GPR-1/2 (regulators of Gα signaling necessary for asymmetric cell division) receives a positional cue from Let-99, resulting in its appropriate distribution around the posterior cortex.
Abstract
Cortical pulling on astral microtubules positions the mitotic spindle in response to PAR polarity cues and G protein signaling in many systems. In Caenorhabditis elegans single-cell embryos, posterior spindle displacement depends on Gα and its regulators GPR-1/2 and LIN-5. GPR-1/2 and LIN-5 are necessary for cortical pulling forces and become enriched at the posterior cortex, which suggests that higher forces act on the posterior spindle pole compared with the anterior pole. However, the precise distribution of cortical forces and how they are regulated remains to be determined. Using spindle severing, single centrosome assays, and centrosome fragmentation, we show that both the anterior and posterior cortices generate more pulling force than the lateral–posterior region. Lateral inhibition depends on LET-99, which inhibits GPR-1/2 localization to produce a bipolar GPR-1/2 pattern. Thus, rather than two domains of cortical force, there are three. We propose that the attenuation of lateral forces prevents counterproductive pulling, resulting in a higher net force toward the posterior that contributes to spindle elongation and displacement.
Introduction
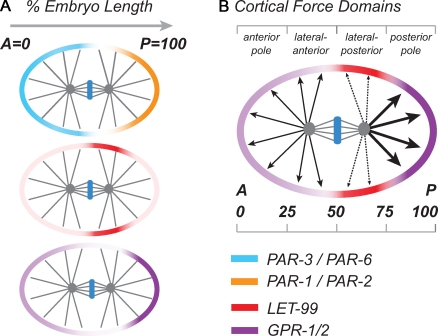
Asymmetric cell division is a fundamental mechanism for organizing developmental patterns and generating cell diversity in all organisms. Successful execution of asymmetric division requires two steps. First, a polarized axis is established, which provides the positional information required for asymmetric distribution of cell fate determinants. Second, the mitotic spindle becomes aligned and asymmetrically positioned along the polarity axis so that determinants are properly segregated into the daughter cells. The use of PAR (partitioning defective) polarity proteins and a Gα signaling pathway are conserved in asymmetric division (for reviews see Cowan and Hyman, 2007; Goldstein and Macara, 2007; Galli and van den Heuvel, 2008; Gönczy, 2008; Segalen and Bellaïche, 2009; Siller and Doe, 2009). In Caenorhabditis elegans embryos, anterior–posterior (AP) polarity is defined at fertilization and depends on segregation of PAR proteins into anterior (PAR-3 and PAR-6) and posterior (PAR-1 and PAR-2) cortical domains (Fig. 1 A). Once polarity is established, several distinct, microtubule-based movements characterize spindle positioning. The male and female pronuclei meet in the posterior and migrate as a unit toward the center of the embryo during prophase (centration). The nuclear–centrosome complex also rotates to align the centrosomes along the AP axis (rotation). During metaphase, the entire spindle shifts to the posterior (Oegema et al., 2001; Labbé et al., 2004) and, together with asymmetric spindle elongation during anaphase, results in posterior spindle displacement. As the anaphase spindle elongates, the posterior pole also undergoes vigorous lateral oscillations, whereas the anterior pole is more stationery.
Figure 1.
Schematic representation of the localizations of proteins that regulate spindle positioning in the single-cell embryo. (A) Localization of PAR proteins, LET-99, and GPR-1/2 in wild-type embryos based on previous work (Gotta and Ahringer, 2001; Colombo et al., 2003; Bringmann et al., 2007; Wu and Rose, 2007; Panbianco et al., 2008; Park and Rose, 2008). In this, and all subsequent figures, 0% embryo length marks the anterior pole, and embryos are oriented with the anterior to the left. (B) Three force domain model for anaphase spindle positioning in wild-type embryos. The thickness of the arrows indicates proposed magnitude of force acting on microtubules from the corresponding cortical region.
Spindle severing and centrosome ablation experiments have shown that dynein-dependent pulling forces from the cell cortex act on astral microtubules to position the spindle (Grill et al., 2001, 2003; Labbé et al., 2004; Pecreaux et al., 2006). Net asymmetry of cortical pulling forces results in asymmetric spindle movements. Early in the cell cycle, higher pulling forces act from the anterior to center and rotate the nuclear–centrosome complex (Labbé et al., 2004). Similarly, an imbalance in force is required for spindle displacement, but in this case, the posterior cortex produces greater net force (Grill et al., 2001; Labbé et al., 2004).
A G protein signaling pathway acts downstream from PAR polarity cues to regulate cortical pulling forces during spindle positioning (Zwaal et al., 1996; Gotta and Ahringer, 2001; Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003; Tsou et al., 2003a; Goulding et al., 2007; Park and Rose, 2008). Embryos depleted of two partially redundant Gα subunits, GOA-1 and GPA-16, or the positive activators, GPR-1/2 and LIN-5, all display similar phenotypes: reduced rates of centration and rotation, absence of spindle displacement and oscillations, and weak spindle pole separation. GPR-1/2 (two nearly identical proteins of the LGN/AGS3 family) and LIN-5 (a coiled-coil protein) may provide a direct link to the spindle, as they can associate with regulators of the microtubule motor dynein (Couwenbergs et al., 2007; Nguyen-Ngoc et al., 2007); NuMA, the vertebrate orthologue of LIN-5, can bind microtubules as well as dynein (Haren and Merdes, 2002). Exactly how such associations result in asymmetric spindle movements remains to be determined, but in several systems, the asymmetric cortical localization of G protein signaling components correlates with force production.
In C. elegans, GPR-1/2 and LIN-5 exhibit a dynamic localization pattern. During the nuclear rotation stage, GPR-1/2 and LIN-5 cortical levels are higher at the anterior compared with the posterior (Panbianco et al., 2008; Park and Rose, 2008). The pattern switches to an enrichment at the posterior cortex by metaphase (Colombo et al., 2003; Gotta et al., 2003; Tsou et al., 2003a; Afshar et al., 2005; Park and Rose, 2008). This pattern coupled with the observation that net posterior forces are high beginning at metaphase led to the model that the entire posterior cortex is pulling more than the anterior (Pecreaux et al., 2006; Kozlowski et al., 2007; Gönczy, 2008). However, recent quantitative analysis shows that GPR-1/2 and LIN-5 are not uniformly distributed throughout the posterior PAR domain. Rather, most embryos exhibit the lowest levels of cortical GPR-1/2 and LIN-5 at the lateral–posterior cortex, with the highest levels of these proteins restricted to a posterior cap (Fig. 1; Park and Rose, 2008); thus, the posterior half of the cortex has regions of both low and high GPR-1/2 at metaphase. The anterior half of the embryo also exhibits higher levels of cortical GPR-1/2 and LIN-5 than the lateral–posterior region, especially as anaphase proceeds, resulting in a bipolar pattern of cortical localization (Park and Rose, 2008). These observations suggest that pulling forces from the cortex at both poles may be higher than those from the lateral–posterior cortex (Fig. 1 B).
The posterior enrichment of GPR-1/2 at metaphase/anaphase depends on the phosphatase PPH-6, the kinases CSNK-1 and PPK-1, and the DEP domain protein LET-99; the latter three are asymmetrically localized by the PARs (Tsou et al., 2002; Panbianco et al., 2008; Afshar et al., 2010). Significantly, LET-99 is enriched in a lateral–posterior cortical band surrounding the single-cell embryo that is largely complementary to the bipolar pattern of GPR1/2 localization (Fig. 1 A). However, the switch from overall anterior enrichment to posterior enrichment at metaphase cannot be explained by cortical LET-99 levels and is likely caused by the action of the other regulators (Park and Rose, 2008; Afshar et al., 2010). Nonetheless, in let-99 mutants, the levels of GPR-1/2 are uniformly high, which indicates that LET-99 inhibits the localization of GPR-1/2 to the cortex (Tsou et al., 2003a; Park and Rose, 2008).
LET-99 is required for both nuclear rotation and asymmetric anaphase movements. In contrast to embryos depleted for Gα, GPR-1/2, or LIN-5, let-99 mutants exhibit dynein-dependent, hyperactive nuclear–centrosome rocking during prophase instead of nuclear rotation. This rocking depends on G protein signaling and suggests that forces acting on centrosomes are higher in let-99 mutants than in wild type (Rose and Kemphues, 1998; Tsou et al., 2002, 2003a). Thus, we originally proposed that LET-99 functions to attenuate lateral–posterior cortical pulling forces in wild-type embryos. Because LET-99 inhibits GPR-1/2 localization and genetic analysis is consistent with LET-99 acting via inhibition of the G protein signaling pathway (Tsou et al., 2003a; Park and Rose, 2008), these observations lead to the model that LET-99 acts to generate the asymmetric localization of GPR-1/2 and LIN-5, and thus asymmetric pulling forces around the posterior cortex. At prophase, inhibition of lateral–posterior forces would allow centering and rotation. However, after alignment of the spindle onto the AP axis, the LET-99 band would contribute to greater net pulling forces acting on the posterior spindle pole by reducing counterproductive lateral–posterior pulling. In particular, during spindle displacement, we propose that the highest pulling forces emanate from the enriched GPR-1/2 region at the absolute posterior cortex, but the adjacent lateral–posterior region with low GPR-1/2 has the lowest forces; the anterior cortex also has high forces (Fig. 1 B). For ease of discussion, we will refer to this as a “three domain model” to distinguish it from models in which the entire posterior cortex exhibits higher pulling forces, but we stress that there is a steep gradient of GPR-1/2 levels between the lateral and posterior domains. The key aspects of this model are that both anterior and posterior pulling forces are higher than lateral posterior forces, and that there is an asymmetry of cortical pulling forces around the posterior spindle pole.
