Interpolar microtubules are sorted by the directional instability resulting from antagonistic molecular motors, not a stable balance of force.
Abstract
During cell division, different molecular motors act synergistically to rearrange microtubules. Minus end–directed motors are thought to have a dual role: focusing microtubule ends to poles and establishing together with plus end–directed motors a balance of force between antiparallel microtubules in the spindle. We study here the competing action of Xenopus laevis kinesin-14 and -5 in vitro in situations in which these motors with opposite directionality cross-link and slide microtubules. We find that full-length kinesin-14 can form microtubule asters without additional factors, whereas kinesin-5 does not, likely reflecting an adaptation to mitotic function. A stable balance of force is not established between two antiparallel microtubules with these motors. Instead, directional instability is generated, promoting efficient motor and microtubule sorting. A nonmotor microtubule cross-linker can suppress directional instability but also impedes microtubule sorting, illustrating a conflict between stability and dynamicity of organization. These results establish the basic organizational properties of these antagonistic mitotic motors and a microtubule bundler.
Introduction
Proper cytoskeleton function requires collective motor protein activities. Various intracellular processes depend on the action of antagonistically acting motors. An important example is the assembly of the bipolar spindle that segregates chromosomes during cell division (Nédélec et al., 2003; Brust-Mascher and Scholey, 2007; Walczak and Heald, 2008). Minus end–directed motors such as dynein and kinesin-14 contribute to focusing minus ends of microtubules at spindle poles with varying relative importance in different experimental systems (Endow et al., 1994; Matthies et al., 1996; Merdes et al., 1996, 2000; Endow and Komma, 1997; Walczak et al., 1997, 1998; Mountain et al., 1999). In contrast to the situation at spindle poles, the plus ends of microtubules are not focused by plus end–directed motors in the spindle center. Instead, interpolar microtubules establish an antiparallel microtubule overlap, which ultimately ensures the bipolarity of the spindle (Sawin et al., 1992; Heck et al., 1993; Mayer et al., 1999). How this overlap is generated is not understood at a mechanistic level. Based on loss-of-function studies, it has been proposed that it is the consequence of balanced activities of motors with opposite directionality (Saunders et al., 1997; Mountain et al., 1999; Sharp et al., 2000; Mitchison et al., 2005). In most systems, the plus end–directed mitotic motor kinesin-5 is thought to push half-spindles apart by antiparallel microtubule sliding in the central overlap region of the spindle (Miyamoto et al., 2004; Shirasu-Hiza et al., 2004), whereas minus end–directed motors such as dynein or kinesin-14 have been proposed to counteract this activity (Saunders et al., 1997; Mountain et al., 1999; Sharp et al., 2000).
Kinesin-5 is a tetrameric motor with a bipolar configuration having kinesin domain dimers at either end of the elongated molecule (a motor–motor cross-linker; Kashina et al., 1996). In contrast, kinesin-14 is a dimeric molecule with motor domains at only one end of the molecule and diffusible nonmotor microtubule binding domains at the other end (a motor–handle cross-linker; Braun et al., 2009; Fink et al., 2009). Despite their different molecular domain compositions, both motors have been shown to be able to cross-link and slide antiparallel microtubules (Kapitein et al., 2005; Oladipo et al., 2007; Braun et al., 2009; Fink et al., 2009). Such cross-linking combined with directional motility is a key property for the capacity of a motor to organize microtubules in space (Nédélec et al., 1997). In addition to the differences in their domain composition, the two motors also differ in their kinetic properties. Kinesin-5 was shown to be weakly processive (Kwok et al., 2006; Valentine et al., 2006; Kapitein et al., 2008). In contrast, kinesin-14 has been demonstrated to be nonprocessive (Case et al., 1997; deCastro et al., 2000).
How mitotic motors with opposite directionality behave in competition when acting collectively on microtubules is largely unknown. Theoretical work has predicted that mixtures of motors can generate unstable states (Badoual et al., 2002; Grill et al., 2005; Müller et al., 2008). This agrees with microtubule gliding experiments with mixtures of immobilized nonmitotic kinesin-1 and dynein in which bidirectional microtubule transport was observed (Vale et al., 1992). Similar observations were made with mixtures of surface-immobilized Drosophila melanogaster kinesin-5 and -14 (Tao et al., 2006). However, as a consequence of motor immobilization, the configuration of a microtubule gliding experiment is artificial as compared with the behavior of microtubule cross-linking motors in cells. Therefore, it is still an open question whether mitotic motors can establish a stable balance of force when connecting antiparallel microtubules.
Previously, mixtures of artificially constructed oligomeric motors and microtubules have been shown to self-organize into microtubule asters in vitro (Nédélec et al., 1997; Surrey et al., 2001). However, such self-organization experiments have not been performed with natural mitotic motors. Consequently, it is unclear whether full-length kinesin-5 or -14 can autonomously focus microtubule ends into poles without other factors and how mixtures of natural antagonistic kinesins and dynamic microtubules self-organize in space.
In this study, we have examined the interplay between the antagonistic motors kinesin-5 and -14 in vitro in two relevant experimental configurations with respect to their role in mitosis. We show that the two motors do not generate a stable balance of force when acting in antiparallel microtubule pairs. Instead, the system shows directional instability. By slowing down microtubule sliding through protein friction, a nonmotile microtubule cross-linker can stabilize overlaps. In self-organization experiments, we demonstrate that natural kinesin-14 has the intrinsic ability to focus microtubule minus ends to asters. This is in contrast to kinesin-5, which did not organize microtubule asters in our in vitro system. A nonmotile microtubule cross-linker impedes motor-mediated self-organization. These results suggest, first, that the properties of the two studied mitotic motors are adapted for their specific organizational tasks in the spindle and, second, that there is an intrinsic conflict between antiparallel microtubule overlap stabilization by a static microtubule bundler and motor protein–driven microtubule organization, which requires dynamic microtubule sorting.
Results
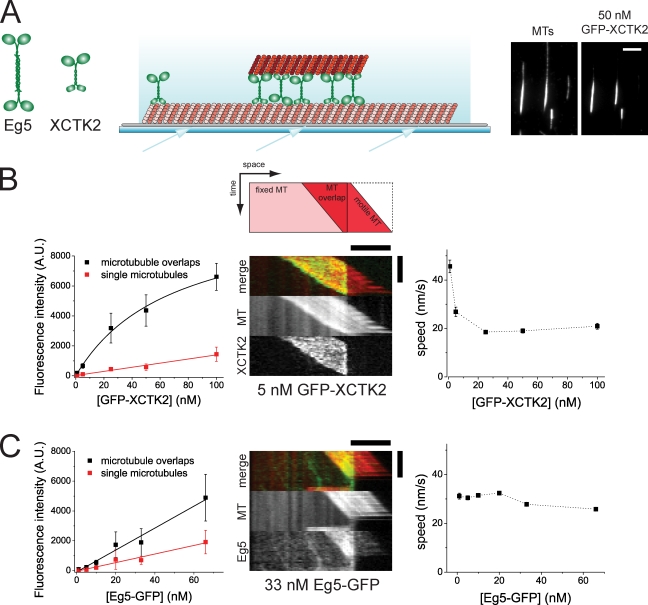
We studied collective antagonistic motor action of purified kinesin-14 and -5 from Xenopus laevis, also called XCTK2 (Walczak et al., 1997) and Eg5 (Sawin et al., 1992), respectively. Using total internal reflection fluorescence (TIRF) microscopy, we first examined how GFP-labeled motors interact in vitro with the pairs of microtubules that they cross-link (Fig. 1). These pairs consisted of one surface-immobilized microtubule that was dimly labeled with Cy5 and a second more strongly labeled, motor-tethered microtubule (Fig. 1 A). Quantification of the fluorescence intensity of GFP-labeled XCTK2 connecting microtubule pairs showed that this motor was strongly enriched in microtubule overlap regions (apparent Kd = 69 nM), where it accumulated at low concentrations around 10 times more strongly than it bound to individual microtubules. (Fig. 1 B, left; and Video 1). This accumulation is an expected consequence of an effectively increased affinity resulting from simultaneous binding of the N-terminal nonmotile microtubule binding site of XCTK2 to one microtubule and of the C-terminal motor part to the second microtubule of a microtubule pair (Jencks, 1981; Butner and Kirschner, 1991).
Figure 1.
GFP-XCTK2 accumulates between microtubule pairs in contrast to Eg5-GFP. (A, left) Schematic representations of the molecular architectures of tetrameric plus end–directed motor Eg5 and dimeric minus end–directed motor XCTK2. Large round and small rectangular symbols indicate motor domains and nonmotor microtubule binding domains, respectively. (middle) Scheme of a microtubule pair in the sliding assay. Long, dimly fluorescently labeled, biotinylated microtubules are attached to a biotin-PEG–coated (gray) glass surface via neutravidin links. Short, brightly fluorescently labeled, unbiotinylated microtubules (top) are bound to the immobilized microtubules via cross-linking motor proteins. The arrows on the bottom indicate the incidence angle of the excitation light, which is totally internally reflected and gives rise to an evanescent field (light blue gradient). (right) TIRF microscopy images of Cy5-labeled microtubule (MT) pairs cross-linked by GFP-XCTK2 present in solution at a concentration of 50 nM, illustrating accumulation of GFP-XCTK2 in microtubule overlaps. (B) Quantification of GFP-XCTK2 in Cy5-microtubule pairs in assay buffer containing 120 mM KCl. (left) Binding curves showing averaged TIRF intensities of GFP-XCTK2 along microtubule overlaps and single microtubules at varying concentrations of GFP-XCTK2. The error bars are standard deviations, indicating the width of the fluorescence intensity distribution. The Kd for the binding of GFP-XCTK2 to the microtubule overlap derived from the fit is 69 ± 15 nM (error is standard error of the mean). (middle) Dual-color kymograph (see also the scheme on top) of XCTK2-driven microtubule pair sliding showing the GFP-XCTK2 signal in green and the Cy5-microtubule signal in red (separate channels are shown individually in bottom panels). The length of the overlap between the two microtubules is decreasing over time. A bright microtubule moving to the right is pushed over the end of an immobilized, dim microtubule; XCTK2 accumulation is confined to the overlap region. (right) Averaged speeds of microtubule sliding at varying concentrations of GFP-XCTK2. Error bars are standard errors of means. (C) Quantification of Eg5-GFP in Cy5-microtubule pairs in assay buffer containing 50 mM KCl. (left) Binding curves showing averaged TIRF intensities of Eg5-GFP along microtubule overlaps and single microtubules at varying concentrations of Eg5-GFP. Error bars are standard deviations. Absolute intensity values are not comparable with B (left) because the excitation intensity was increased. (middle) Dual-color kymograph of Eg5-driven microtubule pair sliding showing the Eg5-GFP signal in green and the Cy5-microtubule signal in red. (right) Averaged speeds of microtubule sliding at varying concentrations of Eg5-GFP. Error bars are standard errors of the mean. A.U., arbitrary unit. Bars: (A, B [horizontal], and C [horizontal]) 5 µm; (B and C, vertical) 120 s.
