Abstract
Evidence indicates that endosomal entry promotes signaling by the Notch receptor, but the mechanisms involved are not clear. In a search for factors that regulate Notch activation in endosomes, we isolated mutants in Drosophila genes that encode subunits of the vacuolar ATPase (V-ATPase) proton pump. Cells lacking V-ATPase function display impaired acidification of the endosomal compartment and a correlated failure to degrade endocytic cargoes. V-ATPase mutant cells internalize Notch and accumulate it in the lysosome, but surprisingly also show a substantial loss of both physiological and ectopic Notch activation in endosomes. V-ATPase activity is required in signal-receiving cells for Notch signaling downstream of ligand activation but upstream of γ-secretase-dependent S3 cleavage. These data indicate that V-ATPase, probably via acidification of early endosomes, promotes not only the degradation of Notch in the lysosome but also the activation of Notch signaling in endosomes. The results also suggest that the ionic properties of the endosomal lumen might regulate Notch cleavage, providing a rationale for physiological as well as pathological endocytic control of Notch activity.
Keywords: Drosophila, V-ATPase, Notch signaling, Endocytosis, ESCRT
INTRODUCTION
Cell-cell signaling via the Notch receptor is used throughout development to regulate multiple cell behaviors, and inappropriate activation of Notch is emerging as a common hallmark of an increasing number of cancers (reviewed by Schweisguth, 2004; Miele et al., 2006; Roy et al., 2007). Thus, resolving the mechanisms by which Notch signaling is regulated is of great importance and of widespread interest in order to understand human development as well as to devise effective anticancer therapies. In response to ligand engagement, the Notch receptor is activated by S3 cleavage, a γ-secretase-mediated intramembrane proteolysis that liberates the Notch intracellular domain from its transmembrane anchor, allowing the soluble form to travel to the nucleus, where it regulates the transcription of a variety of important target genes (Schweisguth, 2004).
Mounting evidence, particularly in Drosophila, has pointed to the unexpected involvement of the endosomal system in regulating activation of the Notch receptor in signal-receiving cells. Importantly, endosomal regulators can either increase or decrease Notch signaling depending on the specific site of the endocytic pathway at which they act. For example, factors that positively regulate traffic from the cell surface to endosomes, including Dynamin (Shibire – FlyBase), the ubiquitin ligase Deltex, the syntaxin Avalanche (Avl; Syntaxin 7), the GTPase Rab5, the Rab5 effector Rabenosyn-5, and the Sec1/Munc18 family protein Vps45, are required to promote signaling. By contrast, Endosomal sorting required for transport (ESCRT) complex components, the C2-domain protein Lethal (2) giant discs 1 (Lgd), and the ubiquitin ligase Su(dx), which subsequently sort cargo within the endosome towards lysosomal degradation, are required to prevent excess signaling (reviewed by Brou, 2009; Fortini and Bilder, 2009; Fürthauer and González-Gaitán, 2009; Tien et al., 2009). Much of this evidence points to early endosomes as important sites of signaling activation. However, the molecules and mechanisms that restrict activation of the Notch receptor to this endosomal compartment remain unknown.
MATERIALS AND METHODS
Genetics
Mosaic eyes were generated as described previously (Tapon et al., 2001). Completely mutant eye discs (referred to in the text as mutant discs) were generated as described (Newsome et al., 2000). Follicle cell clones were generated as described previously (Bilder et al., 2000). Other alleles utilized were: Vha55j2e9 (Davies et al., 1996), VhaSFDEY04644 (Bloomington Stock Center #15758), vps22zz13 (Vaccari et al., 2009), HrsD28 (Lloyd et al., 2002), E(spl)mβ-lacZ (Nellesen et al., 1999) and UAS GFP-Lamp-1 (a gift of T. E. Rusten, Norwegian Radium Hospital, Oslo, Norway). Recombined chromosomes were generated using standard genetic techniques. The lesion in Vha68-2R6 was identified by sequencing PCR products amplified from genomic DNA; at least two independent reactions were sequenced for each allele. Detailed genotypes for each experiment are available upon request; the primers used to sequence Vha68-2 are listed in Table S1 in the supplementary material.