Studies of rotation and centration in the single-cell embryo, including modeling experiments, are consistent with the three domain model (Tsou et al., 2002, 2003b; Goulding et al., 2007; Kimura and Onami, 2007). The situation is less clear at anaphase. Although the hyperactivity of centrosomes in let-99 mutants continues through metaphase, anaphase spindle pole rocking and separation appear reduced compared with wild type; asymmetric anaphase elongation does not occur but cleavage is unequal because the spindle forms in the posterior due to the centration defect (Video 1; Tsou et al., 2002, 2003a). Rather than being hyperactive, these anaphase movements resemble the loss-of-force generation phenotypes exhibited by embryos depleted for Gα, GPR-1/2, or LIN-5, raising questions about the role of LET-99 at this stage. An explanation consistent with our model is that forces are still high in let-99 mutants but that the larger number of astral microtubules present at this stage, coupled with a symmetric distribution of forces, prevents spindle pole oscillations and asymmetric elongation.
In contrast to the three domain model, most proposed mechanisms for posterior spindle displacement in C. elegans have focused on differences in parameters between the anterior and posterior cortical halves, which correspond to the PAR domains (Pecreaux et al., 2006; Kimura and Onami, 2007; Kozlowski et al., 2007). C. elegans has emerged as a premier model for studies of microtubule–cortex interactions, and provides an excellent system for dissecting how cortical pulling forces are asymmetrically regulated. Thus, resolving the distribution of force domains is essential for understanding how polarity cues are translated into spindle positioning mechanics. In this study, using a combination of laser ablation and single centrosome assays, we show that LET-99 provides a positional cue to regulate GPR-1/2–dependent forces around the posterior cortex. This results in an inhibition of pulling forces at the posterior–lateral cortex relative to the posterior-most cortex that we propose is critical for posterior-directed spindle positioning and spindle elongation during asymmetric cell division.
Results
Anterior and posterior net pulling forces are equal in let-99 embryos
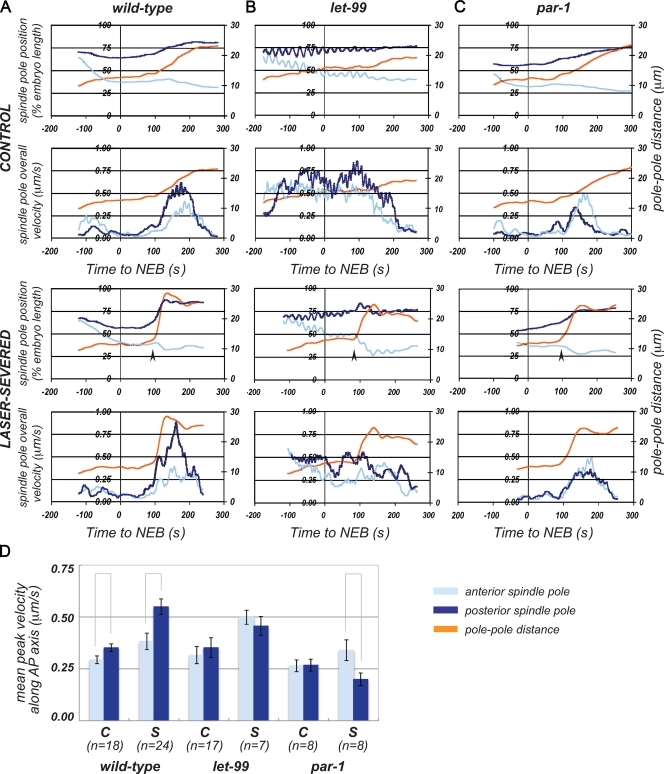
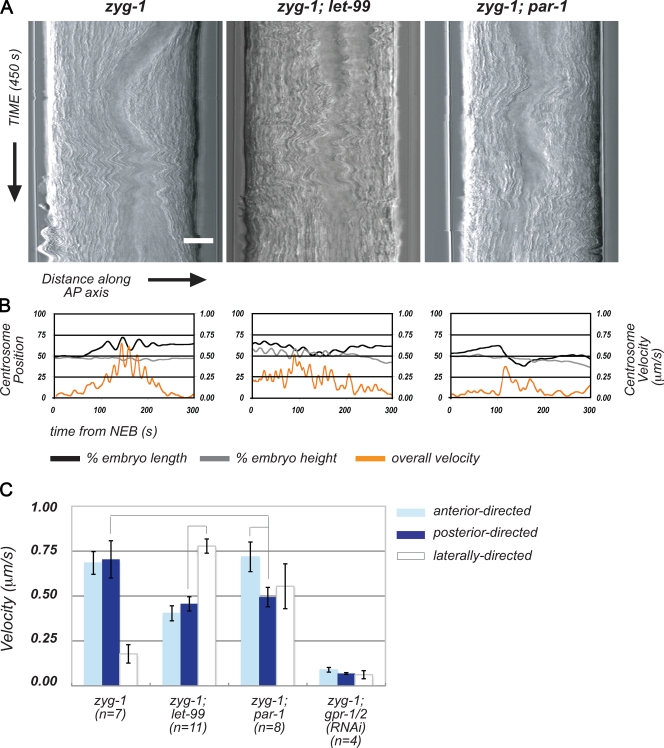
Previous studies support a role for LET-99 in inhibiting cortical pulling forces during nuclear centration and rotation. To address the role of LET-99 in controlling both the levels and asymmetry of net pulling forces during metaphase/anaphase, we conducted spindle-severing experiments. In such experiments, removal of the central spindle allows for each spindle pole to be analyzed independently of each other and of the central spindle; the velocity of spindle pole movements is used as a readout of the forces acting on the spindle (Grill et al., 2001; Colombo et al., 2003; Labbé et al., 2004). We used two methods to remove the central spindle: laser ablation (Fig. 2, Fig. S1, and Video 2) and genetic ablation with the spindle midzone mutant spd-1 (Verbrugghe and White, 2004; Bringmann et al., 2007). Both techniques produced similar results, although rates of pole–pole elongation were lower with spd-1(RNAi) (Table I). For all experiments, the positions of the anterior and posterior centrosomes were tracked over time (Fig. 2, A–C, spindle pole position plots show traces from representative embryos), and centrosome velocities over each 1-s interval were calculated from the tracking data (Fig. 2, A–C, spindle pole overall velocity plots show running averages of overall velocities from representative embryos; Table I shows the mean overall peak velocities). In our analysis of centrosome movements, we found that high centrosome velocities did not always correlate with displacement along the AP axis, particularly in let-99 embryos where centrosomes rocked randomly. We therefore also compared velocities specifically along the AP axis, which is the direction of spindle displacement (Table I and Fig. 2 D, compare mean AP peak velocities).
Figure 2.
LET-99 is required for asymmetry of net spindle positioning forces. (A–C) Analysis of spindle pole movements over time in control and laser-severed spindles for representative wild-type (A), let-99 (B), and par-1 (C) embryos. Top panels for control and severed embryos show spindle pole positions (left y axis, percentage of embryo length) and pole–pole distance (right y axis) plotted against time (x axis). NEB = 0 s. Bottom panels for control and severed embryos show overall spindle pole velocities (left y axis) plotted against time (x axis); the right y axis indicates pole–pole distance. Arrows indicate the time of spindle severing in laser-ablated spindles. (D) Mean spindle pole peak velocities along the AP axis in control (C) and laser-severed spindles (S). Error bars show SEM, lines between columns indicate significance at P = 0.05.
Table I.
Centrosome movements in wild-type, let-99, and par-1
| Parameter measured | Control | Laser severing | Genetic severing | ||||||
| WT (n = 18) | let-99 (n = 17) | par-1 (n = 8) | WT (n = 24) | let-99 (n = 7) | par-1 (n = 8) | spd-1(RNAi) (n = 9) | let-99; spd-1(RNAi) (n = 9) | par-1; spd-1(RNAi) (n = 9) | |
| AP-directed peak velocities | |||||||||
| Anterior centrosome (μm/s) | 0.29 (0.08)ad | 0.32 (0.08)b | 0.26 (0.08)bd | 0.37 (0.18)ad | 0.50 (.08)b | 0.34 (0.05)ad | 0.20 (0.05)ad | 0.25 (0.16)b | 0.20 (0.05)ad |
| Posterior centrosome (μm/s) | 0.35 (0.08)a | 0.35 (0.11)b | 0.27 (0.08)b | 0.55 (0.18)ac | 0.46 (0.11)b | 0.20 (0.08)ac | 0.30 (0.05)ac | 0.24 (0.12)b | 0.16 (0.03)ac |
| Overall peak velocities | |||||||||
| Anterior velocity (μm/s) | 0.58 (0.12)a | 0.53 (0.18)b | 0.83 (0.19)b | 0.56 (0.18)ac | 0.57 (0.07)b | 0.85 (0.18)ac | 0.54 (0.11)ac | 0.40 (0.10)a | 0.89 (0.19)ac |
| Posterior velocity (μm/s) | 0.98 (0.18)a | 0.69 (0.31)b | 0.67 (0.19)b | 0.97 (0.30)ac | 0.78 (0.24)b | 0.65 (0.16)ac | 0.97 (0.18)ac | 0.55 (0.21)a | 0.57 (0.24)ac |
| Maximum amplitude of spindle pole rocking | |||||||||
| Anterior pole (% embryo height) | 14.9 (3.2)a | 12.4 (4.0)a | 20.3 (4.4)b | 13.6 (6.2)a | 25.5 (6.4)b | 24.8 (7.5)a | 18.9 (3.2)a | 12.9 (4.2)a | 20.2 (3.7)a |
| Posterior pole (% embryo height) | 20.4 (4.9)a | 21.7 (4.8)a | 17.4 (5.1)b | 24.8 (10.2)a | 32.1 (7.1)b | 20.0 (6.1)a | 26.7 (3.1)a | 21.1 (2.6)a | 14.2 (4.7)a |
| Spindle measurements | |||||||||
| Minimum spindle length (% embryo length) | 24.2 (2.6) | 24.6 (2.1) | 26.5 (2.9) | 25.7 (6.6) | 26.9 (1.0) | 29.5 (3.6) | 26.6 (2.3) | 25.4 (0.8) | 28.3 (2.0) |
| Maximum spindle length (% embryo length) | 50.3 (2.0) | 39.2 (1.8)e | 49.0 (4.9) | 54.6 (3.1) | 49.7 (2.6)e | 55.6 (4.1) | 55.1 (1.5) | 38.3 (1.1)e | 54.5 (3.4) |
| Spindle elongation rate (μm/s) | 0.06 (0.03) | 0.05 (0.03) | 0.05 (0.02) | 0.49 (0.15) | 0.35 (0.13) | 0.38 (0.21) | 0.12 (0.05) | 0.04 (0.02) | 0.11 (0.05) |
All values are means with SEM given in parentheses.