The bipolar motor Eg5 behaved very differently. GFP-labeled Eg5 generally bound considerably more weakly to microtubule pairs, even after reducing the ionic strength in the buffer. Surprisingly, Eg5-GFP did not accumulate strongly in the microtubule overlap region as XCTK2 did. The fluorescence intensity roughly doubled in the overlap region as compared with single microtubules, simply reflecting the doubled number of binding sites in this region (Fig. 1 C, left; and Video 2). This observation might suggest that the two motor dimers at the ends of the Eg5 molecule can bind with similar probability to binding sites either on a single microtubule or on two different microtubules in a pair.
In agreement with previous studies (Kapitein et al., 2005; Uteng et al., 2008; Braun et al., 2009; Fink et al., 2009), both motors could slide antiparallel microtubules in vitro, leading to a decrease of the length of the overlap region when the sliding microtubule was pushed beyond the end of the immobilized microtubule (Fig. 1, B and C, middle). Strict confinement of accumulated GFP-XCTK2 to the shortening overlap region indicated highly dynamic binding and unbinding. The speed of microtubule sliding driven by GFP-XCTK2 decreased from 45 to 20 nm/s with motor concentrations increasing from 1 to 20 nM (Fig. 1 B, right), possibly because of crowding effects or inhibition as a consequence of mechanical coupling between motors (Bieling et al., 2008). The latter explanation is supported by microtubule gliding assays with surface-immobilized XCTK2, which also show a density-dependent velocity decrease (Fig. S1). A similar decrease in the microtubule sliding velocity was not observed for Eg5-GFP (Fig. 1 C, right), probably because of the lower numbers of Eg5 molecules in microtubule pairs as a consequence of weaker binding.
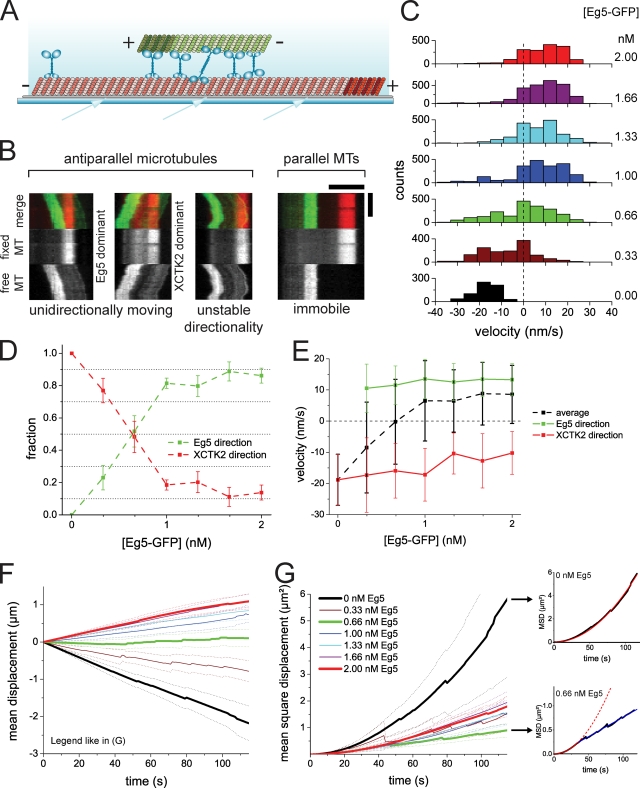
Next, we addressed the question of competition between the two antagonistic motors in microtubule pairs that they cross-link (Fig. 2). We asked, in particular, whether a stable balance of forces can be generated in these microtubule pairs at a certain XCTK2/Eg5 ratio. We used polarity-marked microtubules in these microtubule sliding assays (Fig. 2 A) to be able to distinguish between parallel and antiparallel microtubule pairs. Keeping the XCTK2 concentration constant at 0.5 nM, we varied the Eg5 concentration between 0 and 2 nM (staying well below the apparent Kd for pair binding for both motors). We observed that on average, 48 ± 2% of the microtubule pairs were antiparallel, which was roughly independent of the motor concentration ratio studied. 94 ± 1% of the antiparallel microtubule pairs were motile, whereas 96 ± 1% of the parallel microtubule pairs were immobile (Fig. 2 B, right). This agrees with previous conclusions stating that cross-linking motors move antiparallel but not parallel microtubule pairs with respect to each other (Kapitein et al., 2005; Fink et al., 2009).
Figure 2.
Competition of antagonistic motors in antiparallel microtubule sliding. (A) Scheme of a pair of polarity-marked microtubules in a sliding assay (symbols as in Fig. 1 A). (B) Different motility states of microtubule pairs. Example kymographs of a freely movable Alexa Fluor 568–microtubule (MT; green) bound via motor cross-links to a surface-immobilized Cy5-microtubule (red) in the presence of 0.5 nM XCTK2 and 0.66 nM Eg5-GFP, illustrating Eg5-dominated unidirectional antiparallel sliding, XCTK2-dominated unidirectional antiparallel sliding, movement with unstable directionality, and static parallel binding (from left to right). All kymographs are taken from the same sample. (C) Histograms of the instantaneous velocities of antiparallel sliding microtubules during the competition between 0.5 nM XCTK2 and varying concentrations of Eg5 (indicated to the right). Histograms contain all data of three independent experiments. (D) Fractions of Eg5-GFP– and XCTK2-dominated movement during microtubule sliding, calculated as described in Materials and methods. (E) Mean velocities of all microtubule movements and mean velocities separately determined for only the XCTK2-dominated microtubule movements and for the Eg5-GFP–dominated movements compiled from C. (F) Mean displacement curves of antiparallel sliding microtubules during the competition. Color code as in C and G. The highest, the lowest, and the balance point Eg5 concentrations are emphasized by thick lines. (G, left) MSD curves of antiparallel sliding microtubules. (top right) MSD curve of antiparallel sliding microtubules in the presence of 0.5 nM XCTK2 (black) fitted with a parabolic function (red). (bottom right) MSD curve of antiparallel sliding microtubules in the presence of 0.5 nM XCTK2 and 0.66 nM Eg5-GFP (black) fitted with a parabolic function (from 0 to 36 s; red) and a linear function (from 36 s onward; blue). The dashed lines show continuations of the fit equation in the data range that was not used for the fit. Error bars in D and E are standard errors of the mean. Dotted lines in F and G are 95% confidence intervals. All experiments shown were performed in assay buffer containing 50 mM KCl. Bars: (B, horizontal) 5 µm; (B, vertical) 120 s.
The majority of the sliding and antiparallel microtubules moved unidirectionally, exhibiting occasional pauses (Fig. 2 B, left). Below an Eg5 concentration of 0.66 nM, microtubules moved most of the time with their marked plus ends leading, indicating dominating XCTK2 activity (Fig. 2 C). Increasing the Eg5 concentration above 0.66 nM changed the directionality of movement, indicating that the Eg5 activity dominated. However, the speed of Eg5-driven antiparallel microtubule sliding was reduced in the presence of overlap-accumulated XCTK2 (Fig. 2 C), probably as a consequence of a counter force generated by XCTK2. Further increasing the Eg5 concentration increased the sliding velocity toward the natural Eg5 sliding velocity (unpublished data).
Interestingly, in the transition zone around 0.66 nM Eg5, microtubule pairs sliding in both directions coexisted (Fig. 2 B, left). Occasionally, directional reversals were observed, which were often associated with pausing (Fig. 2 B and Video 3). In this regime of directional instability, the instantaneous velocity distribution was significantly broadened as compared with the cases in which one of the motors dominated (Fig. 2 C), and about half of the antiparallel microtubules moved in either direction (Fig. 2 D). Thus, on average, none of the two motors dominated in this regime, resulting in a mean velocity of close to zero (Fig. 2 E, dotted black line) and essentially no net relative displacement of microtubule pairs (Fig. 2 F, green). Nevertheless, a stable balance of force resulting in static microtubule pairs was not generated. Instead, microtubules moved back and forth for micrometer distances, indicating that in individual microtubule pairs, one or the other motor type can dominate for a certain time. This also became evident in a mean square displacement (MSD) analysis (Fig. 2 G, green curve). In contrast to the parabolic MSD curve for the case of directional motion in the presence of only one motor (Fig. 2 G, top right, red line), a linear MSD curve was obtained for time intervals larger than ∼40 s in the case of the balanced situation with the two antagonistic motors (Fig. 2 G, bottom right, blue line), which is indicative of an effectively diffusive motion for this part of the MSD curve (Tawada and Sekimoto, 1991; Berg, 1993).
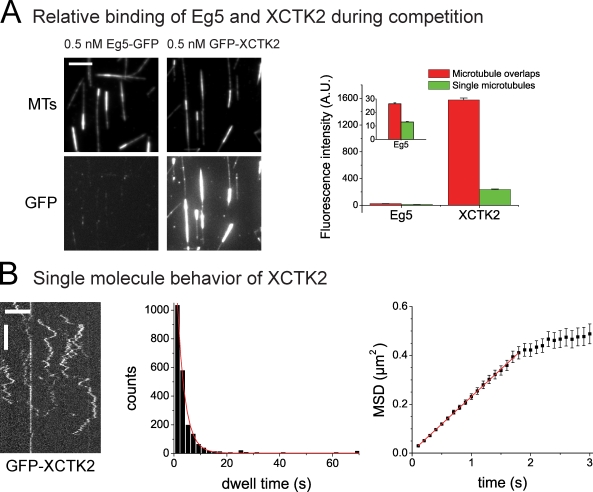
We observed directional instability when similar concentrations of the two motors XCTK2 and Eg5 were present (0.5 nM and 0.66 nM, respectively). However, as a consequence of the differing affinities of the two motors for microtubules, the amount of XCTK2 present in the overlap zone was much higher than that of Eg5, as indicated by a 60 times higher GFP-XCTK2 fluorescence intensity as compared with the Eg5-GFP intensity at equal monomer concentrations (Fig. 3 A). This means that 120 times more XCTK2 dimers bind to the overlap region than Eg5 tetramers. Nevertheless, Eg5 competes efficiently against XCTK2. This is most likely a consequence of the nonprocessivity of the XCTK2 motor and probably also of the diffusive nature of the nonmotor microtubule binding domain of XCTK2 (Fig. 3 B; Fink et al., 2009), suggesting that only a fraction of the motor force of XCTK2 can be transmitted to the second microtubule via its diffusive nonmotor microtubule binding site for the case of antiparallel microtubule sliding. In conclusion, a stable balance of force leading to static antiparallel microtubule pairs is not established in our assay by the two antagonistic motors studied here. Instead, directional instability is generated when their activities are balanced.