Live disc culture assays
Notch internalization assays were performed as described (Vaccari et al., 2008). For Lysotracker incorporation, mosaic eye discs or ovaries were dissected in Drosophila cell medium (M3, Sigma) and incubated in medium containing 1 μM Lysotracker (DND-99, Molecular Probes) for 5 minutes at room temperature, washed in fresh medium, mounted and imaged. For Dextran-uptake assays, mosaic eye discs dissected in M3 medium were incubated for 10 or 40 minutes in Texas Red-Dextran MW 3000, lysine-fixable (Molecular Probes), washed, fixed and counterstained using an antibody to Notch ICD, mounted and imaged.
Immunohistochemistry and optical microscopy
Immunostainings were performed as described (Vaccari et al., 2009). Actin stains used TRITC-conjugated phalloidin (Sigma). Primary antibodies to the following antigens were used: Avl (Lu and Bilder, 2005), Hrs (Lloyd et al., 2002), ubiquitin (FK2; Biomol), Caspase-3 (Decay – FlyBase) (Cell Signaling Technologies), β-gal (Cappell) and Notch ECD, Notch ICD, Mmp1, Hnt, Cut, Elav and Dlg (DSHB). Alexa 488- or Alexa 647-conjugated secondary antibodies (Molecular Probes) were used. All images shown are single confocal sections taken with a TCS microscope (Leica) using 16×/NA 0.5, 40×/NA 1.25 or 63×/NA 1.4 oil-immersion lenses. Images were edited with Adobe Photoshop CS and were assembled with Adobe Illustrator. Quantification was performed using ImageJ (NIH). The data presented are the average of measurements from eight confocal sections for each genotype (Fig. 1H), of two discs for each genotype (Fig. 3L), and of four discs for each genotype (Fig. 4M).
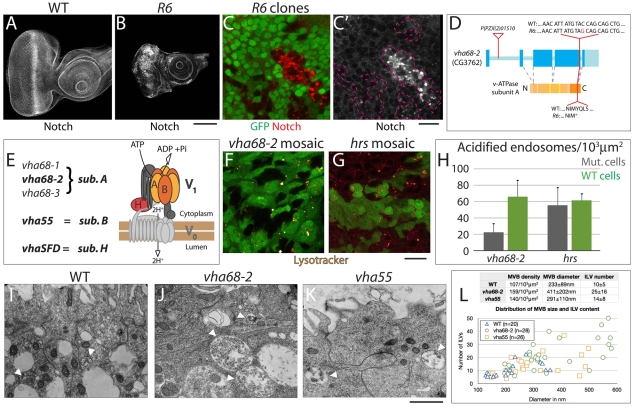
Fig. 1.