Statistically significant difference between anterior and posterior spindle poles within a genotype and treatment, P < 0.05 (Student’s t test).
AP values within a genotype and treatment were tested and are not significantly different from each other.
Statistically significant difference between spindle pole velocities in WT and par-1, anterior versus anterior, and posterior versus posterior within a treatment, P < 0.05 (Student’s t test).
The difference between WT and par-1, anterior versus anterior within a treatment, were tested and are not significantly different from each other.
Statistically significant difference between maximum spindle lengths in let-99 embryos, control versus severed, and severed versus spd-1(RNAi), P < 0.05 (Student’s t test).
Laser severing and spd-1(RNAi) in wild-type embryos revealed a posterior-biased asymmetry in centrosome peak velocities, as previously described (Fig. 2 A, Table I, and Video 2; Grill et al., 2001; Labbé et al., 2004). In each case, the posterior spindle pole also moved farther and faster along the AP axis than the anterior spindle pole (Fig. 2 A and Table I), and oscillated with greater amplitude at anaphase (Table I). Spindle severing also increased both the extent and rate of spindle pole separation compared with unsevered wild-type controls (Table I). In contrast, in severed let-99 spindles, spindle pole velocities along the AP axis were symmetrical (Fig. 2 D, Table I, and Video 3). These results correlate with the uniform localization of GPR-1/2 in let-99 embryos (Park and Rose, 2008) and indicate that net spindle positioning forces are uniform. Importantly, the peak velocities in let-99 embryos were intermediate to those of wild-type anterior and posterior spindle poles (Table I), rather than significantly lower than wild type as reported for embryos depleted of Gα or GPR-1/2 (Colombo et al., 2003). We conclude that LET-99 inhibits force generation to regulate net asymmetry of spindle positioning forces at anaphase, as in earlier stages.
Although the velocities along the AP axis in let-99 embryos were symmetric, we did observe slightly higher overall peak velocities for the posterior spindle pole compared with the anterior pole in individual embryos, due to lateral movements (Fig. 2 B and Table I). This subtle asymmetry did not correlate with centrosome movement along the AP axis, and could reflect forces that are independent of asymmetric cortical GPR-1/2 localization (see Discussion).
In addition to positioning the mitotic spindle, cortically based pulling forces also separate spindle poles at anaphase, which is the major mechanism for chromosome separation in C. elegans early embryos (Grill et al., 2001; Oegema et al., 2001). let-99 mutants display a reduced extent of spindle elongation compared with wild type (Table I), and we previously proposed that lateral inhibition of pulling forces by LET-99 could also contribute to spindle pole separation (Tsou et al., 2002). After either laser or genetic spindle severing in let-99 embryos, the maximum pole–pole distance was lower than that observed in wild-type, severed spindles (Table I). Combined with the observations that peak velocities along the AP axis are high and symmetrical in let-99 embryos, these data suggest that lateral inhibition of cortical pulling forces is required for robust anaphase spindle pole separation.
Posterior pulling forces are decreased in par-1 embryos
To further test the role of LET-99 in regulating cortical pulling forces, we analyzed spindle pole velocities in a par-1 mutant background where the normal pattern of LET-99 localization is perturbed. In par-1 single-cell embryos, LET-99 accumulates to high levels throughout the posterior cortex (Fig. 3; Wu and Rose, 2007). As previously described, par-1 spindles undergo a complex sequence of asymmetrical spindle movements (Wu and Rose, 2007). During metaphase, par-1 spindles initially move toward the posterior, but then shift toward the anterior at a variable time during late metaphase or anaphase; anterior spindle poles also undergo more vigorous anaphase oscillations than posterior spindle poles in par-1 embryos (Fig. 2 C, Table I, and Video 1). The anterior-directed movements could result from a decrease in posterior-based pulling forces caused by the mislocalization of LET-99 at the posterior. Alternatively, anterior forces could be increased. There was no asymmetry in peak velocities along the AP axis of spindle poles in par-1 intact spindles (Fig. 2 C and Table I). Thus, to determine how forces on each spindle pole are altered, we conducted spindle severing experiments.
Figure 3.
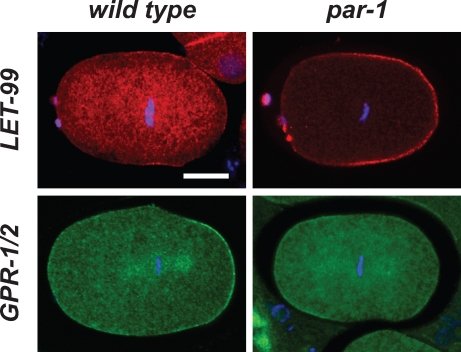
GPR-1/2 is not enriched at the posterior of par-1 embryos during spindle positioning. Representative metaphase embryos stained for LET-99 (red) or GPR-1/2 (green), and DNA (blue). Bar, 10 µm.
In par-1 laser-severed embryos, posterior peak velocities along the AP axis were lower than anterior peak velocities (Fig. 2 D, Table I, and Video 4), and anterior centrosomes oscillated more vigorously at anaphase (Table I). Compared with wild-type severed controls however, anterior centrosome velocities along the AP axis were the same, whereas posterior centrosome AP velocities were reduced in par-1 embryos with severed spindles. Similar results were obtained using the spd-1(RNAi) background (Table I). These data support the view that ectopic localization of LET-99 to the posterior attenuates posterior pulling forces.
Based on our model, LET-99 should be exerting this effect by inhibiting GPR-1/2 localization at the entire posterior cortex. To test this, we stained par-1 embryos for GPR-1/2. As previously reported, in wild-type embryos, GPR-1/2 appeared enriched at the posterior-most cortex in metaphase (4/4) and early anaphase (3/4, 1 had bipolar enrichment). In contrast, none of the par-1 embryos showed posterior enrichment of GPR-1/2 at this stage (n = 13), which is consistent with inhibition by LET-99. Interestingly, some of these par-1 embryos appeared enriched for GPR-1/2 in anterior cortical regions (4/6 metaphase, 2/7 early anaphase; Fig. 3), although staining was not always uniform over the anterior cortex. The failure to detect a pattern complementary to the mislocalized LET-99 in all embryos could be caused by the dynamic localization of GPR-1/2; alternatively, this could be due to other regulators of GPR-1 acting in parallel to LET-99 (Panbianco et al., 2008; Afshar et al., 2010). The anterior enrichment of GPR-1/2 in some par-1 embryos correlates with the anterior movement of the spindle, which occurs variably at metaphase/anaphase, whereas the failure of enrichment of GPR-1/2 at the posterior cortex correlates with the presence of LET-99 and lower pulling forces. Surprisingly though, these observations reveal that the initial posterior movements of the centrosome at metaphase in par-1 embryos are independent of the asymmetric cortical enrichment of GPR-1/2.
Single centrosome assays reveal lateral inhibition of pulling forces in wild-type embryos
The spindle severing experiments in let-99 and par-1 embryos suggest that LET-99 regulates both the magnitude and net asymmetry of spindle positioning forces at metaphase and anaphase. To test this further, we used zyg-1 mutants as a background to track net forces. ZYG-1 is required for centrosome duplication, and loss of paternal ZYG-1 produces a single-cell embryo with a single centrosome and monoaster of radially arranged microtubules (Fig. 4; O’Connell et al., 2001; O’Connell, 2002; Kimura and Onami, 2005). We reasoned that in the geometrically simplified case of a single centrosome, the movement of the aster would reflect the overall net forces generated around the entire cortex. Analysis of zyg-1 embryos can thus reveal the contribution of lateral pulling forces, as well as pulling forces from the anterior and posterior cortices; unlike spindle severing, this allows for tracking of forces throughout the entire cell cycle. This method was used by Kimura and Onami (2005) to investigate the early movements of nuclear centration but was not used to explore the asymmetric movements at metaphase/anaphase.