Figure 3.
Comparison of relative affinity of XCTK2 and Eg5 and XCTK2 single molecule characterization of XCTK2. (A) Comparative quantification of the binding of GFP-XCTK2 and Eg5-GFP to single microtubules (MTs) and microtubule pairs (made with 0.5 nM unlabeled XCTK2) at identical conditions. (left) TIRF microscopy images of Cy5-microtubules (top) and of GFP-labeled motors as indicated (bottom) measured at identical microscopy settings for the two motors. (right) Averaged fluorescence intensities of the two GFP-labeled motors in microtubule overlaps and on single microtubules. A.U., arbitrary unit. (B) Single molecule TIRF microscopy of 0.02 nM GFP-XCTK2 on immobilized microtubules. (left) Kymographs showing binding events on an individual microtubule over time. (middle) Histogram showing the distribution of GFP-XCTK2 dwell times on microtubules and monoexponential fits (red line). The mean dwell time derived from this fit is τ = 2.86 ± 0.08 s. (right) MSD curve generated from 2,186 GFP-XCTK2 binding events and a linear fit (red line; R2 = 0.999). The initial linear increase of the MSD curve shows that XCTK2 does not move in a directional manner but diffuses via its nonmotor binding site on the microtubule. The one-dimensional diffusion coefficient D as derived from the slope of the fit (MSD = v2t2 = 2Dt) is D = 0.112 ± 0.001 µm2/s. (A and B) Stated errors and error bars are standard errors of the mean. All experiments shown were performed in assay buffer containing 50 mM KCl. Bars: (A and B [horizontal]) 5 µm; (B, vertical) 5 s.
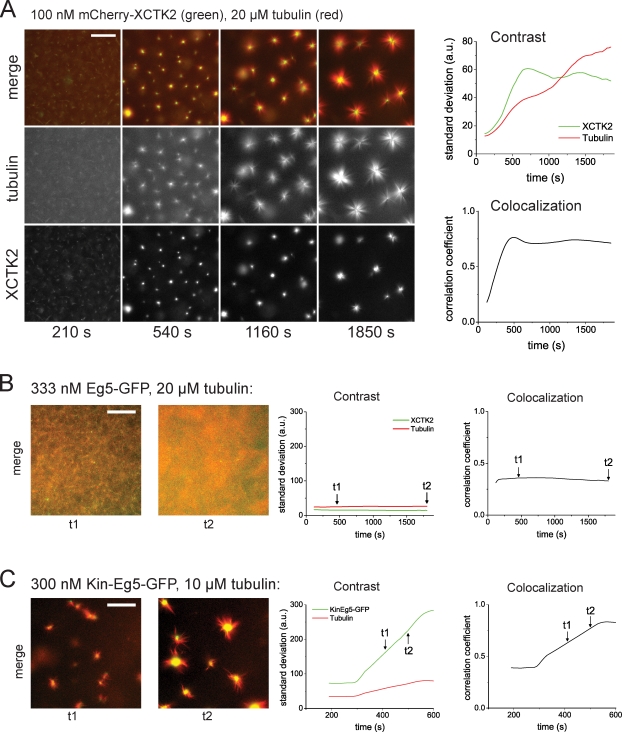
After having established the basic behavior of the antagonistic motors XCTK2 and Eg5 in pairs of stabilized microtubules, we studied the large-scale self-organization of mixtures of these motors with an ensemble of growing microtubules in solution. Because kinesin-14 is important for spindle pole focusing in several organisms (Matthies et al., 1996; Merdes et al., 1996; Endow and Komma, 1997; Walczak et al., 1998), we tested whether this microtubule cross-linking and sliding motor of the type nonprocessive motor-diffusive handle has the intrinsic ability to focus microtubule minus ends into asters. We incubated 100 nM XCTK2 with 20 µM tubulin, ATP, and GTP and observed that asters indeed formed from microtubules nucleated in solution (Fig. 4 A and Video 4), which is reminiscent of structures formed by processive, artificially oligomerized motors in vitro (Nédélec et al., 1997; Surrey et al., 2001) or by mitotic spindles in the absence of Eg5 activity (Sawin et al., 1992; Mayer et al., 1999). Lack of XCTK2 in the self-organization mixture or presence of an XCTK2 fragment lacking the second nonmotor microtubule binding domain did not organize microtubules into asters (Fig. S2 A and not depicted), reflecting the importance of the microtubule cross-linking ability for microtubule/motor self-organization. Our results also show that teams of nonprocessive XCTK2 motors (Case et al., 1997; deCastro et al., 2000) accumulating efficiently between intersecting microtubules can generate effectively processive cross-links and bring microtubule ends together. A previous observation that Drosophila kinesin-14 can move effectively processively when trapped in microtubule bundles supports this notion (Furuta and Toyoshima, 2008).
Figure 4.
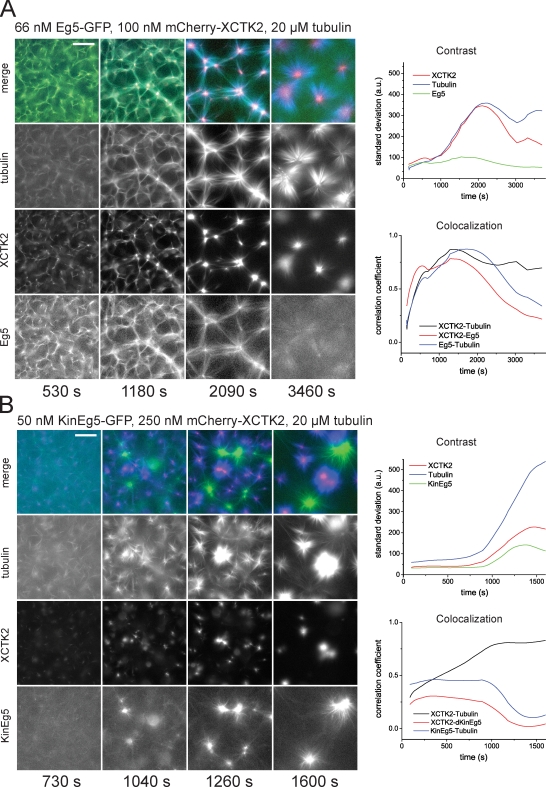
Self-organization of microtubules and mitotic kinesins. (A) Self-organization of microtubules and XCTK2: time course of the formation of microtubule asters in the presence of 100 nM mCherry-XCTK2 and 20 µM tubulin. (left) Epifluorescence images of mCherry-XCTK2 and Cy5-tubulin taken at the indicated times in the individual color channels and as merged images. (right) Time course of organization monitored by image parameters: total contrast (top) for mCherry-XCTK2 images (green) and Cy5-microtubule images (red) as calculated by the standard deviation from the mean; colocalization (bottom) of mCherry-XCTK2 and Cy5-microtubule signals as calculated by Pearson’s correlation. (B) Self-organization of microtubules and Eg5. (left) Early (t1) and late (t2) example images illustrating the development of self-organization with Eg5-GFP (green) and Cy5-microtubules at the concentrations indicated. (right) The time course of self-organization is monitored by the standard deviation, which is indicative of contrast (left), and Pearson’s correlation coefficient, which is indicative of colocalization (right), for the color channels as indicated. The time points of the example images to the left are indicated in the curves with arrows. (C) Self-organization of microtubules and chimeric Kin-Eg5. Composition of the figure is as described in B. All experiments were performed in assay buffer with 50 mM KCl. a.u., arbitrary unit. Bars, 50 µm.
The basic characteristics of the self-organized structures and thus the pathway of self-organization can be expressed using two simple parameters derived from the fluorescence microscopy images. The image contrast (Gonzalez et al., 2004), a measure for compaction of the observed structures (see Materials and methods), reached a stable value after about 10 min (Fig. 4 A, right), which is indicative of steady-state accumulation of the motor in the center of the asters at this time. The Pearson correlation coefficient (Zinchuk and Zinchuk, 2008) between images of Cy5-microtubules and of mCherry-XCTK2, a measure for colocalization (see Materials and methods), reached a plateau at about the same time (Fig. 4 A, right), which is indicative of no relative changes in motor–microtubule colocalization. However, the contrast of the microtubule signal increased continuously during the entire observation time of 30 min (Fig. 4 A, right), demonstrating that a true steady-state is not reached during the observation period. Asters continue to fuse, which is the result of continued microtubule growth and pulling forces generated by motors on overlapping microtubules from neighboring asters.
As observed in previous in vitro self-organizing systems with artificially oligomerized motors (Nédélec et al., 1997; Surrey et al., 2001), the pathway leading to self-organization with natural XCTK2 critically depended on the ratio of motor to tubulin concentration (the latter determining microtubule nucleation efficiency and growth speed; Fig. S2 B and Video 8). In agreement with previous predictions from computer simulations (Nédélec and Surrey, 2001), self-organization of asters after initial microtubule–motor cluster formation was first governed by a competition between microtubule cluster expansion driven by microtubule growth and contraction driven by motor activity. Later, either large interconnected networks that were generated by fast cluster expansion (at high tubulin concentrations) broke apart as a consequence of internal tension or, under conditions of slow cluster expansion and fast contraction (at lower tubulin concentrations), isolated asters fused when aster microtubules began to overlap as a consequence of continued microtubule growth (Fig. S2 B).
Previously, it was shown that mixtures of antagonistic, artificially oligomerized motors could generate microtubule networks with interconnected plus and minus poles provided that they individually could organize microtubules into asters (Surrey et al., 2001). It remained an open question how natural antagonistic motors organize microtubules, especially with respect to the question why Eg5 does not focus microtubules into poles at the center of natural spindles. Using our purified motors, we were then in the position to address this question. First, we tested Eg5 alone in the self-organization assay and found that it behaved very differently from XCTK2. Surprisingly, we did not observe microtubule aster formation (having tested the accessible range of Eg5-GFP concentrations up to 670 nM, which was determined by the solubility limit of the motor; Fig. 4 B; Fig. S3, A and B; and Videos 5 and 9). Instead, only large-scale microtubule density fluctuations were observed.