Altered Notch protein levels and endosomal acidification in Drosophila eye imaginal disc cells mutant for Vha68-2. (A,B) Eye imaginal discs stained to detect Notch. Compared with wild type (WT) (A), eye discs consisting predominantly of R6 mutant cells are smaller and show high levels of mislocalized Notch (B). (C,C′) Notch staining (red) of mosaic R6 eye discs; the absence of GFP expression marks the mutant tissue. Clones of R6 mutant cells (outlined in C′) display various degrees of Notch accumulation in intracellular puncta. (D) Drosophila Vha68-2 gene and protein organization with the locations and nature of mutant alleles shown. Exons and introns are shown in blue and light blue, respectively, and the protein domains and ATP-binding sites are shown in orange and yellow, respectively. The transposon l(2)01510 is inserted in the first intron, whereas the R6 mutation introduces a stop codon in the region encoding the ATP synthase α/β chain (dark orange). (E) List of genes that encode subunits A, B and H and schematic of subunit organization of the V-ATPase proton pump. Subunit A, which is encoded by Vha68-1, -2 and -3 (CG5075 – FlyBase) in Drosophila, is the ATP-binding subunit. (F,G) Lysotracker incorporation in cultured mosaic Vha68-2 (F) and Hrs (G) eye discs. Clones of mutant cells within WT tissue are marked by the absence of GFP expression. Compared with WT cells, Vha68-2 cells incorporate very little Lysotracker, indicating that endomembrane acidification is severely impaired. By contrast, clones of Hrs mutant cells incorporate similar levels of Lysotracker to WT cells. (H) Quantification of the experiment shown in F,G. (I-K) Transmission electron microscopy of WT eye discs (I) and of Vha68-2 (J) and Vha55 (K) completely mutant discs (see Materials and methods). A portion of two to three eye disc cells facing the peripodial membrane is shown in each micrograph. Multivesicular bodies (MVBs) are marked by arrowheads. (L) Quantification of the experiment shown in I-K. Average MVB density, diameter and internal luminal vesicle (ILV) content per genotype are presented in the table, and the distribution in size of representative individual MVBs and their ILV content are plotted. Compared with WT, Vha68-2 and Vha55 cells contain more and larger MVBs, which are filled with an higher number of ILVs. Extrapolation of volumes from the linear data suggests that Vha68-2 mutant cells contain up to three times more MVBs, and that these are up to five times larger than those of WT cells. Scale bars: 100 μm in A,B; 10 μm in C,C′,F,G; 1 μm in I-K.
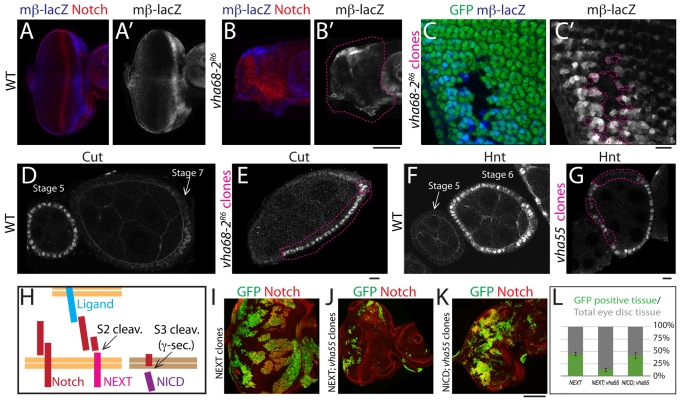
Fig. 3.
V-ATPase activity is required to activate the Notch receptor downstream of ligand binding in epithelial tissue. (A-C′) Drosophila eye discs stained to detect the Notch reporter mβ-lacZ and Notch (single mβ-lacZ channel is shown in A′-C′). Compared with WT (A), mβ-lacZ expression is reduced in predominantly mutant Vha68-2R6 discs (B) and in clones of Vha68-2R6 mutant cells located posterior to the morphogenetic furrow in mosaic discs (C). Mutant tissue is outlined in B′,C′. The regions of the tissue that accumulate Notch the most appear to express mβ-lacZ the least (compare B with B′). In C, mβ-lacZ is expressed at high level in WT developing photoreceptors but is absent in clones of mutant cells. (D-G) Egg chambers at stages 5-7 of oogenesis stained for Cut (D,E) and Hnt (F,G). In WT cells, Cut expression is subjected to Notch-dependent downregulation after stage 5 (D), whereas Hnt is expressed in a Notch-dependent manner from stage 6 onwards (F). In Vha68-2 mutant FCs, Cut expression is maintained after stage 5 (E; for GFP channel see Fig. S4C in the supplementary material), whereas in Vha55 mutant FCs, Hnt expression is reduced after stage 5 (G; for GFP channel see Fig. S4D in the supplementary material), in both cases indicating impairment in Notch signaling activation in V-ATPase mutant FCs. (H) Schematic of the Notch cleavage events preceding signaling activation. The full-length Notch is cleaved at the S2 site to generate the γ-secretase cleavage substrate NEXT. NEXT is subsequently cleaved by γ-secretase at the S3 site to generate the active cytoplasmic NICD fragment. (I-K) Mosaic eye discs generated using the MARCM system stained to detect NICD. GFP-positive cells expressing NEXT overproliferate to form very large clones (I). By contrast, GFP-positive cells expressing NEXT that lack V-ATPase activity underproliferate and form small clones (J). In GFP-positive cells that express NICD and lack V-ATPase activity, normal proliferation is restored (K). (L) Quantification of the experiment shown in I-K. Scale bars: 100 μm in A-B′,I-K; 10 μm in C-G.