Figure 4.
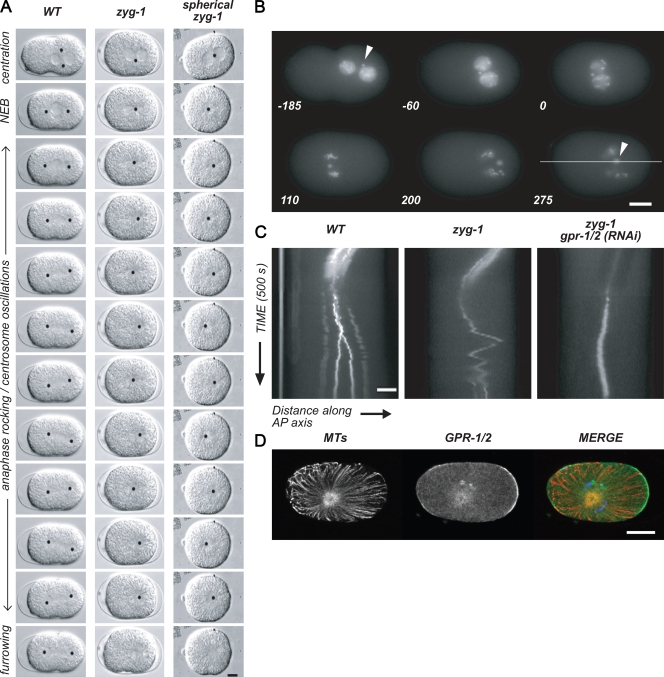
zyg-1 single centrosomes undergo posterior-directed asymmetric transitions along the AP axis. (A) Still frames from time-lapse sequences of wild-type (WT), zyg-1, and spherical zyg-1 embryos. Black dots mark centrosomes. The cell cycle stage is indicated on the left. (B) Still frames from a time-lapse sequence of a zyg-1 embryo expressing GFP::γ-tubulin and GFP::histone. Arrowheads mark the centrosome. The line marks the plane of focus for kymographs in C. Times are given in seconds to NEB. (C) Kymographs from time-lapse videos of representative embryos. WT and zyg-1 embryos express GFP::γ-tubulin and GFP::histone. zyg-1; gpr-1/2(RNAi) embryo expresses GFP::γ-tubulin only for clarity. (D) Fixed WT and zyg-1 embryos stained for MTs, DNA, and GPR-1/2. MTs, microtubules. Bars, 10 µm.
To establish a baseline for analysis, we first analyzed zyg-1 single centrosome movements in an otherwise wild-type background. After pronuclear meeting, the nuclear–centrosome complex migrated to the center (Fig. 4, Table II, and Video 5). Although rotation cannot be scored in single-centrosome embryos, the centrosome moved from a lateral or posterior position to become centrosome leading (Kimura and Onami, 2005), which correlates with the anterior cortical enrichment of GPR-1/2 and higher anterior net forces at this stage. After nuclear envelope breakdown (NEB), the single aster moved toward the posterior of the embryo at a time consistent with the initial posterior shift of bipolar spindles. Interestingly, however, the single centrosome did not exhibit lateral oscillations in the posterior. Rather, zyg-1 asters underwent several transitions along the AP axis. There was no difference between the peak velocities achieved in the anterior- versus posterior-directed movements in zyg-1 embryos (Fig. 5 C), but the transitions were asymmetric with respect to the AP axis, occurring between 50% and 70% embryo length (Table II). After completing a mean of five transitions, the aster came to rest at a final position of ∼60% egg length, which is similar to the final midpoint of the spindle in wild-type embryos (Table II). Significantly, the single aster never oscillated perpendicularly to the AP axis. In all zyg-1 embryos, we observed cortical ruffling and invaginations at the time of cytokinesis, but cleavage ultimately failed. These results support the view that net forces in zyg-1 embryos lie primarily at the anterior and posterior, with lower forces in the lateral regions.
Table II.
Parameters of zyg-1 centrosome movements in various backgrounds
| Parameter measured | zyg-1 (b1) (n = 12) | zyg-1 (oj7) (n = 7) | zyg-1 (oj7); gpr-1/2(RNAi) (n = 4) | zyg-1 (b1) spherical (n = 5) | zyg-1 (b1); par-3 (n = 7) | zyg-1 (oj7); let-99 (n = 11) | zyg-1 (oj7); par-1 (n = 8) |
| Initial centrosome position at NEB (% embryo length) | 52.1 (2.3) | 52.7 (1.9) | 57.9 (2.8) | 52.3 (2.8) | 50.1 (2.8) | 64.5 (1.1) | 55.2 (3.2) |
| Final centrosome position at end of cell cycle (% embryo length) | 63.7 (5.5) | 60.0 (1.3) | 51.3 (1.7) | 61.7 (3.2) | 50.9 (3.4) | 67.4 (1.4) | 42.1 (2.7) |
| Anterior-most centrosome position after NEB (% embryo length) | 50.7 (3.4) | 51.2 (1.9) | 49.8 (1.3) | 49.3 (1.9) | 46.5 (4.2) | 55.0 (0.9) | 38.9 (2.5) |
| Posterior-most centrosome position after NEB (% embryo length) | 70.1 (3.3) | 72.2 (1.0) | 58.0 (2.9) | 67.3 (3.5) | 59.3 (2.5) | 71.8 (0.9) | 61.2 (2.0) |
| Total number of complete AP transits | 5.2 (2.3) | 4.0 (.32) | 0 | 5.0 (1.6) | 3.1 (1.2) | 0 | 0 |
All values are means with SEM given in parentheses.
Figure 5.
Single centrosome movements are randomized in let-99 embryos and anterior-directed in par-1 embryos. (A) Kymographs from time-lapse sequences of representative embryos. The x axis is a cross section along the AP axis. The y axis indicates time. Bar, 10 µm. (B) Plots of centrosome positions and velocities from kymographs in A. The left y axis gives centrosome position in percentage of embryo length (AP-directed movement) or percentage of embryo height (laterally directed movement). The right y axis indicates velocity. The x axis indicates time from NEB. (C) Mean centrosome peak velocities by direction of centrosome travel. The y axis is velocity. Error bars indicate SEM; lines between columns indicate significance at P = 0.05.
To confirm that AP transitions in zyg-1 embryos reflect the normal cues that regulate spindle positioning, we examined the localization of GFP::PAR-6 and GFP::PAR-2. Both markers localized normally to the anterior and posterior cortices, respectively, of zyg-1 embryos (Fig. S2, n = 5 each). In addition, we examined zyg-1; par-3 embryos. In this background, transitions still occurred, though at a central position along the AP axis (Table II); this was consistent with previous studies showing that par-3 embryos exhibit equal forces on both spindle poles (Grill et al., 2001). Further, in zyg-1; gpr-1/2(RNAi) embryos, the nuclear–centrosome complex did not center until after NEB, and the centrosome remained in the center of the embryo during the time of normal spindle displacement, as seen for the bipolar spindle in gpr-1/2(RNAi) embryos (Fig. 4 C and Table II). GPR-1/2 localized normally in zyg-1 embryos (n = 4, Fig. 4 D). Finally, because the elongated shape of the eggshell also imposes an AP constraint, chitinase treatment was used to remove the eggshell and generate spherical embryos (Tsou et al., 2003b). As in untreated embryos, single centrosomes in spherical zyg-1 embryos underwent transitions along the AP axis that were biased toward the posterior (Fig. 4 A and Table II). Significantly, we observed no lateral movement of the centrosome in the spherical embryos, which indicates that AP transitions in zyg-1 embryos are independent of cell shape. Together, these observations indicate that the AP transitions exhibited by zyg-1 embryos reflect the normal wild-type forces that are under control of polarity dependent, G protein signaling cues. Thus, these results are consistent with the model that in wild type, LET-99 inhibits forces in the lateral–posterior domain during centration, rotation, and spindle displacement, such that nuclear and spindle-positioning forces lie along the AP axis of the embryo.
Single centrosome movements are randomized in let-99 embryos and biased toward the anterior in par-1 mutants
To directly test the model that LET-99 inhibits lateral forces, we generated zyg-1; let-99 embryos and tracked the position of the single centrosome throughout the first cell cycle. As expected, the single centrosome rocked vigorously after pronuclear meeting and failed to center (Fig. 5, Table II, and Video 6). After NEB, the centrosome did not transit along the AP axis but continued rocking in the posterior half of the embryo, between 55 and 70% embryo length (Table II). Although the rocking was largely random in direction, centrosome velocities were highest during laterally directed movements (Fig. 5 C). These observations support the view that lateral inhibition by LET-99 is required for AP transitions in zyg-1 embryos.
To test how the balance of net AP forces is altered in par-1 embryos, we analyzed zyg-1; par-1 embryos. The nuclear–centrosome complex was centered in zyg-1; par-1 embryos. After nuclear envelope breakdown, the centrosome shifted toward the posterior, followed by a sudden movement toward the anterior (Fig. 5 and Video 7). The timing of the initial posterior-directed movement in zyg-1; par-1 embryos corresponds to the onset of AP oscillations in zyg-1 single mutant embryos, which suggests that this movement reflects the onset of posterior-directed pulling forces in both single and double mutants. Peak velocities during anterior-directed movement were consistently and significantly faster than the initial posterior-directed shift. Compared with zyg-1 single mutants, however, anterior-directed velocities are not higher; rather, posterior-directed velocities are lower in zyg-1; par-1 embryos (Fig. 5 C). Consistent with spindle severing data from par-1 embryos, these results provide evidence that mislocalization of LET-99 causes a reduction in posterior-directed pulling forces rather than increased anterior pulling. Furthermore, in contrast to zyg-1 embryos, in some zyg-1; par-1 embryos, the centrosome showed some lateral movements after reaching the anterior but did not transit back to the posterior. We hypothesize that zyg-1; par-1 embryos do not transit along the AP axis because lateral forces are not inhibited.