To test how two mitotic motors with opposite directionality organize microtubules, we mixed 66 nM Eg5-GFP with 100 nM mCherry-XCTK2 and found that Eg5 slowed down initial cluster contraction by XCTK2, leading then first to the formation of a large network to which both antagonistic motors bound rather uniformly (Fig. 5 A and Video 6). Then XCTK2 started to dominate visibly and to accumulate in aster centers, whereas Eg5 failed to form poles. Instead, Eg5 was finally rather evenly distributed, as theory predicts for a motor walking toward the outside of an aster (Nédélec et al., 2001). These dissimilar distributions of the motors are reflected by their colocalization curves (Fig. 5 A, right). Although Eg5 lost the competition with XCTK2, its presence nevertheless influenced the pathway of self-organization. Its competitive action initially slowed down the local XCTK2 accumulation driving aster formation through an interconnected network state. These networks transformed into local clusters by motor and microtubule sorting without going through network rupturing. Mixing Eg5 at higher concentrations with 100 nM XCTK2 did further slow down self-organization as a result of competing motor action but did eventually also lead to XCTK2 asters following a similar pathway (Fig. S3 C and Video 10). XCTK2 always dominated over Eg5 (Fig. S3, C and D). We did not find a regime under which Eg5 focused microtubule plus ends efficiently into plus poles as observed in earlier experiments with artificially oligomerized kinesin-1 oligomers (Nédélec et al., 1997; Surrey et al., 2001).
Figure 5.
Self-organization of microtubules and antagonistic mitotic motors. (A) Competition between XCTK2 and wild-type Eg5 in self-organization of microtubules and motors. (left) Time course of the self-organization of mCherry-XCTK2 (red), Eg5-GFP (green), and Cy5-tubulin (blue) into asters with centrally accumulated mCherry-XCTK2; concentrations are as indicated. (right) Time course of self-organization as monitored by image parameters as in Fig. 4 A for the color channels as indicated. (B) Competition between XCTK2 and chimeric Kin-Eg5 in self-organization of microtubules and motors. (left) Time course of the self-organization of mCherry-XCTK2 (red), Kin-Eg5–GFP (green), and Cy5-tubulin (blue) into separate mCherry-XCTK2–containing asters and Kin-Eg5–GFP-containing asters; concentrations are as indicated. (right) Time course of self-organization as monitored by image parameters. All experiments shown were performed in assay buffer containing 50 mM KCl. a.u., arbitrary unit. Bars, 50 µm.
To test whether the specific motor properties of Eg5 are responsible for it being an inefficient pole organizer, we used a chimeric Eg5 molecule in which the Eg5 motor domain and its neck-linker were replaced by the motor domain and the neck-linker of kinesin-1, a fast and processive motor. We showed previously that this chimeric Kin-Eg5 retains the tetrameric architecture of Eg5 but has kinetic properties similar to those of native kinesin-1 (Cahu and Surrey, 2009). Interestingly, when mixed with tubulin, this chimeric motor was able to organize microtubules into asters (Fig. 4 C and Video 5), which is reminiscent of the asters generated previously by artificially oligomerized kinesin-1 molecules that were shown to have plus poles (Nédélec et al., 1997; Surrey et al., 2001). Mixing both the chimeric Kin-Eg5–GFP and mCherry-XCTK2 with microtubules produced now a new state, namely a mixture of asters in which exclusively either Kin-Eg5 or XCTK2 were accumulated, indicating opposite aster polarity (Fig. 5 B and Video 6).
Our results suggest that the properties of the natural Eg5 motor are optimized to allow efficient antiparallel microtubule sliding and to avoid focusing microtubules into plus poles. The asymmetric situation generated by the different organizational properties of the natural antagonistic motors XCTK2 and Eg5 favored pole formation of microtubule minus ends and separation of microtubule plus ends without focusing plus ends into poles.
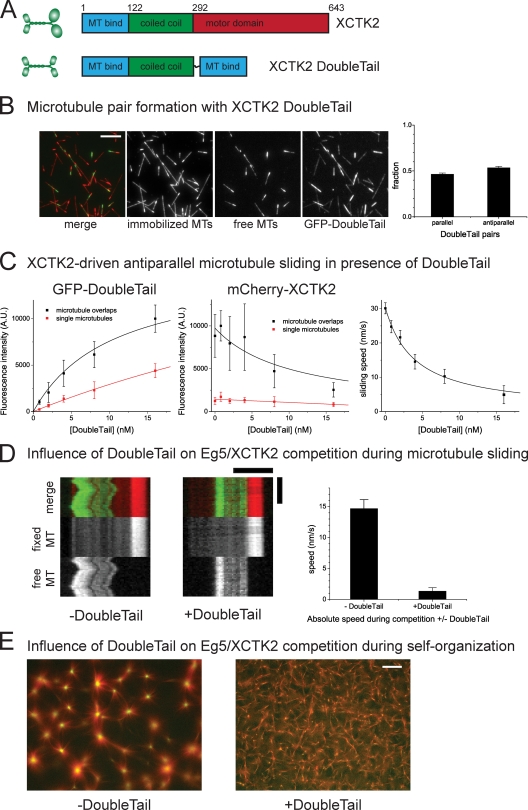
Next, we asked how nonmotile microtubule cross-linkers affect directional instability of antiparallel microtubule pair sliding and self-organization in the presence of opposing motor activities. To construct a model microtubule cross-linker that cross-links parallel and antiparallel microtubules without any preference, we fused the two nonmotor microtubule binding domains of XCTK2 (lacking the motor domain) inserting a flexible linker (Fig. 6 A). This XCTK2 DoubleTail construct was indeed able to cross-link microtubules and to create microtubule pairs without a preference for the microtubule orientation (Fig. 6 B). When added to microtubule pairs in combination with XCTK2, it bound with higher affinity to microtubule pairs than to individual microtubules, as expected (Fig. 6 C, left). DoubleTail was observed to compete with XCTK2 for binding sites (Fig. 6 C, middle) and to decrease the speed of XCTK2-driven microtubule sliding in a dose-dependent manner (Fig. 6 C, right). The braking effect of DoubleTail can be well described assuming protein friction (Fig. 6 C, right, black line; see Materials and methods).
Figure 6.
Effect of a nonmotor microtubule cross-linking protein on microtubule sliding and self-organization. (A) Schematic representations of XCTK2 and the DoubleTail construct. Numbers indicate amino acid position of the XCTK2 coding sequence. (B) GFP-DoubleTail cross-links microtubules (MTs). (left) Merged microtubule channels and individual color channels of microtubule pairs formed with 0.8 nM XCTK2 GFP-DoubleTail. (right) Fraction of microtubule pairs with parallel and antiparallel orientation. (C) Addition of XCTK2 GFP-DoubleTail to an antiparallel microtubule sliding assay with 1 nM mCherry-XCTK2 motor. (left) Binding of GFP-DoubleTail to single microtubules and to microtubule overlaps. (middle) GFP-DoubleTail competes with mCherry-XCTK2 for binding to microtubules. Black lines are global fits to the DoubleTail binding and competitive inhibition curves for microtubule overlaps with the shared fit parameter Kd for the DoubleTail binding to microtubule pairs, yielding a Kd of 10.4 ± 3.0 nM (error is standard error of the mean). (right) Increasing concentrations of GFP-DoubleTail decreases the sliding velocity. The black line is a fit assuming protein friction exerted by DoubleTail (see Materials and methods). A.U., arbitrary unit. (D, left) Typical kymographs of microtubule pairs in a motor competition experiment with 0.66 nM Eg5 and 0.5 nM XCTK2 in the absence or presence of 16 nM GFP-DoubleTail. (right) Quantification of the mean absolute sliding velocity of antiparallel microtubules under these conditions. (E) Self-organization of microtubules and antagonistic kinesins (100 nM mCherry-XCTK2 and 66 nM Eg5-GFP) in the absence or presence of 1 µM Z-GFP-DoubleTail. Merged images of microtubules (red) and mCherry-XCTK2 (green) after 36 min. Entire recorded time course is shown in Video 10. Error bars are standard errors of the mean (B, C [right], and D) or standard deviations (C, left and middle). Bars: (B) 10 µm; (D, horizontal) 5 µm; (D, vertical) 120 s; (E) 50 µm.
To test how this braking activity affects directional instability of microtubule pair sliding in the presence of both XCTK2 and Eg5 at concentrations of 0.5 nM and 0.66 nM, respectively, we also added 16 nM DoubleTail and imaged microtubule pairs over time. We observed that the mean velocity of antiparallel microtubule pair sliding was greatly decreased (Fig. 6 D), which is similar to the effect DoubleTail had on the sliding of microtubule pairs driven by XCTK2 alone. A considerable fraction of antiparallel microtubule pairs even appeared to be immobile (Fig. 6 D). This demonstrates that additional protein friction can, in principle, stabilize antiparallel microtubule overlaps.
To test how microtubule cross-linking by DoubleTail affects large scale self-organization of microtubules, we added 1 µM DoubleTail to self-organization experiments with the two motors XCTK2 and Eg5 at concentrations of 100 nM and 66 nM, respectively. DoubleTail prevented sorting of motors and eventual XCTK2-driven aster formation, which occurs under these conditions in the absence of DoubleTail (Fig. 6 E and Video 7). These experiments suggest that there is an intrinsic conflict between antiparallel microtubule overlap stabilization by a simple microtubule cross-linker and motor-driven microtubule organization that requires relative microtubule movements and motor sorting.
Discussion
Several loss-of-function studies in living cells have suggested that a stable balance of force generated by antagonistic motor proteins is involved in the maintenance of spindle shape and bipolarity (Saunders et al., 1997; Mountain et al., 1999; Sharp et al., 2000; Mitchison et al., 2005). To which extent microtubule cross-linking motor proteins alone can produce such a balance of force is poorly understood. We have studied here in vitro the competition between the two mitotic antagonistic microtubule motors kinesin-5 and -14 in situations in which these microtubule cross-linking motors connect either two individual microtubules or a large population of microtubules that they organize in space.
We have shown in this study that directional instability of antiparallel microtubule pair sliding is generated by the two antagonistic mitotic motors when their activities are balanced (Fig. 2, D and E). This phenomenon was previously also observed in microtubule gliding assays with mixtures of surface-immobilized kinesin-1 and dynein (Vale et al., 1992) and Klp61F and Ncd, the kinesin-5 and -14 orthologues from Drosophila (Tao et al., 2006). In the directional instability regime, microtubules moving in both directions coexist together with briefly pausing microtubules. The failure to establish a stable antiparallel overlap with antagonistic motors is expected to arise as a consequence of force-dependent motor dissociation and indicates that motors rather detach than stall when experiencing a counteracting force (weak motors; Jülicher and Prost, 1995; Badoual et al., 2002; Nédélec, 2002; Grill et al., 2005; Müller et al., 2008; Civelekoglu-Scholey et al., 2010). Single molecule force measurements have indeed shown that Eg5 has a higher probability to detach than to reach stall under load (Valentine et al., 2006; Korneev et al., 2007). In a situation of balancing forces between two opposing teams of weak motors, stochastic fluctuations in motor numbers are expected to invariably lead to increasing detachment of those motors that belong to the team that suffered a stochastic reduction of motor number, explaining why the balanced situation is unstable for motors with such properties. A mathematical model taking such stochastic fluctuations and force-dependent motor unbinding into account has recently provided support for this interpretation for the situation of microtubule gliding assays with mixtures of surface-immobilized Drosophila kinesin-5 and -14 (Civelekoglu-Scholey et al., 2010). In the future, it will be interesting to test whether a stable balance of forces can be generated under more complex conditions, for example at increased motor crowding in microtubule pairs or when multiple microtubule pairs are coupled mechanically.