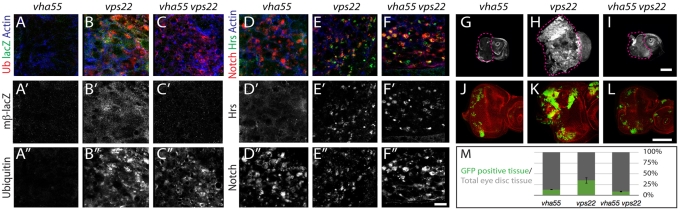
Fig. 4.
Impaired V-ATPase activity prevents the signaling activation and overproliferation observed in ESCRT endocytic tumors. (A-C′) Ubiquitin staining of Vha55, vps22 and Vha55 vps22 double-mutant Drosophila eye discs expressing mβ-lacZ. As in Vha68-2R6, mβ-lacZ expression is low in Vha55 discs and ubiquitin accumulation is not observed (A-A′). By contrast, vps22 mutants express high levels of mβ-lacZ and display accumulation of ubiquitin (B-B′). The Vha55 vps22 double mutant expresses levels of mβ-lacZ that are similar to those of Vha55 mutants, but displays an accumulation of ubiquitin that is similar to that of vps22 mutants (C-C′). (D-F′) Vha55, vps22 and Vha55 vps22 double-mutant eye discs colabeled to the detect the ICD of Notch and Hrs. As in Vha68-2R6, Notch does not colocalize with Hrs in Vha55 discs (D-D′). By contrast, in both vps22 mutants (E-E′) and Vha55 vps22 double mutants (F-F′) partial colocalization of Notch and Hrs is observed. (G-I) mβ-lacZ expression and proliferation in eye discs. As in Vha68-2R6, Vha55 mutant discs underproliferate and express low levels of mβ-lacZ (G), whereas vps22 mutants express high levels of mβ-lacZ and overproliferate (H). The Vha55 vps22 double mutant expresses levels of mβ-lacZ and exhibits levels of proliferation that are similar to those of Vha55 mutants (I). (J-L) Mosaic eyes generated by MARCM stained to detect cortical actin. Compared with GFP-positive vps22 mutant cells, which display Notch-dependent overgrowth (K), GFP-positive Vha55 mutant cells (J) and GFP-positive Vha55 vps22 double-mutant cells (L) do not overgrow. (M) Quantification of the experiment shown in J-L, indicating that lack of V-ATPase function rescues the Notch-dependent overproliferation observed in vps22 mutant cells. Scale bars: 10 μm in A-F′; 100 μm in G-L.