Pulling forces are asymmetrically distributed around the posterior cortex in wild-type embryos
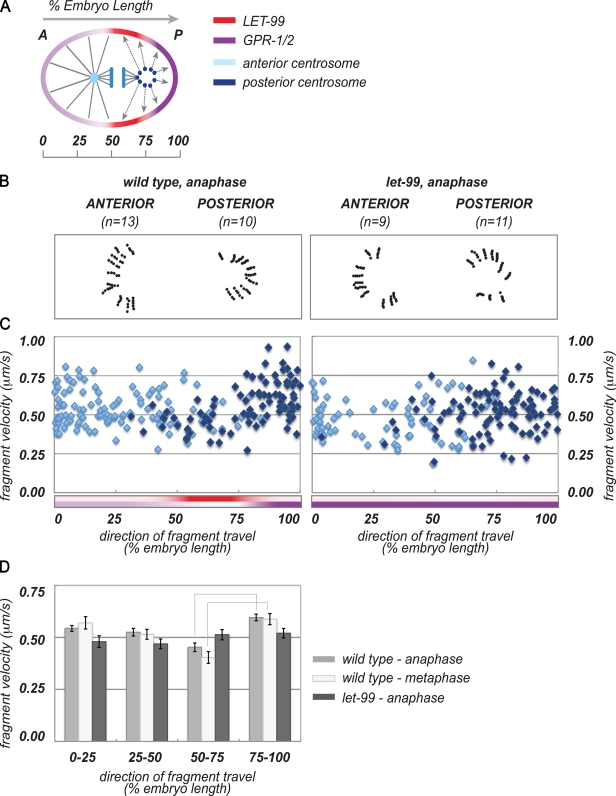
Our results show that LET-99 is required for asymmetry of net forces acting on the spindle, and support the model that the lateral–posterior region has the lowest forces. Spindle-severing and zyg-1 single-centrosome assays both test the balance of net forces acting on centrosomes, but do not address the precise distribution of forces around the cortex. Therefore, to map spindle-positioning forces to specific cortical domains, we conducted centrosome fragmentation assays. In this technique, laser disintegration of the pericentriolar material produces centrosome fragments that remain connected to the cortex by astral microtubules. Measuring the directional velocity of these fragments thus reveals the relative levels of force produced by different regions of the cortex (Grill et al., 2003).
If LET-99 acts to reduce lateral–posterior cortical pulling forces, in wild-type embryos, the velocities of fragments directed toward the lateral–posterior cortex should be low. To test this hypothesis, we fragmented wild-type centrosomes at anaphase and tracked the velocity and direction of fragment travel (Fig. 6 and Video 8). We grouped fragments based on their projected traveling path toward different cortical regions. The posterior–lateral region corresponds to the cortical region with the highest LET-99 levels, whereas the posterior corresponds to the region of highest GPR-1/2 (Figs. 1 B and 6 A; Park and Rose, 2008). Analysis by scatter plot suggested that more fragments from the posterior spindle pole exhibited a trajectory toward the absolute posterior pole than toward the lateral posterior region (Fig. 6 C). Fragments from the anterior spindle pole had a more uniform distribution of trajectories. Quantification of mean velocities for fragments with trajectories toward these regions showed that posterior spindle pole fragments traveled with a slightly higher velocity than anterior pole fragments, which is consistent with published work. Significantly, however, the velocity of fragments moving toward the lateral–posterior region were lower than those of fragments moving toward either the posterior or anterior regions, which is consistent with the three-domain model (Fig. 6, C and D). We next conducted centrosome fragmentation experiments at metaphase, when posterior enrichment of GPR-1/2 is first evident and spindle displacement begins. As with anaphase, the posterior-directed fragments traveled significantly faster than lateral–posterior–directed fragments (Fig. 6 D).
Figure 6.
LET-99 is required for spatial asymmetry of cortical pulling forces during spindle positioning. (A) Schematic of centrosome fragmentation analysis in a WT embryo. Gray lines extending to the cortex indicate astral MTs linking centrosome fragments with cortical force domains. Arrows show the direction of fragment travel. (B) Tracking data from individual centrosomes in representative embryos. (C) Complete dataset from centrosome fragmentation at anaphase. Tracking velocities (y axis) are plotted against the direction of fragment travel (x axis). Posterior centrosome data points are dark blue; anterior, light blue. Colored bars show the extent of LET-99 and GPR-1/2 cortical domains. (D) Velocity data binned by direction of travel, and averaged for cortical domains. Error bars show SEM; lines between columns highlight statistically significant differences between lateral–posterior and absolute posterior mean velocities in wild type (P = 0.05). The following comparisons were also significant. WT (anaphase): 0–25 versus 75–100, 0–25 versus 50–75. WT (metaphase): 0–25 versus 50–75.
To determine if LET-99 is required for these wild-type asymmetries as predicted, we conducted fragmentation experiments in let-99 embryos (Video 9). We were unable to fragment centrosomes at metaphase in let-99 embryos because of the rapid rocking at this stage; in addition, centrosomes were often still misaligned. Thus, we examined let-99 in anaphase, when spindle pole rocking had slowed and poles were aligned along the AP axis. Scatter plot analysis showed a relatively uniform distribution of fragment trajectories for both spindle poles (Fig. 6 C). Further, fragments traveling toward the lateral posterior cortices achieved the same velocities as those traveling toward the anterior and posterior poles of the embryo (Fig. 6, C and D). These results show that spindle positioning forces remain active at anaphase in let-99 embryos, which is consistent with our spindle severing experiments and the model that LET-99 inhibits G protein–mediated forces throughout the cell cycle.
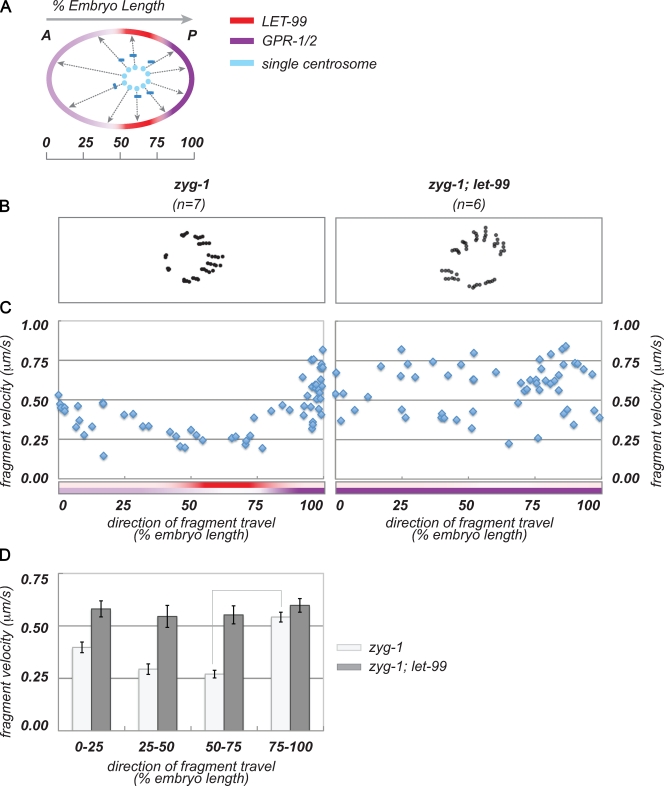
These data also support our model that lateral posterior cortical forces are inhibited at both metaphase/anaphase. These results are consistent with the study by Grill et al. (2003), which showed lower force vectors oriented toward what we define as the lateral–posterior cortex, compared with the posterior pole. However, one caveat to this interpretation is that such a bias in direction and velocity could result from interference by the spindle structure itself, or from the lateral oscillatory behavior of the posterior spindle pole in wild type at anaphase. We did not observe such a strong directional bias for anterior centrosomes, or for either pole in let-99 mutants (Fig. 6, C and D). Nonetheless, to more directly rule out effects of centrosome position and oscillations, we repeated fragmentation experiments in zyg-1 mutants, which have a radial array of microtubules and undergo AP-directed movements at anaphase as described (Fig. 7 and Video 10).
Figure 7.
LET-99 is required for spatial asymmetry of cortical pulling forces in zyg-1 embryos. (A) Schematic of centrosome fragmentation analysis in a zyg-1 embryo, as in Fig. 7. (B) Tracking data from representative embryos. (C) Complete dataset from centrosome fragmentation experiments. Tracking velocities (y axis) are plotted against the direction of fragment travel (x axis). Colored bars show the extent of LET-99 and GPR-1/2 cortical domains. (D) Velocity data binned by direction of travel, and averaged for cortical domains. Error bars show SEM; lines between columns highlight statistically significant differences between lateral–posterior and absolute posterior mean velocities in wild type (P = 0.05). The following comparisons were also significant: zyg-1: 0–25 versus 75–100, 0–25 versus 50–75.
