafish mutants to identify secondary target effects of acetylcholine esterase inhibitors. Toxicol. Sci. 77:325–333 10.1093/toxsci/kfh020 [DOI] [PubMed] [Google Scholar]
- Blanc V., Davidson N.O. 2003. C-to-U RNA editing: mechanisms leading to genetic diversity. J. Biol. Chem. 278:1395–1398 10.1074/jbc.R200024200 [DOI] [PubMed] [Google Scholar]
- Bolaños-Jiménez F., Bordais A., Behra M., Strähle U., Mornet D., Sahel J., Rendón A. 2001. Molecular cloning and characterization of dystrophin and Dp71, two products of the Duchenne Muscular Dystrophy gene, in zebrafish. Gene. 274:217–226 10.1016/S0378-1119(01)00606-0 [DOI] [PubMed] [Google Scholar]
- Cheng L., Guo X.F., Yang X.Y., Chong M., Cheng J., Li G., Gui Y.H., Lu D.R. 2006. Delta-sarcoglycan is necessary for early heart and muscle development in zebrafish. Biochem. Biophys. Res. Commun. 344:1290–1299 10.1016/j.bbrc.2006.03.234 [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Thomas C.J., Petersen-Mahrt S.K., Neuberger M.S. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22:367–377 10.1093/molbev/msi026 [DOI] [PubMed] [Google Scholar]
- Costa M.L., Escaleira R.C., Rodrigues V.B., Manasfi M., Mermelstein C.S. 2002. Some distinctive features of zebrafish myogenesis based on unexpected distributions of the muscle cytoskeletal proteins actin, myosin, desmin, alpha-actinin, troponin and titin. Mech. Dev. 116:95–104 10.1016/S0925-4773(02)00149-1 [DOI] [PubMed] [Google Scholar]
- Crow M.T., Stockdale F.E. 1986. Myosin expression and specialization among the earliest muscle fibers of the developing avian limb. Dev. Biol. 113:238–254 10.1016/0012-1606(86)90126-0 [DOI] [PubMed] [Google Scholar]
- D’Amico A., Bertini E. 2008. Congenital myopathies. Curr. Neurol. Neurosci. Rep. 8:73–79 10.1007/s11910-008-0012-3 [DOI] [PubMed] [Google Scholar]
- Deniziak M., Thisse C., Rederstorff M., Hindelang C., Thisse B., Lescure A. 2007. Loss of selenoprotein N function causes disruption of muscle architecture in the zebrafish embryo. Exp. Cell Res. 313:156–167 10.1016/j.yexcr.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Etard C., Behra M., Fischer N., Hutcheson D., Geisler R., Strähle U. 2007. The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev. Biol. 308:133–143 10.1016/j.ydbio.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Etard C., Roostalu U., Strähle U. 2008. Shuttling of the chaperones Unc45b and Hsp90a between the A band and the Z line of the myofibril. J. Cell Biol. 180:1163–1175 10.1083/jcb.200709128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge L., Diiorio P., Sagerström C.G. 2002. A zebrafish unc-45-related gene expressed during muscle development. Dev. Dyn. 224:457–460 10.1002/dvdy.10123 [DOI] [PubMed] [Google Scholar]
- Flinn L., Bretaud S., Lo C., Ingham P.W., Bandmann O. 2008. Zebrafish as a new animal model for movement disorders. J. Neurochem. 106:1991–1997 10.1111/j.1471-4159.2008.05463.x [DOI] [PubMed] [Google Scholar]
- Guyon J.R., Kudryashova E., Potts A., Dalkilic I., Brosius M.A., Thompson T.G., Beckmann J.S., Kunkel L.M., Spencer M.J. 2003a. Calpain 3 cleaves filamin C and regulates its ability to interact with gamma- and delta-sarcoglycans. Muscle Nerve. 28:472–483 10.1002/mus.10465 [DOI] [PubMed] [Google Scholar]
- Guyon J.R., Mosley A.N., Zhou Y., O’Brien K.F., Sheng X., Chiang K., Davidson A.J., Volinski J.M., Zon L.I., Kunkel L.M. 2003b. The dystrophin associated protein complex in zebrafish. Hum. Mol. Genet. 12:601–615 10.1093/hmg/12.6.601 [DOI] [PubMed] [Google Scholar]
- Guyon J.R., Mosley A.N., Jun S.J., Montanaro F., Steffen L.S., Zhou Y., Nigro V., Zon L.I., Kunkel L.M. 2005. Delta-sarcoglycan is required for early zebrafish muscle organization. Exp. Cell Res. 304:105–115 10.1016/j.yexcr.2004.10.032 [DOI] [PubMed] [Google Scholar]
- Guyon J.R., Steffen L.S., Howell M.H., Pusack T.J., Lawrence C., Kunkel L.M. 2007. Modeling human muscle disease in zebrafish. Biochim. Biophys. Acta. 1772:205–215 [DOI] [PubMed] [Google Scholar]