Electron microscopy
Wild-type (WT), Vha55 and Vha68-2 completely mutant eye discs were placed in 0.1 M cacodylate buffer containing 2.5% glutaraldehyde for 3 hours at room temperature. Samples were post-fixed in osmium tetroxide (Electron Microscopy Science, Hatfield, PA, USA) for 2 hours and in uranyl acetate (Electron Microscopy Science) for 1 hour at room temperature. Samples were then dehydrated through a graded ethanol series with propylene oxide as a transition fluid (TAAB Laboratories Equipment, Aldermaston, UK) and embedded in epoxy resin (Poly-Bed; Polysciences, Warrington, PA, USA) overnight at 42°C then for 2 days at 60°C. Subsequent examination of the region of interest (the eye disc epithelium underneath the peripodial membrane) was performed on semi-thin sections (500 nm) stained with Toluidine Blue. Ultrathin sections (50 nm) were then cut and stained with uranyl acetate and lead citrate and images were taken with a CM10 (Philips, Eindhoven, The Netherlands) or Tecnai 12-G2 (FEI, Eindhoven, The Netherlands) electron microscope.
Quantification of the multivesicular body (MVB) compartment within ultrathin sections was performed using iTEM software (FEI) (Fig. 1L). We determined the density of MVBs as the total number of MVBs in 187, 195 and 186 μm2 of WT, Vha68-2 and Vha55 mutant tissue, respectively, and normalized each to 1000 μm2. We measured MVB diameter as the length of the major axis, measuring only those completely enclosed in the micrograph.
RESULTS
V-ATPase function controls Notch protein levels, endosomal acidification and overall tissue size and morphology of Drosophila epithelial organs
To identify genes involved in endocytic control of the Notch receptor, we screened a collection of Drosophila mutants enriched for endocytic regulators (Menut et al., 2007) to identify those that display altered Notch localization. We generated eye imaginal discs consisting predominantly of mutant cells (see Materials and methods) and immunostained using Notch antibodies. We found that discs mutant for the single allele complementation group MENE2L-R6 accumulate high levels of Notch as compared with wild-type (WT) discs (Fig. 1A,B). Clones of MENE2L-R6 mutant cells generated in the context of a mosaic eye disc also displayed substantial accumulation of Notch, which was found in large intracellular puncta (Fig. 1C). MENE2L-R6 discs were consistently smaller than WT discs and also displayed aberrant morphology, compromised epithelial polarity, and upregulated Matrix metalloprotease 1 (Mmp1) production (Fig. 1A,B; see Fig. S1A-B′ in the supplementary material). Together, these data suggest that R6 disrupts a gene that regulates Notch trafficking as well as the proper growth and organization of imaginal disc tissue.
We mapped the R6 mutant by complementation to a deficiency that removes the chromosomal region 34A3. Tests with lethal transposon insertions within the deficiency region revealed that R6 fails to complement P[PZ]l(2)01510, which is inserted in the first intron of the predicted gene CG3762 (Fig. 1D). CG3762 is also known as Vha68-2, the most widely expressed of the three Drosophila genes that encode the A subunit of the multiprotein vacuolar ATPase (V-ATPase) proton pump (Allan et al., 2005) (Fig. 1E). The A and B subunits form a hexamer that binds nucleotide in the peripheral V1 sector; nucleotide binding and hydrolysis are essential for transmembrane proton conductance through the integral membrane V0 sector (reviewed by Nishi and Forgac, 2002) (Fig. 1E). Sequencing of Vha68-2 in R6 mutants revealed a nonsense mutation in the region encoding the C-terminal domain. Based on the crystal structure of the related A-ATPase subunit A (Maegawa et al., 2006), the truncated C-terminal domain of the R6 mutant protein, even if expressed, is likely to lack part of the region that contacts subunits D and F, which are involved in torque generation (Fig. 1D,E). Since torque generation is essential for proton transport and thus endomembrane acidification, we tested the ability of Vha68-2 mutant cells to incorporate the vital dye Lysotracker, which accumulates in acidified lysosomes. Compared with WT cells or cells mutant for the endosomal component Hrs, we found that Vha68-2 mutant cells incorporate very low levels of Lysotracker, indicating impairment of the ability to acidify endocytic organelles (Fig. 1F-H).