Centrosome fragmentation experiments in zyg-1 embryos were performed after AP transits began, when the centrosome was at approximately the same position as the posterior wild-type spindle pole during metaphase (65% embryo length). After ablation, fragments traveled outwards toward all cortical regions simultaneously, with highest velocities in the direction of the poles. In particular, fragments traveling toward the posterior pole achieved the highest velocities, whereas we observed the lowest velocities in laterally directed fragments (Fig. 7, C and D). In zyg-1; let-99 double mutant embryos (Video 11), laterally-directed fragments exhibited the same high velocities as those traveling toward the posterior pole (Fig. 7, C and D), which indicates that forces are symmetrically distributed around the anterior and posterior cortices in let-99 embryos. These results support our findings in wild-type embryos and suggest that low laterally directed velocities are not caused by interference by the central spindle. Altogether, the centrosome fragmentation experiments strongly support the model that in wild-type embryos, the inhibition of GPR-1/2 by LET-99 at the lateral-posterior cortex results in an inhibition of cortical forces relative to the extreme posterior cortex.
Discussion
We have shown that LET-99 is required for asymmetry of cortical forces during posterior spindle displacement in the single-cell C. elegans embryo. In particular, the results of centrosome fragmentation experiments in wild type revealed that there is an asymmetrical distribution of cortical pulling forces around the posterior spindle pole, with lateral–posterior forces being lower than those at either the posterior or anterior cortical regions. The AP transitions observed in single-centrosome zyg-1 embryos also support a three-force domain regimen, where both anterior and posterior poles generate higher cortical pulling forces than lateral regions. The presence of the cortical LET-99 band is necessary for the down-regulation of lateral forces in both cases, based on our analysis of let-99 and par-1 mutant backgrounds. Thus, even though models involving only two force domains can account for asymmetrical spindle movements, our results clearly support the existence of three cortical force domains. Because LET-99 is required for both the lateral inhibition of GPR-1/2 and its posterior enrichment, it is not possible to distinguish the relative contributions of lateral inhibition versus increased posterior forces to spindle displacement at this time. However, our findings also indicate that lateral inhibition of forces by LET-99 correlates with more efficient spindle elongation. We propose that lateral inhibition of forces contributes to the asymmetry of anaphase spindle elongation and thus spindle displacement. Inhibition of lateral forces could be especially critical in certain cell types to attenuate counterproductive pulling forces produced by large overlapping astral microtubule arrays, or in situations where anaphase B plays a major role in chromosome segregation, as it does in early C. elegans embryos.
GPR-1/2 and LIN-5 are positively required for spindle positioning forces, and recent evidence shows that these proteins associate with the dynein light chain DYRB-1 (Couwenbergs et al., 2007) and the dynein regulator LIS-1 (Nguyen-Ngoc et al., 2007). These observations suggest a mechanistic framework for spindle positioning, and together with the findings presented here, support the view that the cortical accumulation of GPR-1/2 and LIN-5 largely determines cortical pulling forces. G protein signaling is a conserved feature of spindle positioning in several systems. It remains to be determined whether the band pattern of LET-99, and the corresponding bipolar pattern of GPR and LIN-5, is also widely conserved, or whether this pattern is specially adapted for spindle displacement movements that occur in an opposite direction to nuclear rotation movements, as in the C. elegans P lineage cells. Interestingly however, the Drosophila LIN-5 orthologue Mud is present in a bipolar pattern by anaphase (Siller et al., 2006). There is no obvious orthologue of LET-99 in the predicted proteome of Drosophila, but LET-99 has homology to the DEPDC1 family of proteins, which are present in many organisms including sea urchins, zebrafish, and mammals. In addition to spindle positioning, LET-99 has been shown to play a role in cytokinesis in C. elegans, as part of an astral signal that functions redundantly with a midzone signal to induce cytokinetic furrow formation (Bringmann et al., 2007). It will be of interest to determine if LET-99 orthologues are localized in asymmetric band patterns and function in spindle positioning, elongation, or cytokinesis in other systems.
Although the pattern of GPR-1/2 localization closely correlates with the force domains in wild-type embryos, our analysis of spindle movements in let-99 and par-1 backgrounds also uncovered movements that appear to be independent of asymmetric cortical GPR-1/2 enrichment. Although let-99 embryos showed a dramatic loss in asymmetry of centrosome movements along the AP axis, as expected given the uniform cortical GPR-1/2 localization in these embryo, we did observe subtle asymmetries. In particular, the posterior spindle pole had slightly higher overall velocities due to lateral movements compared with the anterior spindle pole. In addition, we observed a posterior-directed movement in both zyg-1; let-99 and zyg-1; par-1 embryos (Fig. 5 B) that occurred at the onset of cortical ruffling, which is late relative to the zyg-1 AP transits. Finally, posterior spindle poles still flatten somewhat in let-99 embryos (unpublished data). Some of these asymmetries could be caused by the closer proximity of the posterior spindle pole to the cortex because nuclear centration is incomplete in let-99 embryos. In addition, actin is required for posterior spindle pole flattening, and PAR-2 and PAR-3 act to restrict flattening to the posterior spindle pole (Severson and Bowerman, 2003). Thus the actin-based forces that flatten the posterior centrosome could account for the residual posterior asymmetries in let-99 and let-99; zyg-1 embryos.
We also observed GPR-1/2 localization-independent asymmetries in par-1 embryos. In this background, the spindle shifts first to the posterior then to the anterior. Interestingly, GPR-1/2 was not enriched at the posterior cortex in par-1 embryos even at the time of the posterior shift, and some embryos exhibited anterior GPR-1/2 localization. One explanation for the par-1 phenotype involves the spindle positioning tether: although posterior pulling begins before metaphase in wild-type embryos, the spindle is held in place by an anterior tether that anchors the anterior spindle pole without concomitant pulling (Labbé et al., 2004). The molecular nature of the tether is unknown but recent data indicates it is cell-cycle regulated (McCarthy Campbell et al., 2009). If the tether prevents active anterior-directed pulling by GPR-1/2–mediated forces, this would produce an asymmetry of pulling forces at metaphase that requires GPR-1/2 activity but not its asymmetric enrichment; however, when the tether is released and anterior pulling forces become active, the lower levels of GPR-1/2 at the posterior in par-1 embryos would result in anterior-directed spindle displacement. If this explanation is correct, it suggests that the tether acts independently of asymmetrical cortical GPR-1/2 localization but does not rule out an effect of the tether on GPR-1/2 or LIN-5 activity. Further, as we observed such an asymmetry in the onset of posterior and anterior pulling in both wild type and par-1, this suggests that PAR-1 does not regulate the spindle tether.
C. elegans is a major model system for elucidating the mechanisms by which the PAR proteins and G protein signaling lead to changes in microtubule-to-cortex interactions that regulate spindle positioning. Beyond a thorough understanding of spindle movements in C. elegans embryos, our determination of the cortical force domains has direct implications for mechanistic models of cortical force generation. Several studies in C. elegans embryos have analyzed parameters such as microtubule dynamics in the anterior versus posterior PAR domains in an attempt to understand how high posterior cortical pulling forces are generated (Labbé et al., 2003; Pecreaux et al., 2006; Kozlowski et al., 2007). In particular, the finding that microtubules contacting the posterior cortical region are more dynamic overall than those contacting the anterior region could suggest that highly dynamic microtubules allow for search and capture by cortically based microtubule motors, and thus higher force generation at the posterior (Labbé et al., 2003). These results were interpreted generally in terms of two domains of force, but because of the curvature of the embryo, both regions analyzed were lateral, rather than at the extreme anterior and posterior poles. Based on the recent analysis of GPR-1/2 (Park and Rose, 2008) and our centrosome fragmentation results, the lateral–posterior region of more dynamic microtubules actually corresponds to the region of lowest cortical force and GPR-1/2 and LIN-5 levels. We therefore propose that the more dynamic microtubules correlate with reduced engagement of astral microtubules with cortical force generators in the lateral–posterior region. This would be consistent with the model proposed by Pecreaux et al. (2006) in which higher levels of GPR-1/2 result in lower rates of detachment of force generators, and thus higher pulling forces; the lateral–posterior cortex with low levels of GPR-1/2 would have correspondingly higher detachment rates and more dynamic microtubules. Further studies examining microtubule dynamics and cortical localizations of regulators with respect to the newly defined force domains will thus allow for refined models of the mechanics of spindle positioning.
Materials and methods
Strains and genetics
Worms were cultured using standard protocols (Brenner, 1974; Church et al., 1995). The following strains were used in this study: N2, wild-type Bristol variant; RL19, unc-22(e66) let-99(or81)/nT1 [unc (n754) let] IV; RL169, unc-22(e66) let-99(ok1403)/nT1 [unc (n754) let] IV, V; RL175, unc-22(e66) let-99(dd17)/nT1 [unc (n754) let] IV, V; KK288 (provided by K. Kemphues, Cornell University, Ithica, NY), rol-4(sc8) par-1(b274)/nT1 [unc (n754) let] IV, V; WH30 (provided by K. O’Connell, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD), zyg-1(oj7) II; OC10, zyg-1(b1) II; KK653 (provided by K. Kemphues), par-3(it71) unc-32(e189)/qC1 III; RL142, par-3(it71) unc-32(e189)/qC1 III; zyg-1(b1) II; OD44 (provided by K. Oegema, University of California, San Diego, San Diego, CA), unc-119(ed3); ltIs2 [pIC27-4; pie-1/GFP-TEV-Stag::tbg01 cDNA; unc-119(+)]; TH32 (provided by K. Oegema), unc-119(ed3); ruIs32[pAZ132; pie-1::GFP;Histone H2B] III; ddIs6[GFP::tbg-1; unc-119 (+)]; JH1623 (provided by G. Seydoux, Johns Hopkins University, Baltimore, MD), unc-119 (ed3); axIs1182[pie-1:gfp::par-2 + unc-119(+)]; JH1512 (provided by G. Seydoux), axIs1137 [pRF-4; pJH7.04 pie-1::gfp::par-6]. Strains for which a provider is not specified were obtained from the Caenorhabditis Genetics Center at the University of Minnesota or created in our laboratory.