- Guyon J.R., Goswami J., Jun S.J., Thorne M., Howell M., Pusack T., Kawahara G., Steffen L.S., Galdzicki M., Kunkel L.M. 2009. Genetic isolation and characterization of a splicing mutant of zebrafish dystrophin. Hum. Mol. Genet. 18:202–211 10.1093/hmg/ddn337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.E., Bryson-Richardson R.J., Berger S., Jacoby A.S., Cole N.J., Hollway G.E., Berger J., Currie P.D. 2007. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc. Natl. Acad. Sci. USA. 104:7092–7097 10.1073/pnas.0700942104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., Malim M.H. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell. 113:803–809 10.1016/S0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- Hawkins T.A., Haramis A.P., Etard C., Prodromou C., Vaughan C.K., Ashworth R., Ray S., Behra M., Holder N., Talbot W.S., et al. 2008. The ATPase-dependent chaperoning activity of Hsp90a regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development. 135:1147–1156 10.1242/dev.018150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C.A., McNulty I.M., Durst W.A., Munchel S.E., Amacher S.L. 2005. Interactions between muscle fibers and segment boundaries in zebrafish. Dev. Biol. 287:346–360 10.1016/j.ydbio.2005.08.049 [DOI] [PubMed] [Google Scholar]
- Hoppe T., Cassata G., Barral J.M., Springer W., Hutagalung A.H., Epstein H.F., Baumeister R. 2004. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell. 118:337–349 10.1016/j.cell.2004.07.014 [DOI] [PubMed] [Google Scholar]
- Ingham P.W. 2009. The power of the zebrafish for disease analysis. Hum. Mol. Genet. 18(R1):R107–R112 10.1093/hmg/ddp091 [DOI] [PubMed] [Google Scholar]
- Janiesch P.C., Kim J., Mouysset J., Barikbin R., Lochmüller H., Cassata G., Krause S., Hoppe T. 2007. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat. Cell Biol. 9:379–390 10.1038/ncb1554 [DOI] [PubMed] [Google Scholar]
- Karlsson J., von Hofsten J., Olsson P.E. 2001. Generating transparent zebrafish: a refined method to improve detection of gene expression during embryonic development. Mar Biotechnol (NY). 3:522–527 10.1007/s1012601-0053-4 [DOI] [PubMed] [Google Scholar]
- Knott T.J., Pease R.J., Powell L.M., Wallis S.C., Rall S.C., Jr., Innerarity T.L., Blackhart B., Taylor W.H., Marcel Y., Milne R., et al. 1986. Complete protein sequence and identification of structural domains of human apolipoprotein B. Nature. 323:734–738 10.1038/323734a0 [DOI] [PubMed] [Google Scholar]
- König H., Matter N., Bader R., Thiele W., Müller F. 2007. Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell. 131:718–729 10.1016/j.cell.2007.09.043 [DOI] [PubMed] [Google Scholar]
- Kudo H., Amizuka N., Araki K., Inohaya K., Kudo A. 2004. Zebrafish periostin is required for the adhesion of muscle fiber bundles to the myoseptum and for the differentiation of muscle fibers. Dev. Biol. 267:473–487 10.1016/j.ydbio.2003.12.007 [DOI] [PubMed] [Google Scholar]
- Liao W., Hong S.H., Chan B.H., Rudolph F.B., Clark S.C., Chan L. 1999. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem. Biophys. Res. Commun. 260:398–404 10.1006/bbrc.1999.0925 [DOI] [PubMed] [Google Scholar]
- Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 424:99–103 10.1038/nature01709 [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Marusawa H., Endo Y., Ueda Y., Matsumoto Y., Chiba T. 2006. Expression of APOBEC2 is transcriptionally regulated by NF-kappaB in human hepatocytes. FEBS Lett. 580:731–735 10.1016/j.febslet.2005.12.081 [DOI] [PubMed] [Google Scholar]
- Mikl M.C., Watt I.N., Lu M., Reik W., Davies S.L., Neuberger M.S., Rada C. 2005. Mice deficient in APOBEC2 and APOBEC3. Mol. Cell. Biol. 25:7270–7277 10.1128/MCB.25.16.7270-7277.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Anant S., Lee R.M., Kennedy S., Viskochil D., Davidson N.O. 2002. C—>U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am. J. Hum. Genet. 70:38–50 10.1086/337952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Chang B., Albert S., Fischer N., Tora L., Strähle U. 1999. Intronic enhancers control expression of zebrafish sonic hedgehog in floor plate and notochord. Development. 126:2103–2116 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476 10.1074/jbc.274.26.18470 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Navaratnam N., Sarwar R. 2006. An overview of cytidine deaminases. Int. J. Hematol. 83:195–200 10.1532/IJH97.06032 [DOI] [PubMed] [Google Scholar]