Failure of Lysotracker incorporation into Vha68-2 mutant cells is not due to absent endocytic structures, as staining with endosomal markers and electron microscopy revealed that both early and late endosomes, as well as lysosomes, are present in mutant cells (Fig. 1I,J,L; Fig. 2E-G). In addition, ultrastructural analysis of the morphology of the endocytic compartment revealed that Vha68-2 mutant cells contain, compared with WT, more multivesicular bodies (MVBs), a degradative endosomal organelle. These MVBs were also enlarged and contained a high number of internal luminal vesicles (ILVs) (Fig. 1I,J,L). Consistent with a role for acidification in activating low-pH lipid hydrolases (Soyombo et al., 2006), these data suggest that Vha68-2 is required for lysosomal degradation of ILVs. Overall, the Lysotracker incorporation and the ultrastructural analyses indicate that Vha68-2 causes a substantial loss of proton transport activity.
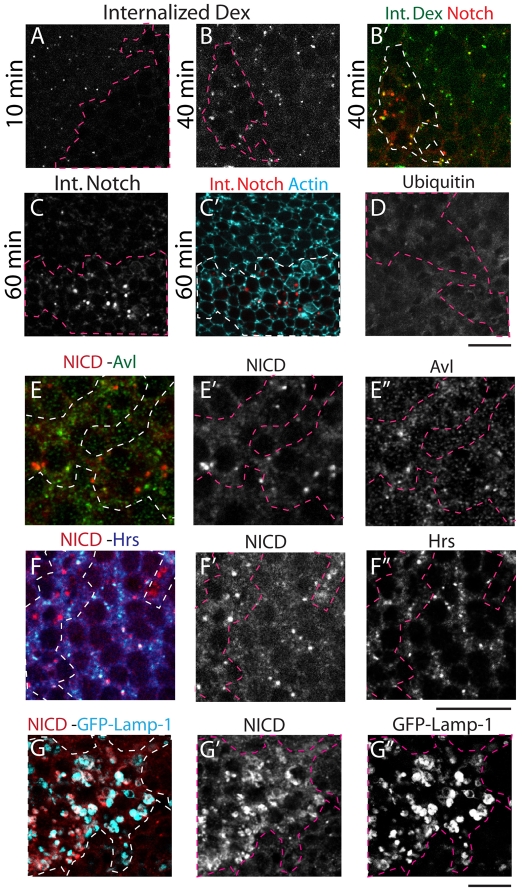
Fig. 2.
V-ATPase activity controls lysosomal degradation of endocytic cargoes. (A-B′) Dextran uptake in live mosaic Vha68-2R6 Drosophila eye discs. Discs were counterstained to detect the intracellular domain of Notch (NICD) to assess colocalization (B′). Mutant tissue is outlined. Compared with WT tissue, mutant tissue internalizes less Dextran after 10 minutes (A), whereas after 40 minutes the amount of Dextran internalized is similar (B). Note that some of the internalized Dextran at 40 minutes colocalizes with the accumulated Notch (B′). (C,C′) Notch internalization assay in live imaginal discs. In WT tissue (C), surface-bound anti-Notch extracellular domain (NECD) is internalized into endosomes and degraded after 60 minutes. In Vha68-2R6 tissue (C′), anti-NECD is internalized but is not degraded, accumulating intracellularly (actin staining is used to demarcate cell boundaries). (D) Vha68-2R6 mosaic eye imaginal disc stained to detect ubiquitin. No ubiquitin accumulation is observed in the mutant tissue. (E-F′) Vha68-2R6 mosaic eye imaginal discs stained with anti-NICD and anti-Avl (E) or anti-Hrs (F); separate channels are shown in E′,E′,F′,F′. No colocalization of Notch is observed with either endosomal marker. (G-G′) Vha68-2R6 mosaic eye imaginal discs expressing GFP-Lamp-1 and stained to detect NICD (G); separate channels are shown in G′,G′. Partial colocalization in the mutant tissue is observed. Scale bars: 10 μm.