Strains carrying the zyg-1 mutation were grown at permissive temperature (15°C), and strains expressing GFP were maintained at 25°C for optimal transgene expression. All other strains were grown at 20°C. Embryos were filmed at room temperature (23–25°C). Three different alleles of let-99 (or81, dd17, and ok1403), all putative nulls (Tsou et al., 2002; Bringmann et al., 2007; Wu and Rose, 2007), behaved similarly in all analyses of intact spindles and were used interchangeably for laser and single-centrosome experiments. For zyg-1 experiments, single-centrosome embryos were produced by shifting zyg-1(b1) or (oj7) L3 hermaphrodites to 25°C until the adult stage, or by mating zyg-1(oj7) males raised at restrictive temperature to wild-type or mutant hermaphrodites. In some cases, the hermaphrodites also carried transgenes expressing GFP markers as indicated in the text. RL142 segregating zyg-1; par-3 double mutants were constructed by crossing OC10 with KK653. For zyg-1; gpr-1/2(RNAi) analysis, zyg-1 crosses were performed on gpr-1/2(RNAi) feeding plates.
RNAi
All RNAi experiments were performed by bacterial feeding using the following constructs from the Ahringer RNAi library: par-1 (V-9E06), gpr-2 (III-5C03), and spd-1 (I-7D17) (Kamath et al., 2003). RNAi treatments were performed at 20°C, and worms were fed for 36 h before analysis, except in the case of RNAi experiments in the zyg-1 background, which were conducted at 25°C for 24 h. After feeding, embryos were screened for published phenotypes by differential interference contrast time-lapse microscopy. In the case of spd-1(RNAi), loss of the spindle midzone was determined by premature spindle pole separation and the presence of yolk granules between spindle poles.
Time-lapse microscopy and laser ablations
For live imaging experiments, embryos were cut from gravid hermaphrodites and mounted on 2% agarose pads with sufficient water to prevent desiccation. Agar pads flatten embryos, which facilitates laser ablations; wild-type and mutant embryos examined using the hanging drop method to prevent flattening (Rose and Kemphues, 1998) showed no differences in phenotypes. Spindles or centrosomes were irradiated with multiple pulses of 436-nm light (Coumarin 440) from a 2-mW pulsed nitrogen laser (VSL-337ND-S; Laser Science, Inc.) mounted on a microscope (BX60; Olympus), and focused through a Plan-Fluor 100×, 1.3 NA objective lens (Olympus).
Spindle midzones were severed at metaphase/early anaphase, just before posterior spindle pole displacement using laser ablation as in Grill et al. (2001). In addition, genetic ablation with the spindle midzone mutant spd-1 was used (Verbrugghe and White, 2004; Bringmann et al., 2007). Although previous studies used klp-7(RNAi) to ablate the midzone (Grill et al., 2001), subsequent work revealed changes in microtubule dynamics in klp-7 (RNAi) embryos (Srayko et al., 2005). Further, we observed greater amplitudes in spindle pole oscillations in klp-7(RNAi) embryos, as well as interactions with mutant phenotypes that were not seen using spd-1(RNAi) or laser severing.
For centrosome fragmentation experiments, the laser was focused in the yolk-free zone of the pericentriolar material. Irradiation resulted in outward expansion of the pericentriolar material; centrosome fragments were visible after irradiation as yolk-free regions traveling toward the cell cortex (Grill et al., 2003). For anaphase experiments in wild-type embryos, anterior or posterior centrosomes were irradiated after posterior spindle pole displacement, after one complete oscillation of the posterior spindle pole. Centrosomes were ablated immediately before posterior spindle pole displacement for metaphase experiments. In let-99 embryos, centrosome fragmentation at anaphase was conducted after the spindle was aligned along the AP axis and spindle rocking had slowed. Centrosomes were fragmented in zyg-1 single centrosome embryos after NEB, after AP transits had started, or, in zyg-1; let-99 embryos, at a similar time after NEB during centrosome rocking. In all cases, differential interference contrast images were captured at 1-s intervals using OpenLab (PerkinElmer) software. Ablation success was assayed by monitoring spindle or centrosome morphology after irradiation.
Quantification of laser severing experiments and centrosome fragmentation
Images and time-lapse sequences were processed using ImageJ software (http://rsbweb.nih.gov/ij/). Prior to tracking, spindle severing and centrosome fragmentation time-lapse sequences were first aligned using the StackReg plugin for ImageJ (http://bigwww.epfl.ch/thevenaz/stackreg/). Aligned stacks were tracked using the Manual Tracking plugin for ImageJ (http://rsbweb.nih.gov/ij/plugins/track/track.html); data were transferred to Excel (Microsoft) for processing.
For spindle severing analysis, relative forces acting on anterior and posterior spindle poles were estimated by calculating peak velocities of centrosomes from tracking data (Grill et al., 2001). To minimize tracking error, videos were tracked three times, and averages of these three replicates were used for subsequent analyses. Control and severed spindle pole trace data from severing experiments were smoothed by applying a running average, and centrosome velocities were calculated over 1-s intervals. All velocities were calculated relative to the direction of centrosome travel. AP-directed velocities refer to movement along the AP axis and were calculated by considering only the change in x from tracking data. Overall velocity includes lateral or rocking movements, and incorporates both the x and y components from tracking data. Peak velocities were calculated for each embryo by taking the maximum velocity achieved by each centrosome during anaphase spindle pole separation in control spindles, or after severing in laser-ablated spindles. Individual peak velocities were then grouped and averaged within a genotype and/or treatment to generate mean peak velocities as shown in Fig. 2 D and Table I. Amplitude measurements in Table I are peak-to-peak amplitude measurements that reflect the lateral range of spindle pole oscillations.
Centrosome fragments after laser fragmentation were tracked as described previously (Grill et al., 2003) by following yolk granules closely associated with expanding pericentriolar material around the ablated centrosome in a 5-s interval immediately after irradiation. 10 yolk granules around the centrosome were tracked for each fragmentation, and a mean velocity was calculated for each track. Only yolk granules that could be followed over the entire interval were included. To correlate fragment velocity with direction of travel, the track produced over the 5-s interval was extended to the cell cortex. The cortical target point was then expressed as a percentage of embryo length and plotted against fragment velocity. To determine mean fragment velocities toward cortical polarity domains, velocities were grouped by direction of travel. Data points derived from anterior centrosome fragmentation were averaged for regions 0–25% (anterior pole) and 25–50% (lateral–anterior); data from posterior centrosomes was averaged for regions 50–75% (lateral–posterior) and 75–100% (posterior pole; Figs. 1 A, 6 A, and 7 A).
In situ immunolocalization
Immunolocalization was performed using standard freeze-fracture methods, followed by a 20-min fixation in −20°C methanol (Tsou et al., 2003a). Briefly, gravid hermaphrodites were rinsed in ddH20, mounted onto polylysine-coated slides, and cut to release embryos. All primary antibodies were diluted in PBS with 0.1% Tween-20 (PBST) at the following ratios: rabbit anti-GPR (1:50; Tsou et al., 2003a; Park and Rose, 2008), rat anti-LET-99 (1:2; Wu and Rose, 2007), rabbit anti-LET-99 (1:50; Tsou et al., 2002), and monoclonal DM1A anti–α-tubulin (1:400; Sigma-Aldrich). Secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were diluted 1:200 in PBST. Primary and secondary antibodies were preabsorbed with acetone powders of GST-expressing bacteria and wild-type worms, respectively. Embryos were stained with DAPI to visualize DNA and determine cell cycle stage, then mounted in Vectashield (Vector Laboratories) mounting medium. The position of the polar body was used to determine the anterior pole of the embryo. For imaging of fixed embryos after laser ablation, individual single-cell embryos were mounted in ddH20 on polysine-coated slides. After laser ablation, slides were transferred immediately to liquid N2 and fixed as above.
Confocal sections were acquired on an Olympus Fv1000 Fluoview Laser Scanning Confocal Microscope using a 60× Planpon NA 1.42 objective lens. For each embryo, z series of five confocal sections, 0.2 µm apart, were taken at the mid-embryo focal plane, with below-saturation acquisition settings. Images shown in panels are maximum intensity projections made in ImageJ (Park and Rose, 2008).
All figures were assembled in Illustrator (Adobe).
Online supplemental material
Fig. S1 shows that laser or genetic ablation removes the central spindle. Fig. S2 demonstrates that PAR proteins are localized normally in zyg-1 embryos. Video 1 shows the spindle positioning phenotypes of wild-type, let-99, and par-1 embryos during the first cell cycle. Videos 2–4 show the phenotype after laser ablation of the central spindle in those genotypes. Video 5 shows that the single centrosome in zyg-1 embryos undergoes AP-directed transits. Video 6 shows that the single centrosome in zyg-1; let-99 embryos undergoes random movements. Video 7 shows that the single centrosome in zyg-1; par-1 embryos moves anteriorly instead of undergoing transits. Video 8 is an example of particle tracking after fragmentation of the posterior centrosome in a wild-type embryo, where the posteriorly directly fragments had the highest velocities. Video 9 shows tracking after centrosome fragmentation in a let-99 embryo, where the laterally and posteriorly directed particles moved at similar high velocities. Video 10 is an example of tracking after centrosome fragmentation in a zyg-1 embryo, where the particles moving toward the anterior or posterior had the highest velocities. Video 11 shows tracking after centrosome fragmentation in a zyg-1; let-99 embryo, where particles moving laterally had high velocities. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001115/DC1.