- North K. 2008. What’s new in congenital myopathies? Neuromuscul. Disord. 18:433–442 10.1016/j.nmd.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Nyamsuren O., Faggionato D., Loch W., Schulze E., Baumeister R. 2007. A mutation in CHN-1/CHIP suppresses muscle degeneration in Caenorhabditis elegans. Dev. Biol. 312:193–202 10.1016/j.ydbio.2007.09.033 [DOI] [PubMed] [Google Scholar]
- Oxtoby E., Jowett T. 1993. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 21:1087–1095 10.1093/nar/21.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa I., Astier C., Kwiatek O., Raynaud F., Bonnal C., Lebart M.C., Roustan C., Benyamin Y. 1999. Alpha actinin-CapZ, an anchoring complex for thin filaments in Z-line. J. Muscle Res. Cell Motil. 20:187–197 10.1023/A:1005489319058 [DOI] [PubMed] [Google Scholar]
- Parsons M.J., Campos I., Hirst E.M., Stemple D.L. 2002. Removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos. Development. 129:3505–3512 [DOI] [PubMed] [Google Scholar]
- Prochnow C., Bransteitter R., Klein M.G., Goodman M.F., Chen X.S. 2007. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 445:447–451 10.1038/nature05492 [DOI] [PubMed] [Google Scholar]
- Raeker M.O., Su F., Geisler S.B., Borisov A.B., Kontrogianni-Konstantopoulos A., Lyons S.E., Russell M.W. 2006. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev. Dyn. 235:2018–2029 10.1002/dvdy.20812 [DOI] [PubMed] [Google Scholar]
- Robu M.E., Larson J.D., Nasevicius A., Beiraghi S., Brenner C., Farber S.A., Ekker S.C. 2007. p53 activation by knockdown technologies. PLoS Genet. 3:e78 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottbauer W., Wessels G., Dahme T., Just S., Trano N., Hassel D., Burns C.G., Katus H.A., Fishman M.C. 2006. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ. Res. 99:323–331 10.1161/01.RES.0000234807.16034.fe [DOI] [PubMed] [Google Scholar]
- Schafer D.A., Waddle J.A., Cooper J.A. 1993. Localization of CapZ during myofibrillogenesis in cultured chicken muscle. Cell Motil. Cytoskeleton. 25:317–335 10.1002/cm.970250403 [DOI] [PubMed] [Google Scholar]
- Sewry C.A. 2008. Pathological defects in congenital myopathies. J. Muscle Res. Cell Motil. 29:231–238 10.1007/s10974-008-9155-8 [DOI] [PubMed] [Google Scholar]
- Sewry C.A., Jimenez-Mallebrera C., Muntoni F. 2008. Congenital myopathies. Curr. Opin. Neurol. 21:569–575 10.1097/WCO.0b013e32830f93c7 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Skuse G.R., Cappione A.J., Sowden M., Metheny L.J., Smith H.C. 1996. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res. 24:478–485 10.1093/nar/24.3.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikakulam R., Winkelmann D.A. 2004. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 117:641–652 10.1242/jcs.00899 [DOI] [PubMed] [Google Scholar]
- Teng B., Burant C.F., Davidson N.O. 1993. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 260:1816–1819 10.1126/science.8511591 [DOI] [PubMed] [Google Scholar]
- Thornhill P., Bassett D., Lochmüller H., Bushby K., Straub V. 2008. Developmental defects in a zebrafish model for muscular dystrophies associated with the loss of fukutin-related protein (FKRP). Brain. 131:1551–1561 10.1093/brain/awn078 [DOI] [PubMed] [Google Scholar]
- Wedekind J.E., Dance G.S., Sowden M.P., Smith H.C. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19:207–216 10.1016/S0168-9525(03)00054-4 [DOI] [PubMed] [Google Scholar]
- Westerfield M. 1993. The Zebrafish Book. University of Oregon Press, Eugene, OR: 300 pp [Google Scholar]
- Yamanaka S., Poksay K.S., Balestra M.E., Zeng G.Q., Innerarity T.L. 1994. Cloning and mutagenesis of the rabbit ApoB mRNA editing protein. A zinc motif is essential for catalytic activity, and noncatalytic auxiliary factor(s) of the editing complex are widely distributed. J. Biol. Chem. 269:21725–21734 [PubMed] [Google Scholar]
- Yamanaka S., Poksay K.S., Arnold K.S., Innerarity T.L. 1997. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 11:321–333 10.1101/gad.11.3.321 [DOI] [PubMed] [Google Scholar]
- Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435–442 10.1038/nsmb758 [DOI] [PubMed] [Google Scholar]