To confirm that the Notch trafficking, growth and architecture phenotypes observed in Vha68-2 imaginal disc cells are due to a lack of V-ATPase activity, we analyzed the phenotypes of mutations that disrupt other V-ATPase subunits. Vha55j2e9 is a transposon insertion that creates a strong mutant in the single Drosophila gene encoding the B subunit of V-ATPase (Davies et al., 1996) (Fig. 1E). We found that, similar to Vha68-2R6 discs, Vha55j2e9 mutant discs display Notch accumulation, Mmp1 expression, small size and aberrant morphology; they also showed ultrastructural defects similar to, although less strong than, those of Vha68-2 (Fig. 1K,L; see Fig. S1C and Fig. S2C,D in the supplementary material). In addition, we found that eye discs homozygous for EY04644, a transposon insertion in the 5′UTR of VhaSFD, the single Drosophila gene encoding V-ATPase subunit H (Allan et al., 2005), phenocopy Vha68-2 and Vha55 mutant discs (see Fig. S2E,F in the supplementary material). Moreover, a portion of the cells mutant for any of the three genes is eliminated from the disc, especially when surrounded by WT cells (see Fig. S3 in the supplementary material). These data strongly suggest that the common phenotypes observed in Vha55, Vha68-2 and VhaSFD mutant cells are caused by impaired V-ATPase function. Taken together, they indicate that the V-ATPase controls endosomal acidification and is also a major regulator of Notch localization, epithelial organization and growth in imaginal disc cells.
Lysosomal degradation, but not endocytic trafficking to late endosomes/lysosomes, is impaired in cells lacking acidic compartments
The high levels of intracellular Notch seen in V-ATPase mutant cells could in theory result from impaired exocytosis or endocytosis. To test whether V-ATPase controls endocytic transport in imaginal cells, we first followed the uptake of Dextran over time in mosaic eye discs containing clones of Vha68-2 mutant cells that were cultured ex vivo. Compared with WT cells, internalization of Dextran was reduced at the 10-minute time point in Vha68-2 mutant cells, but after 40 minutes Vha68-2 cells accumulated similar amounts of Dextran to WT cells (Fig. 2A,B). These data indicate that although its rate may be reduced, endocytic trafficking is still functional upon impairment of Vha68-2 expression. To directly test endocytic trafficking of Notch, we used an ex vivo assay for Notch internalization and found that, compared with WT cells, Notch is internalized but fails to be degraded in Vha68-2 mutant cells (Fig. 2C). In mutant cells, Notch accumulated in large and distinct intracellular puncta (Fig. 2C′). These intracellular puncta resemble those seen in discs treated with chloroquine, a compound that prevents lysosomal acidification (Vaccari and Bilder, 2005).
To determine the specific site of endocytic Notch accumulation in Vha68-2 mutant cells, we colabeled eye disc cells for Notch and Avl or Hrs, two early endosomal markers, or GFP-Lamp-1 (CG3305), a late endosomal/lysosomal marker. We found that in Vha68-2 mutant cells, Notch rarely colocalizes with either Hrs or Avl, but frequently colocalizes with GFP-Lamp-1 (Fig. 2E-G). In addition, we observed colabeling of Notch with internalized Dextran at the 40-minute time point, a time sufficient to deliver Dextran to late endosomes (Fig. 2B′). Consistent with Notch accumulation in the late endosomal/lysosomal compartment, Vha68-2 mutant cells did not accumulate ubiquitin (Fig. 2D), a phenotype that is associated with Notch trapping during endosomal sorting and is not observed in late endosomal mutants such as fab1 (Rusten et al., 2006). Taken together, these data indicate that endocytic trafficking in V-ATPase mutant cells can sort Notch within the endosome but leads to accumulation in a post-sorting endocytic compartment that has characteristics of the late endosome/lysosome.