Acknowledgments
We thank the laboratories of K. Kemphues, J. White, K. O’Connell, K. Oegema, and G. Seydoux for strains. Other strains were provided by the Caenorhabditis Genetics Center (funded by the National Institutes of Health [NIH] National Center for Research Resources). We are indebted to Noelle L’Etoile for use of her laser. We thank Dan Starr, Frank McNally and the reviewers for comments on the manuscript, and the Rose, McNally, and Scholey laboratories for helpful discussions.
This research was supported by NIH grant R01GM68744 (partially funded by the American Recovery and Reinvestment Act) to L.S. Rose, and by National Science Foundation and NIH Predoctoral Fellowships to L.E. Krueger.
Footnotes
Abbreviations used in this paper:
- AP
- anterior–posterior
- NEB
- nuclear envelope breakdown
References
- Afshar K., Willard F.S., Colombo K., Siderovski D.P., Gönczy P. 2005. Cortical localization of the Galpha protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division. Development. 132:4449–4459 10.1242/dev.02039 [DOI] [PubMed] [Google Scholar]
- Afshar K., Werner M.E., Tse Y.C., Glotzer M., Gönczy P. 2010. Regulation of cortical contractility and spindle positioning by the protein phosphatase 6 PPH-6 in one-cell stage C. elegans embryos. Development. 137:237–247 10.1242/dev.042754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H., Cowan C.R., Kong J., Hyman A.A. 2007. LET-99, GOA-1/GPA-16, and GPR-1/2 are required for aster-positioned cytokinesis. Curr. Biol. 17:185–191 10.1016/j.cub.2006.11.070 [DOI] [PubMed] [Google Scholar]
- Church D.L., Guan K.L., Lambie E.J. 1995. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 121:2525–2535 [DOI] [PubMed] [Google Scholar]
- Colombo K., Grill S.W., Kimple R.J., Willard F.S., Siderovski D.P., Gönczy P. 2003. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 300:1957–1961 10.1126/science.1084146 [DOI] [PubMed] [Google Scholar]
- Couwenbergs C., Labbé J.C., Goulding M., Marty T., Bowerman B., Gotta M. 2007. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J. Cell Biol. 179:15–22 10.1083/jcb.200707085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C.R., Hyman A.A. 2007. Acto-myosin reorganization and PAR polarity in C. elegans. Development. 134:1035–1043 10.1242/dev.000513 [DOI] [PubMed] [Google Scholar]
- Galli M., van den Heuvel S. 2008. Determination of the cleavage plane in early C. elegans embryos. Annu. Rev. Genet. 42:389–411 10.1146/annurev.genet.40.110405.090523 [DOI] [PubMed] [Google Scholar]
- Goldstein B., Macara I.G. 2007. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 13:609–622 10.1016/j.devcel.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2008. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9:355–366 10.1038/nrm2388 [DOI] [PubMed] [Google Scholar]
- Gotta M., Ahringer J. 2001. Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat. Cell Biol. 3:297–300 10.1038/35060092 [DOI] [PubMed] [Google Scholar]
- Gotta M., Dong Y., Peterson Y.K., Lanier S.M., Ahringer J. 2003. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol. 13:1029–1037 10.1016/S0960-9822(03)00371-3 [DOI] [PubMed] [Google Scholar]
- Goulding M.B., Canman J.C., Senning E.N., Marcus A.H., Bowerman B. 2007. Control of nuclear centration in the C. elegans zygote by receptor-independent Gα signaling and myosin II. J. Cell Biol. 178:1177–1191 10.1083/jcb.200703159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., Gönczy P., Stelzer E.H., Hyman A.A. 2001. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 409:630–633 10.1038/35054572 [DOI] [PubMed] [Google Scholar]
- Grill S.W., Howard J., Schäffer E., Stelzer E.H., Hyman A.A. 2003. The distribution of active force generators controls mitotic spindle position. Science. 301:518–521 10.1126/science.1086560 [DOI] [PubMed] [Google Scholar]
- Haren L., Merdes A. 2002. Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J. Cell Sci. 115:1815–1824 [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kimura A., Onami S. 2005. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev. Cell. 8:765–775 10.1016/j.devcel.2005.03.007 [DOI] [PubMed] [Google Scholar]
- Kimura A., Onami S. 2007. Local cortical pulling-force repression switches centrosomal centration and posterior displacement in C. elegans. J. Cell Biol. 179:1347–1354 10.1083/jcb.200706005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski C., Srayko M., Nedelec F. 2007. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 129:499–510 10.1016/j.cell.2007.03.027 [DOI] [PubMed] [Google Scholar]
- Labbé J.C., Maddox P.S., Salmon E.D., Goldstein B. 2003. PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr. Biol. 13:707–714 10.1016/S0960-9822(03)00251-3 [DOI] [PubMed] [Google Scholar]
- Labbé J.C., McCarthy E.K., Goldstein B. 2004. The forces that position a mitotic spindle asymmetrically are tethered until after the time of spindle assembly. J. Cell Biol. 167:245–256 10.1083/jcb.200406008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy Campbell E.K., Werts A.D., Goldstein B. 2009. A cell cycle timer for asymmetric spindle positioning. PLoS Biol. 7:e1000088 10.1371/journal.pbio.1000088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T., Afshar K., Gönczy P. 2007. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9:1294–1302 10.1038/ncb1649 [DOI] [PubMed] [Google Scholar]
- O’Connell K.F. 2002. The ZYG-1 kinase, a mitotic and meiotic regulator of centriole replication. Oncogene. 21:6201–6208 10.1038/sj.onc.1205776 [DOI] [PubMed] [Google Scholar]
- O’Connell K.F., Caron C., Kopish K.R., Hurd D.D., Kemphues K.J., Li Y., White J.G. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558 10.1016/S0092-8674(01)00338-5 [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A.A. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panbianco C., Weinkove D., Zanin E., Jones D., Divecha N., Gotta M., Ahringer J. 2008. A casein kinase 1 and PAR proteins regulate asymmetry of a PIP(2) synthesis enzyme for asymmetric spindle positioning. Dev. Cell. 15:198–208 10.1016/j.devcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.H., Rose L.S. 2008. Dynamic localization of LIN-5 and GPR-1/2 to cortical force generation domains during spindle positioning. Dev. Biol. 315:42–54 10.1016/j.ydbio.2007.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecreaux J., Röper J.C., Kruse K., Jülicher F., Hyman A.A., Grill S.W., Howard J. 2006. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr. Biol. 16:2111–2122 10.1016/j.cub.2006.09.030 [DOI] [PubMed] [Google Scholar]
- Rose L.S., Kemphues K. 1998. The let-99 gene is required for proper spindle orientation during cleavage of the C. elegans embryo. Development. 125:1337–1346 [DOI] [PubMed] [Google Scholar]
- Segalen M., Bellaïche Y. 2009. Cell division orientation and planar cell polarity pathways. Semin. Cell Dev. Biol. 20:972–977 10.1016/j.semcdb.2009.03.018 [DOI] [PubMed] [Google Scholar]
- Severson A.F., Bowerman B. 2003. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J. Cell Biol. 161:21–26 10.1083/jcb.200210171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., Doe C.Q. 2009. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11:365–374 10.1038/ncb0409-365 [DOI] [PubMed] [Google Scholar]
- Siller K.H., Cabernard C., Doe C.Q. 2006. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8:594–600 10.1038/ncb1412 [DOI] [PubMed] [Google Scholar]
- Srayko M., Kaya A., Stamford J., Hyman A.A. 2005. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell. 9:223–236 10.1016/j.devcel.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Srinivasan D.G., Fisk R.M., Xu H., van den Heuvel S. 2003. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 17:1225–1239 10.1101/gad.1081203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M.F., Hayashi A., DeBella L.R., McGrath G., Rose L.S. 2002. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development. 129:4469–4481 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Hayashi A., Rose L.S. 2003a. LET-99 opposes Galpha/GPR signaling to generate asymmetry for spindle positioning in response to PAR and MES-1/SRC-1 signaling. Development. 130:5717–5730 10.1242/dev.00790 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Ku W., Hayashi A., Rose L.S. 2003b. PAR-dependent and geometry-dependent mechanisms of spindle positioning. J. Cell Biol. 160:845–855 10.1083/jcb.200209079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbrugghe K.J., White J.G. 2004. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr. Biol. 14:1755–1760 10.1016/j.cub.2004.09.055 [DOI] [PubMed] [Google Scholar]
- Wu J.C., Rose L.S. 2007. PAR-3 and PAR-1 inhibit LET-99 localization to generate a cortical band important for spindle positioning in Caenorhabditis elegans embryos. Mol. Biol. Cell. 18:4470–4482 10.1091/mbc.E07-02-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal R.R., Ahringer J., van Luenen H.G., Rushforth A., Anderson P., Plasterk R.H. 1996. G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell. 86:619–629 10.1016/S0092-8674(00)80135-X [DOI] [PubMed] [Google Scholar]