An interesting difference between previous surface gliding assays (Tao et al., 2006) and our antiparallel microtubule sliding assays is that the balance point of directional instability is reached at roughly equal motor densities in the case of surface gliding, whereas we found that a much smaller number of Eg5 molecules can efficiently compete against a large number of XCTK2 molecules in antiparallel microtubule pairs. This suggests that Eg5 is optimized for force production between antiparallel overlaps. In contrast, the diffusive nonmotor microtubule binding site of XCTK2 apparently causes rather inefficient force production between two microtubules that are exactly antiparallel, as in our microtubule pair sliding experiments. This is probably a consequence of XCTK2’s nonmotor binding site diffusing along protofilaments. In contrast, Eg5 with its two motor domains at each end of the molecule does probably not diffuse when cross-linking and sliding two microtubules (Kapitein et al., 2008), allowing it to produce force efficiently between two antiparallel microtubules, which is the configuration found in the center of bipolar spindles.
In vivo, the minus motors kinesin-14 and dynein are important for spindle pole focusing (Matthies et al., 1996; Merdes et al., 1996; Endow and Komma, 1997; Heald et al., 1997; Walczak et al., 1998; Mountain et al., 1999; Zhu et al., 2005). Whereas dynein appears to require additional partners for its pole-focusing activity (Gaglio et al., 1996; Merdes et al., 1996), our results show that kinesin-14 can act, in principle, as an autonomous pole-forming motor not requiring additional proteins. This might indeed be the dominant mechanism by which centrosome-free meiotic spindles are focused, for example, in Drosophila (Matthies et al., 1996; Sköld et al., 2005).
However, Eg5 appears to be optimized for preventing focusing of microtubule plus ends into poles (Fig. 4, A and B). But why didn’t Eg5 generate plus poles in our self-organization experiments, although it has been reported to be weakly processive on its own (Valentine et al., 2006) and to form stable links between two individual static microtubule plus ends (Kapitein et al., 2005; Uteng et al., 2008)? In comparison with XCTK2, Eg5 accumulates surprisingly much less efficiently between microtubules (Fig. 3 A). Furthermore, its intrinsic mechanical velocity is in the range of the growth velocity of microtubule plus ends in our experiment (which is 29.2 ± 2.5 nm/s in the beginning of our self-organization experiments; unpublished data), probably making it more difficult for Eg5 than for XCTK2 to reach microtubule ends and to bring them to other microtubule ends, which is a prerequisite for pole forming activity (Nédélec and Surrey, 2001; Surrey et al., 2001). In vertebrate cells, this difference between the velocity of Eg5 motility and microtubule growth is even enhanced as a consequence of the higher microtubule growth velocity in vivo as compared with our in vitro situation. In some spindles, such as in Xenopus eggs, a major fraction of the Eg5 population is even actively transported toward the spindle poles in a dynein/dynactin-dependent manner (Kapoor and Mitchison, 2001; Uteng et al., 2008), a process whose biological function is still poorly understood. The importance of the velocity of Eg5 for its organizational properties is emphasized by our finding that a chimeric kinesin-1–Eg5 motor, which is about 10 times faster than Eg5 (Cahu and Surrey, 2009), can focus microtubule plus ends into poles (Fig. 4 C). The recent observation that this chimeric motor (Fig. 4 C) destroys bipolar spindles in Xenopus egg extract (Cahu and Surrey, 2009) emphasizes the importance of preventing plus pole formation during spindle assembly.
As a consequence of directional instability, the formation of stable antiparallel microtubule overlaps in vivo as they are found in the center of spindles probably requires additional activities. Chromosomes and other microtubule-associated proteins such as microtubule-bundling proteins might play important additional roles (Mollinari et al., 2002; Kurasawa et al., 2004; Vernì et al., 2004; Janson et al., 2007; Khmelinskii et al., 2007). We found that a model microtubule cross-linking protein without any preference for the microtubule orientation had a strong influence on the behavior of the mixtures of motors with opposite directionality. It acted like a static cross-linker. These experiments illustrated that an intrinsic conflict exists between contributing to overlap stabilization and allowing motor and microtubule sorting that is required for self-organization. In this context, it is interesting to see that the microtubule cross-linker PRC1 is activated in vertebrates only in anaphase and that mutants with a regulation defect perturb spindles by excessive bundling during prometaphase (Mollinari et al., 2002). In contrast to the model cross-linker we used here in our experiments, Ase1, the yeast orthologue of PRC1, has been shown to have the interesting property of binding preferentially to antiparallel microtubules (Janson et al., 2007). In the future, it will be interesting to test which consequences this selectivity has for motor/microtubule self-organization and antiparallel overlap stabilization. However, in metazoans, PRC1 is not crucial for spindle organization before anaphase (Mollinari et al., 2002), leaving still the open question of how antiparallel microtubules are stabilized in metaphase spindles. Interestingly, in many metazoans, most prominently in Xenopus eggs, spindles show continuous antiparallel microtubule flux combined with coordinated local microtubule polymerization in the spindle center and depolymerization at spindle poles (Kwok and Kapoor, 2007). This probably represents another more dynamic mechanism for central spindle organization, requiring other activities in addition to antagonistic motors.
In conclusion, the properties of the mitotic motors kinesin-5 and -14 appear to be adapted to their function in the spindle. Mixtures of the two motors display an inherent asymmetry of behavior with respect to the different microtubule ends, favoring minus pole formation over plus pole formation. When balanced, mixtures of these two motors alone can generate directional instability. This could promote motor protein and microtubule sorting, which is important for self-organization being able to occur. Static microtubule cross-linkers can suppress both directional instability and self-organization. The cell cycle–regulated use of such cross-linkers might provide a mechanism for antiparallel overlap stabilization after self-organization has been initiated. However, it appears likely that additional activities are needed for the spatial organization of stable antiparallel microtubule overlaps in spindle centers. In vitro assays such as the ones presented in this study will, in the future, allow testing systematically the effects of additional spindle components on self-organization and to define the minimal set of mechanochemical activities required for the establishment of stable bipolarity.
Materials and methods
Protein purifications
Full-length Eg5 and Eg5-GFP were cloned and purified essentially as described previously (Cahu et al., 2008; Uteng et al., 2008). Sf9 cells (from 700 ml culture) expressing Eg5 or Eg5-GFP were lysed using a Microfluidizer (M110L; Microfluidics Corporation) in lysis/wash buffer (buffer A [50 mM K2HPO4, 250 mM KCl, 1 mM ATP, 1 mM MgCl2, 5 mM EDTA, 10 mM 2-mercapto ethanol (ME), and complete protease inhibitors (Roche), pH 7] supplemented with 5 mM imidazole). The lysed cells were centrifuged for 30 min at 257,000 g, and the supernatant was loaded (0.5 ml/min) on an equilibrated column with 5 ml Protino Ni-TED resin (Macherey-Nagel). After extensive washing, the protein was eluted with elution buffer (buffer A supplemented with 300 mM imidazole) at concentrations of around 1 mg/ml. The eluted protein was dialyzed overnight into gel filtration buffer (20 mM K2HPO4, 50 mM KCl, 0.5 mM EGTA, 50 mM imidazole, 5 mM ME, 10% glycerol, and 10% sucrose). After centrifugation for 10 min at 278,800 g, part of the supernatant was frozen in small aliquots in liquid nitrogen, and the remaining part was concentrated using Vivaspin 6 filters (Sartorius). After a second ultracentrifugation, the protein was gel filtered using a Superose 6 column (GE Healthcare) equilibrated in gel filtration buffer. The protein eluted as a single peak with a final concentration of 0.2 mg/ml. The peak fractions were united, in small aliquots, frozen, and stored in liquid nitrogen.
The coding sequence of full-length XCTK2 was PCR amplified from a plasmid (a gift from C. Walczak, Indiana University, Bloomington, IN; Walczak et al., 1997) and cloned into pFastBac HTc via the NotI and XhoI sites. Using the EcoRI and NotI sites, coding sequences of mCherry or GFP were inserted upstream of the XCTK2 sequence separated by a penta-Gly linker sequence, generating mCherry-XCTK2 and GFP-XCTK2, respectively. A predicted (Lupas et al., 1991) dimeric fragment of XCTK2 (corresponding to aa 122–643) without the N-terminal tail was PCR amplified and cloned into the pETM-Z, yielding Z-XCTK2(122–643), which was used for controls. The sequences of all plasmids were verified by DNA sequencing of the inserts. Full-length XCTK2 constructs were purified from Sf9 cells as described for Eg5 proteins but with the following modifications: lysis/wash buffer was buffer B (25 mM Hepes, 250 mM KCl, 2 mM MgCl2, and 0.5 mM ME) supplemented with 30 mM imidazole, 1 mM ATP, and complete protease inhibitors, pH 7.5; elution buffer was buffer B supplemented with 300 mM imidazole, 0.1 mM ATP; and gel filtration buffer was 25 mM Hepes, 250 mM KCl, 2 mM KCl, and 1 mM ME, pH 7.5. After gel filtration, XCTK2 proteins had a final concentration of 0.5 mg/ml. mCherry-XCTK2 always copurified with a smaller protein (Fig. S4), which was determined by immunoblotting to be XCTK2 lacking the N-terminal His tag. Thus, there is a fraction of mixed dimers that lack in part the N-terminal tag but contain the full-length motor protein. Z-XCTK2(122–643) was expressed overnight in BL21-RIL cells and purified like full-length XCTK2. It also contained a small fraction of partially degraded XCTK2.
A construct consisting of the two tail domains of XCTK2 (aa 1–121) fused to GFP was cloned into a pFastBac HTc vector. The coding sequence of GFP followed by the coding sequence for a penta-Gly linker was PCR amplified and inserted between the EcoRI and NotI site of the vector. The coding sequence for XCTK2 (aa 1–291) was PCR amplified and inserted between the NotI and XhoI site of the vector. The coding sequence for a Gly4SerGly4 linker followed by the coding sequence for XCTK2 (aa 1–121) and a stop codon was PCR amplified and inserted between the XhoI and KpnI sites of the vector. After sequencing, the construct was excised and inserted into pETM-Z using the NcoI and KpnI sites. This DoubleTail construct was expressed in BL21-RIL cells and purified as described for full-length XCTK2 (yielding Z-GFP-DoubleTail). In some preparations, the Z tag was cleaved with GST-TEV protease (EMBL), the protease was subsequently removed with glutathione Sepharose (GE Healthcare), and the cleaved Z tag was removed by gel filtration (yielding GFP-DoubleTail).
All proteins eluted earlier from the Superose 6 column than expected from their molecular weight probably because of the presence of elongated coiled-coil domains. Their Stokes radii were determined by comparison with protein standards of known Stokes radii (Gel Filtration Calibration kit HMW; GE Healthcare). The obtained values (Table S1) are in good agreement with previously published values for kinesin-5 and -14s (Sharp et al., 1999; Kapoor and Mitchison, 2001; Kwok et al., 2006; Furuta et al., 2008; Furuta and Toyoshima, 2008; Cahu and Surrey, 2009). All kinesin concentrations in this study are monomer concentrations unless stated otherwise.
Chimeric Kin-Eg5–GFP was prepared by J. Cahu (European Molecular Biology Laboratory, Heidelberg, Germany) as described previously (Cahu and Surrey, 2009). Pig brain tubulin was purified and labeled with Alexa Fluor 568–N-hydroxysuccinimide (NHS; Invitrogen), Cy5-NHS (GE Healthcare), or biotin-NHS (Thermo Fisher Scientific) using standard methods (Hyman et al., 1991; Castoldi and Popov, 2003). Unlabeled tubulin was stored in liquid nitrogen at a concentration of 200 µM, and labeled tubulin was stored at concentrations of 150 µM. Tubulin concentrations are tubulin dimer concentrations.
Glass treatment
Tris-NTA (nitrilotriacetic acid)-PEG (polyethylene glycol)–coated glass and methoxy-PEG-2000-amine–coated glass were prepared essentially as previously described (Lata et al., 2005; Bieling et al., 2008). Biotin-PEG–coated glass was prepared by mixing 95% hydroxy-PEG-3000-amine and 5% biotin-PEG-3000-amine (both from Rapp Polymere) and coupling this mixture to glass. To passivate the counter glass of the flow chambers, poly-l-Lys (PLL)-PEG was dried on a glass surface and then washed extensively.
Antiparallel microtubule sliding assay
In this assay, short, brightly Cy5-labeled microtubules (29% labeled tubulin) bind via motor proteins to long, biotinylated, surface-immobilized, dimly Cy5-labeled microtubules (9% labeled tubulin) and can be transported by these motors along the immobilized microtubules. To stabilize the microtubules, they were prepared using GMP-CPP as previously described (Varga et al., 2006; Bieling et al., 2007).
Flow chambers of ∼5 µl volume were assembled from biotin-PEG coverslips, double-sticky tape (Tesa), and PLL-PEG glass slides. The chamber was loaded with the following solutions: (a) 20 µl of 5% pluronic F-127 in water for 5 min; (b) 50 µl assay buffer AB (20 mM Pipes, pH 6.85, 1 µM ATP, 1 mM EGTA, 50 or 120 mM KCl, 2.25 mM MgCl2, 1 mM ME, 30.6 mM glucose, 200 mM sucrose, 3 µM taxol, and 250 µg/ml β-casein); (c) 0.125 mg/ml neutravidin in AB for 2 min; (d) 50 µl AB; (e) 20 µl long biotinylated microtubules diluted in AB for 5 min; (f) 50 µl AB; (g) 0.5 nM unlabeled XCTK2 diluted in AB for 2 min (except for Fig. 1 C, in which 10 nM unlabeled Eg5 was used instead of XCTK2); (h) 50 µl AB; (i) 20 µl of short microtubule seeds diluted in AB for 5 min; (j) 20 µl AB; and (k) 20 µl AB supplemented with final mix of motor proteins (5% [vol/vol] each, concentrations as indicated in the figure legends), 5 mM ATP-Mg, and 2.5% (vol/vol) oxygen scavengers (40 mg/ml glucose oxidase [Serva], and 20 mg/ml catalase [Sigma-Aldrich] dissolved in BRB80, separately snap frozen in small aliquots and stored at −80°C, thawed before use, mixed, incubated on ice for 1 h, and clarified for 15 min at 278,800 g). Only gel-filtered proteins were used for this assay. The motors were prediluted in their own storage buffers to ensure that the final buffer was always identical in composition. XCTK2 sliding was performed in AB containing 120 mM KCl, and Eg5 and competition sliding were performed in AB containing 50 mM KCl. In some experiments, the DoubleTail construct was additionally included. Final protein concentrations were as indicated in the figure legends. The chamber was sealed with lens oil, and microtubule sliding was observed at 24°C by TIRF microscopy using a setup as described previously (Telley et al., 2009). Samples were excited at 488, 532, and 639 nm, and time-lapsed images were recorded at 6-s time intervals with 100-ms exposure times. Two to three independent experiments were performed for each condition.
To determine the directionality of microtubule movement (Fig. 2), we used polarity-marked microtubules prepared as follows: long, biotinylated, dimly labeled Cy5-microtubules (6% labeled tubulin) were plus end marked by incubating them for 10 min at 37°C with 60% (vol/vol) BRB80/DTT (80 mM Pipes, 1 mM MgCl2, 1 mM EGTA, and 1 mM DTT, pH 6.85) and 20% (vol/vol) bright elongation mix (10 µM tubulin [37% labeled tubulin], 8 µM N-ethylmaleimide–GMP-CPP–tubulin and 0.5 mM GMP-CPP in BRB80/DTT). Short, unbiotinylated, dimly labeled Alexa Fluor 568–microtubule seeds (7% labeled tubulin) were polarity marked by incubating them with 81% (vol/vol) BRB80/DTT and 17% (vol/vol) bright elongation mix. After polymerization of the plus end marks, the microtubules were centrifuged for 5 min at 20,800 g at room temperature and resuspended in BRB80 containing 20 µM taxol. Using taxol was crucial to prevent fusion of the polarity-marked microtubules. An estimate for the quality of the polarity marking is given in the Data analysis section.
Single molecule imaging
Single molecules of GFP-XCTK2 (added at 0.02 nM) on immobilized, GMP-CPP–stabilized microtubules were imaged essentially as described previously for kinesin (Telley et al., 2009) except using here AB containing 50 mM KCl. The fluorescence intensities of GFP-XCTK2 molecules were similar to the signal from single dimeric GFP-tagged kinesins (Telley et al., 2009), indicating that individual XCTK2 dimers were observed. To generate TIRF microscopy videos, a continuous stream with 100-ms exposure times per frame was recorded at 24°C for 70 s. At least two independent experiments per condition were performed.
Gliding assay
For microtubule surface gliding assays with XCTK2, Tris-NTA-PEG–functionalized glass was used and loaded with nickel ions as previously described (Bieling et al., 2008). XCTK2 was diluted in XCTK2 buffer at the indicated concentrations and allowed to bind to the surface for 10 min on ice. Imaging occurred in assay buffer containing 120 mM KCl supplemented with taxol-stabilized fluorescent Cy5-labeled microtubules. Microtubule gliding was observed at 24°C by TIRF microscopy using a setup as described previously (Telley et al., 2009). Samples were excited at 639 nm, and time-lapsed images were recorded at 5-s time intervals with 100-ms exposure times. The set of experiments was performed with three different XCTK2 constructs as shown in the graph. Mean speeds were calculated using ∼50 gliding microtubules per data point.
Self-organization assay
Self-organization experiments were performed in 5-µl flow chambers assembled from a methoxy-PEG-2000–coated coverslip, double sticky tape (Tesa), and a glass slide treated with PLL-PEG. The chamber was placed on an ice-cold metal block and equilibrated with 50 µl self-organization buffer (prepared as a 2× stock and diluted to 1× with water to the following final concentrations: 20 mM Pipes, 1 mM EGTA, 7.25 mM MgCl2, 5 mM ATP, 1.5 mM GTP, 1 mM ME, 50 mM KCl, 30.6 mM glucose, 200 mM sucrose, 2 µM taxol, and 250 µg/ml β-casein, pH 6.85). The chamber was then loaded with self-organization mix and sealed with candle wax. The self-organization mix consisted of 50% (vol/vol) of 2× self-organization buffer, 2.5% (vol/vol) oxygen scavengers, 10% (vol/vol) Cy5-labeled tubulin (5% labeled tubulin), 5% (vol/vol) GFP-Eg5 (prediluted in Eg5 gel filtration buffer), 5% (vol/vol) mCherry-XCTK2 (prediluted in XCTK2 gel filtration buffer), and water (ad 100%). At tubulin concentrations higher than 20 µM, up to 20% (vol/vol) tubulin was used. The motor and tubulin concentrations are indicated in the figure legends. All motor proteins used in the assay were gel filtered, with the exception of experiments with Eg5-GFP at concentrations higher than 66 nM, in which non–gel-filtered Eg5-GFP was used.
For imaging, a microscope (Axiovert 200; Carl Zeiss, Inc.) with a 20× NA 0.8 air objective, an Exfo X-Cite 120 light source, and a camera (AxioCam MRm; Carl Zeiss, Inc.) was used. The microscope was equipped with an environment box (EMBL workshops), which was heated to 32°C. Microtubule polymerization and self-organization started (time point t = 0 s) when the temperature increased after placing the slide into the environment box. Images were recorded using 2 × 2 binning every 10 s in the Cy5, GFP, and/or mCherry channels for up to 1 h, with exposure times of 50 (Cy5), 300 (GFP), and 250 ms (mCherry). For some videos with high fluorescence intensities as a consequence of high kinesin concentrations, exposure times were reduced to avoid overexposure.
Data analysis
Fluorescence intensities of GFP-XCTK2 and Eg5-GFP in the microtubule pair sliding experiments were quantified using a self-written Matlab (MathWorks) program in the following way: using local adaptive thresholding (G. Xiong, Matlab Central), two masks were interactively defined for each Cy5 microtubule video, marking the regions of high intensity as microtubule overlap and the low intensity regions as single microtubules. These masks were applied to the corresponding GFP-motor video, obtaining GFP fluorescence values for the regions of microtubule pairs, single microtubules, and background. The signals were background subtracted and averaged over the entire video, yielding mean values. The mean intensities were plotted as a function of the motor concentrations. For each data point, six videos were analyzed and averaged. We assume proportionality between fluorescence intensity and GFP-protein density in all experiments. To extract the Kd for the binding of XCTK2 to microtubule overlaps, Origin (OriginLab) was used to fit the equation I = Imax × c/(c + Kd). Binding curves in the linear range were fitted by linear regression. Note that the arbitrary intensity units are only comparable within one set of experiments, not between different experiments, as the camera and laser settings were always adjusted to make use of the full dynamic range of the camera.
The reduction of the XCTK2-driven antiparallel microtubule sliding velocity v by the DoubleTail construct (Fig. 6 C, right) was assumed to be caused by protein friction caused by DoubleTail. For the fit to the data in Fig. 6 C, we used the equation
| (1) |
This equation is derived as follows: using a linear force–velocity relationship, the total force Ftot generated by nXCTK2 motors is Ftot = nXCTK2 × (1 − v/v0) × Fs, with v0 being the zero-load velocity and Fs being the stall force of the motor.
The drag force Fd exerted by nDoubleTail DoubleTail molecules is Fd = −nDoubleTail × γ × v, with γ being the friction coefficient. For force balance, the velocity is
| (2) |
For competitive binding of XCTK2 and of DoubleTail to microtubules under conditions in which the number of binding sites is small as compared with the number of binders in the experiment (as in our assay), the ratio of the binders is
| (3) |
i.e., the ratio of the concentrations of the binders in the experiments multiplied by the ratio of equilibrium dissociation constants. Inserting Eq. 3 in Eq. 2 leads to Eq. 1 with the fit parameter Q = (1/[XCTK2]) × (Kd,XCTK2/Kd,DoubleTail) × (γ × v0/Fs).
For the fit of the binding curve of DoubleTail in Fig. 6 C (middle), we used I = Imax × [DoubleTail]/([DoubleTail] + Kd,DoubleTail), with I being the fluorescence intensity of GFP-DoubleTail, Imax being the fluorescence intensity at saturation, and Kd the dissociation constant. For the fit of the curve of competitive XCTK2 and DoubleTail binding (Fig. 6 C, right), we used I = I0 − I0 × [DoubleTail]/(Kd,DoubleTail + [DoubleTail]), with I being the measured intensity of mCherry-XCTK2, I0 being the intensity of mCherry-XCTK2 in the absence of DoubleTail, and Kd the dissociation constant of DoubleTail. This equation is valid for competitive binding at conditions when the bound (as compared with free) fractions of both XCTK2 and also DoubleTail are negligibly small and when the XCTK2 concentration is much smaller than its dissociation constant for microtubule binding, which is the case in our experiment (see for example Segel, 1975). We made a global fit to the DoubleTail binding curve and to the XCTK2 displacement curve sharing the parameter for Kd,DoubleTail. All fits were made using Origin.
Microtubule transport speeds in microtubule pair sliding experiments were measured from kymographs that were produced in ImageJ (National Institutes of Health; Seitz and Surrey, 2006) or using a self-written Matlab function. 20–35 sliding microtubules (26 on average) were measured per condition to calculate averaged speeds.
In experiments with polarity-marked microtubules, all Alexa Fluor 568–microtubules in one video (i.e., the microtubules that were bound via motors to the surface-immobilized Cy5 microtubules) were counted manually and classified as parallel or antiparallel. The ratio of antiparallel nonmotile microtubule pairs plus parallel motile microtubule pairs divided by all microtubule pairs calculated over all experiments was 1.9 ± 0.3%, indicating a very low ratio of wrong polarity marks.
From each antiparallel microtubule pair, a kymograph was created, and the trace of the moving microtubule end was divided into segments with constant (or zero) sliding velocity. From these segmented traces, the spatial position of the microtubule end for each time point was obtained, and using a self-written Matlab function, displacement curves for each individual motile microtubule were generated. From this dataset, the instantaneous velocities were extracted (here defined as the speed between two frames of the recorded time-lapse video, i.e., for a time interval of 6 s) and a histogram (containing the pooled data of three independent sets of experiments) was created. From the individual displacement curves, mean displacement and MSD curves were created. The mean speeds for microtubules moving in plus or minus direction were calculated from the histograms for each condition. The percentages of microtubules moving in plus or minus direction (Fig. 2 D) were calculated by averaging the percentages gained from the instantaneous velocity distributions of three independent experiments. Microtubules moving slower than 3 nm/s were considered immotile and not included into this analysis.
Single molecule streams of GFP-XCTK2 on immobilized microtubules were processed using Kalaimoscope motion tracker software (Transinsight). The tracks were exported, and dwell time and MSD plots were generated using a self-written Matlab function. Fits to these plots were again obtained with Origin. Dwell time was defined as the mean lifetime of the single motor on the microtubule, which can be obtained by an exponential fit to a histogram of the temporal track lengths (measured by Kalaimoscope). The slope of the MSD curve is obtained by a linear fit to the linear region of the MSD curve (Qian et al., 1991; Telley et al., 2009).
For the analysis of the dynamics of self-organization of microtubules and motors, the standard deviation for all pixels in a frame of a video (a measure for the contrast; Gonzalez et al., 2004)
(with In being the raw intensity values of the image) and the Pearson correlation coefficient for two corresponding frames recorded in different color channels (a measure for the degree of colocalization (Zinchuk and Zinchuk, 2008)
(with I1 and I2 being the corresponding intensity values in images from different color channels, normalized to values between 0 and 1) were calculated from the raw images using a self-written Matlab program. These standard deviations and the correlation coefficients were plotted versus the time after the start of polymerization. To minimize the effect of focusing artifacts, the obtained curves were filtered using a sliding window averaging with a window size of 29 time points. Two to five of these curves were averaged to obtain the averaged curves shown in Fig. S2 B.
Online supplemental material
Fig. S1 shows microtubule gliding driven by surface-immobilized XCTK2. Fig. S2 shows how the pathway of microtubule organization by XCTK2 depends on the motor/microtubule ratio. Fig. S3 shows self-organization experiments with Eg5-GFP alone and with mixtures of Eg5 and XCTK2. Fig. S4 shows an SDS gel. Videos 1–3 show microtubule sliding experiments with 50 nM GFP-XCTK2 (Video 1), with 10 nM Eg5-GFP (Video 2), and with 0.66 nM Eg5-GFP and 0.5 nM XCTK2 (Video 3). Video 4 shows the self-organization of 100 nM mCherry-XCTK2 and 20 µM tubulin. Video 5 shows how 333 nM Eg5-GFP and 20 µM tubulin self-organize in comparison with 300 nM Kin-Eg5–GFP and 10 µM tubulin. Video 6 shows how 66 nM Eg5-GFP, 100 nM mCherry-XCTK2, and 20 µM tubulin self-organize in comparison with 50 nM chimeric Kin-Eg5–GFP, 250 nM mCherry-XCTK2, and 20 µM tubulin. Video 7 shows the self-organization of 66 nM Eg5-GFP, 100 nM mCherry-XCTK2, and 20 µM tubulin in the absence or presence of 1 µM Z-GFP-DoubleTail. Video 8 shows how varying concentrations of tubulin affect self-organization in the presence of 100 nM mCherry-XCTK2. Video 9 shows inefficient organization in the presence of varying Eg5 concentrations. Video 10 shows the self-organization of 333 or 10 nM Eg5-GFP with 100 nM mCherry-XCTK2 and 20 µM tubulin. Table S1 lists gel filtration data of the purified proteins used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910125/DC1.
Acknowledgments
We thank Mathias Utz for technical assistance, Julie Cahu for chimeric Eg5, Claire Walczak for the XCTK2 clone, Ivo Telley for helpful discussions about image and data analysis, Marianne Uteng for helpful discussions concerning the microtubule sliding assay, and Antje Fischer for illustrations.
This work was funded by the German Research Foundation (DFG) and the European Union Marie Curie Research Training Network “Spindle Dynamics.”
Footnotes
Abbreviations used in this paper:
- ME
- 2-mercapto ethanol
- MSD
- mean square displacement
- NHS
- N-hydroxysuccinimide
- PEG
- polyethylene glycol
- PLL
- poly-l-Lys
- TIRF
- total internal reflection fluorescence
References
- Badoual M., Jülicher F., Prost J. 2002. Bidirectional cooperative motion of molecular motors. Proc. Natl. Acad. Sci. USA. 99:6696–6701 10.1073/pnas.102692399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H.C. 1993. Random Walks in Biology. Expanded edition Princeton University Press, Princeton, N.J: 152 pp [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E.L., Sandblad L., Dogterom M., Brunner D., Surrey T. 2007. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 450:1100–1105 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Bieling P., Telley I.A., Piehler J., Surrey T. 2008. Processive kinesins require loose mechanical coupling for efficient collective motility. EMBO Rep. 9:1121–1127 10.1038/embor.2008.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M., Drummond D.R., Cross R.A., McAinsh A.D. 2009. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat. Cell Biol. 11:724–730 10.1038/ncb1878 [DOI] [PubMed] [Google Scholar]
- Brust-Mascher I., Scholey J.M. 2007. Mitotic spindle dynamics in Drosophila. Int. Rev. Cytol. 259:139–172 10.1016/S0074-7696(06)59004-7 [DOI] [PubMed] [Google Scholar]
- Butner K.A., Kirschner M.W. 1991. Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 115:717–730 10.1083/jcb.115.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahu J., Surrey T. 2009. Motile microtubule crosslinkers require distinct dynamic properties for correct functioning during spindle organization in Xenopus egg extract. J. Cell Sci. 122:1295–1300 10.1242/jcs.044248 [DOI] [PubMed] [Google Scholar]
- Cahu J., Olichon A., Hentrich C., Schek H., Drinjakovic J., Zhang C., Doherty-Kirby A., Lajoie G., Surrey T. 2008. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS One. 3:e3936 10.1371/journal.pone.0003936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case R.B., Pierce D.W., Hom-Booher N., Hart C.L., Vale R.D. 1997. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 90:959–966 10.1016/S0092-8674(00)80360-8 [DOI] [PubMed] [Google Scholar]
- Castoldi M., Popov A.V. 2003. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32:83–88 10.1016/S1046-5928(03)00218-3 [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Tao L., Brust-Mascher I., Wollman R., Scholey J.M. 2010. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J. Cell Biol. 188:49–68 10.1083/jcb.200908150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCastro M.J., Fondecave R.M., Clarke L.A., Schmidt C.F., Stewart R.J. 2000. Working strokes by single molecules of the kinesin-related microtubule motor ncd. Nat. Cell Biol. 2:724–729 10.1038/35036357 [DOI] [PubMed] [Google Scholar]
- Endow S.A., Komma D.J. 1997. Spindle dynamics during meiosis in Drosophila oocytes. J. Cell Biol. 137:1321–1336 10.1083/jcb.137.6.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A., Chandra R., Komma D.J., Yamamoto A.H., Salmon E.D. 1994. Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J. Cell Sci. 107:859–867 [DOI] [PubMed] [Google Scholar]
- Fink G., Hajdo L., Skowronek K.J., Reuther C., Kasprzak A.A., Diez S. 2009. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat. Cell Biol. 11:717–723 10.1038/ncb1877 [DOI] [PubMed] [Google Scholar]
- Furuta K., Toyoshima Y.Y. 2008. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr. Biol. 18:152–157 10.1016/j.cub.2007.12.056 [DOI] [PubMed] [Google Scholar]
- Furuta K., Edamatsu M., Maeda Y., Toyoshima Y.Y. 2008. Diffusion and directed movement: in vitro motile properties of fission yeast kinesin-14 Pkl1. J. Biol. Chem. 283:36465–36473 10.1074/jbc.M803730200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J.B., Hasbani M.J., Gill S.R., Schroer T.A., Compton D.A. 1996. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135:399–414 10.1083/jcb.135.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R.C., Woods R.E., Eddins S.L. 2004. Digital Image Processing Using MATLAB. Pearson/Prentice Hall, Upper Saddle River, NJ: 609 pp [Google Scholar]
- Grill S.W., Kruse K., Jülicher F. 2005. Theory of mitotic spindle oscillations. Phys. Rev. Lett. 94:108104 10.1103/PhysRevLett.94.108104 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Habermann A., Karsenti E., Hyman A. 1997. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138:615–628 10.1083/jcb.138.3.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck M.M., Pereira A., Pesavento P., Yannoni Y., Spradling A.C., Goldstein L.S. 1993. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J. Cell Biol. 123:665–679 10.1083/jcb.123.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A., Drechsel D., Kellogg D., Salser S., Sawin K., Steffen P., Wordeman L., Mitchison T. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485 10.1016/0076-6879(91)96041-O [DOI] [PubMed] [Google Scholar]
- Janson M.E., Loughlin R., Loïodice I., Fu C., Brunner D., Nédélec F.J., Tran P.T. 2007. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 128:357–368 10.1016/j.cell.2006.12.030 [DOI] [PubMed] [Google Scholar]
- Jencks W.P. 1981. On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. USA. 78:4046–4050 10.1073/pnas.78.7.4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jülicher F., Prost J. 1995. Cooperative molecular motors. Phys. Rev. Lett. 75:2618–2621 10.1103/PhysRevLett.75.2618 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Peterman E.J., Kwok B.H., Kim J.H., Kapoor T.M., Schmidt C.F. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Kwok B.H., Weinger J.S., Schmidt C.F., Kapoor T.M., Peterman E.J. 2008. Microtubule cross-linking triggers the directional motility of kinesin-5. J. Cell Biol. 182:421–428 10.1083/jcb.200801145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Mitchison T.J. 2001. Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 154:1125–1133 10.1083/jcb.200106011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina A.S., Baskin R.J., Cole D.G., Wedaman K.P., Saxton W.M., Scholey J.M. 1996. A bipolar kinesin. Nature. 379:270–272 10.1038/379270a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177:981–993 10.1083/jcb.200702145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev M.J., Lakämper S., Schmidt C.F. 2007. Load-dependent release limits the processive stepping of the tetrameric Eg5 motor. Eur. Biophys. J. 36:675–681 10.1007/s00249-007-0134-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa Y., Earnshaw W.C., Mochizuki Y., Dohmae N., Todokoro K. 2004. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23:3237–3248 10.1038/sj.emboj.7600347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok B.H., Kapoor T.M. 2007. Microtubule flux: drivers wanted. Curr. Opin. Cell Biol. 19:36–42 10.1016/j.ceb.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Kwok B.H., Kapitein L.C., Kim J.H., Peterman E.J., Schmidt C.F., Kapoor T.M. 2006. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat. Chem. Biol. 2:480–485 10.1038/nchembio812 [DOI] [PubMed] [Google Scholar]
- Lata S., Reichel A., Brock R., Tampé R., Piehler J. 2005. High-affinity adaptors for switchable recognition of histidine-tagged proteins. J. Am. Chem. Soc. 127:10205–10215 10.1021/ja050690c [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Matthies H.J., McDonald H.B., Goldstein L.S., Theurkauf W.E. 1996. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134:455–464 10.1083/jcb.134.2.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer T.U., Kapoor T.M., Haggarty S.J., King R.W., Schreiber S.L., Mitchison T.J. 1999. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 286:971–974 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., Cleveland D.W. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W.C., Cleveland D.W. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149:851–862 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J., Maddox P., Gaetz J., Groen A., Shirasu M., Desai A., Salmon E.D., Kapoor T.M. 2005. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell. 16:3064–3076 10.1091/mbc.E05-02-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto D.T., Perlman Z.E., Burbank K.S., Groen A.C., Mitchison T.J. 2004. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol. 167:813–818 10.1083/jcb.200407126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C., Kleman J.P., Jiang W., Schoehn G., Hunter T., Margolis R.L. 2002. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157:1175–1186 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Ando A., Schatten G., Compton D.A. 1999. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147:351–366 10.1083/jcb.147.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.J., Klumpp S., Lipowsky R. 2008. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc. Natl. Acad. Sci. USA. 105:4609–4614 10.1073/pnas.0706825105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédélec F. 2002. Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J. Cell Biol. 158:1005–1015 10.1083/jcb.200202051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédélec F., Surrey T. 2001. Dynamics of microtubule aster formation by motor complexes. C. R. Acad. Sci. Paris Ser. IV. 2:841–847 10.1016/S1296-2147(01)01227-6 [DOI] [Google Scholar]
- Nédélec F.J., Surrey T., Maggs A.C., Leibler S. 1997. Self-organization of microtubules and motors. Nature. 389:305–308 10.1038/38532 [DOI] [PubMed] [Google Scholar]
- Nédélec F., Surrey T., Maggs A.C. 2001. Dynamic concentration of motors in microtubule arrays. Phys. Rev. Lett. 86:3192–3195 10.1103/PhysRevLett.86.3192 [DOI] [PubMed] [Google Scholar]
- Nédélec F., Surrey T., Karsenti E. 2003. Self-organisation and forces in the microtubule cytoskeleton. Curr. Opin. Cell Biol. 15:118–124 10.1016/S0955-0674(02)00014-5 [DOI] [PubMed] [Google Scholar]
- Oladipo A., Cowan A., Rodionov V. 2007. Microtubule motor Ncd induces sliding of microtubules in vivo. Mol. Biol. Cell. 18:3601–3606 10.1091/mbc.E06-12-1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Sheetz M.P., Elson E.L. 1991. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 60:910–921 10.1016/S0006-3495(91)82125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M.A. 1997. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 8:1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., LeGuellec K., Philippe M., Mitchison T.J. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Segel I.H. 1975. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. Wiley-Interscience, New York: 957 pp [Google Scholar]
- Seitz A., Surrey T. 2006. Processive movement of single kinesins on crowded microtubules visualized using quantum dots. EMBO J. 25:267–277 10.1038/sj.emboj.7600937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., McDonald K.L., Brown H.M., Matthies H.J., Walczak C., Vale R.D., Mitchison T.J., Scholey J.M. 1999. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144:125–138 10.1083/jcb.144.1.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., Brown H.M., Kwon M., Rogers G.C., Holland G., Scholey J.M. 2000. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 11:241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu-Hiza M., Perlman Z.E., Wittmann T., Karsenti E., Mitchison T.J. 2004. Eg5 causes elongation of meiotic spindles when flux-associated microtubule depolymerization is blocked. Curr. Biol. 14:1941–1945 10.1016/j.cub.2004.10.029 [DOI] [PubMed] [Google Scholar]
- Sköld H.N., Komma D.J., Endow S.A. 2005. Assembly pathway of the anastral Drosophila oocyte meiosis I spindle. J. Cell Sci. 118:1745–1755 10.1242/jcs.02304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey T., Nédélec F., Leibler S., Karsenti E. 2001. Physical properties determining self-organization of motors and microtubules. Science. 292:1167–1171 10.1126/science.1059758 [DOI] [PubMed] [Google Scholar]
- Tao L., Mogilner A., Civelekoglu-Scholey G., Wollman R., Evans J., Stahlberg H., Scholey J.M. 2006. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr. Biol. 16:2293–2302 10.1016/j.cub.2006.09.064 [DOI] [PubMed] [Google Scholar]
- Tawada K., Sekimoto K. 1991. Protein friction exerted by motor enzymes through a weak-binding interaction. J. Theor. Biol. 150:193–200 10.1016/S0022-5193(05)80331-5 [DOI] [PubMed] [Google Scholar]
- Telley I.A., Bieling P., Surrey T. 2009. Obstacles on the microtubule reduce the processivity of Kinesin-1 in a minimal in vitro system and in cell extract. Biophys. J. 96:3341–3353 10.1016/j.bpj.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uteng M., Hentrich C., Miura K., Bieling P., Surrey T. 2008. Poleward transport of Eg5 by dynein–dynactin in Xenopus laevis egg extract spindles. J. Cell Biol. 182:715–726 10.1083/jcb.200801125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D., Malik F., Brown D. 1992. Directional instability of microtubule transport in the presence of kinesin and dynein, two opposite polarity motor proteins. J. Cell Biol. 119:1589–1596 10.1083/jcb.119.6.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine M.T., Fordyce P.M., Krzysiak T.C., Gilbert S.P., Block S.M. 2006. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 8:470–476 10.1038/ncb1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A.A., Tanaka T.U., Howard J. 2006. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8:957–962 10.1038/ncb1462 [DOI] [PubMed] [Google Scholar]
- Vernì F., Somma M.P., Gunsalus K.C., Bonaccorsi S., Belloni G., Goldberg M.L., Gatti M. 2004. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr. Biol. 14:1569–1575 10.1016/j.cub.2004.08.054 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Heald R. 2008. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 265:111–158 10.1016/S0074-7696(07)65003-7 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Verma S., Mitchison T.J. 1997. XCTK2: a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 136:859–870 10.1083/jcb.136.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., Heald R. 1998. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8:903–913 10.1016/S0960-9822(07)00370-3 [DOI] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J.D., Bossy-Wetzel E., Fan J.B., Abraham R.T., Jiang W. 2005. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 16:3187–3199 10.1091/mbc.E05-02-0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchuk V., Zinchuk O. 2008. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr. Protoc. Cell Biol. Chapter 4:Unit 4.19 10.1002/0471143030.cb0419s39 [DOI] [PubMed] [Google Scholar]