V-ATPase activity is required for Notch activation in signal-receiving cells prior to γ-secretase-dependent S3 cleavage
Since V-ATPase mutant cells possess high levels of undegraded Notch, we asked whether this phenotype affects Notch signaling. Because other mutants that trap Notch in late endosomes, such as fab1, do not alter Notch signaling (Rusten et al., 2006; Vaccari et al., 2008), we expected Notch activity to be unaffected in cells lacking V-ATPase activity. Surprisingly, we found that expression of the Notch signaling reporter mβ-lacZ is significantly reduced in Vha68-2 mutant disc cells, as compared with the stereotyped levels and patterns seen in WT discs (Fig. 3A,B) (Nellesen et al., 1999). Interestingly, the degree of reduction in reporter expression paralleled the degree of Notch accumulation in mutant cells (Fig. 3B′). Similarly, compared with WT, mβ-lacZ expression was absent in most Vha68-2 mutant eye disc cells (Fig. 3C). As also observed for Notch accumulation (Fig. 1C), this phenotype was variably penetrant, with some cells showing mild phenotypes. This could reflect a functional redundancy of Vha68-2 with other Vha68 genes or hypomorphy of the allele. Despite the phenotypic variability, these results suggest that V-ATPase activity is in fact positively required for Notch signaling.
Lack of Notch signaling in V-ATPase mutant tissue could be due to effects on Notch ligands or the Notch receptor. To discriminate between these possibilities, we utilized Drosophila ovaries. We first asked whether V-ATPase mutant somatic follicle cells (FCs) show the acidification and Notch degradation defect observed in V-ATPase mutant eye disc cells. We made genetic mosaic ovaries in which some FCs were mutant for Vha68-2. Similarly to mosaic eye discs, we found that, compared with neighboring WT cells, mutant FCs failed to incorporate Lysotracker and accumulated high amounts of Notch intracellularly (see Fig. S4A,B in the supplementary material).
In ovaries, the Notch ligand Delta is upregulated in the germ cells during stages 6-7 of oogenesis to activate Notch signaling in the adjacent FCs (Deng et al., 2001; Lopez-Schier and St Johnston, 2001), leading to downregulation of target genes such as cut and to activation of target genes such as hindsight (hnt; pebbled – FlyBase) (Fig. 3D,F) (Sun and Deng, 2005; Sun and Deng, 2007). Thus, to assay the extent of Notch signaling activation in Vha68-2 mutant FCs, we immunostained mosaic ovaries to detect expression of Cut and Hnt. These ovaries lack V-ATPase function in mutant FCs but retain normal V-ATPase activity in Delta-expressing germ cells. We found that Cut expression is maintained beyond stage 6 in Vha68 mutant FCs (Fig. 3E; see Fig. S4C in the supplementary material). A similar phenotype was observed in VhaSFD mutant FCs (see Fig. S4E in the supplementary material). Consistent with an impairment in Notch signaling activation in V-ATPase mutant FCs, we also found a substantial reduction in Hnt expression in Vha55 mutant FCs (Fig. 3G; see Fig. S4D in the supplementary material). Taken together, these data indicate that V-ATPase activity is required cell-autonomously in signal-receiving cells for activation of the Notch receptor.
Activation of the Notch receptor following ligand binding requires a proteolytic cleavage called S3, which is mediated by γ-secretase (reviewed by Schweisguth, 2004). To test whether V-ATPase function is required for this cleavage event, we compared WT and mutant cells expressing either of two activated signaling forms of Notch. Notch extracellular truncation (NEXT) is a membrane-tethered form of Notch that is a constitutive substrate for S3 cleavage by γ-secretase in WT cells; Notch intracellular domain (NICD) represents the soluble cleavage product of this reaction (Fig. 3H). Clones of WT imaginal disc cells expressing NEXT overproliferate due to unrestrained Notch signaling activation (Fig. 3I). However, Vha55 mutant cells expressing NEXT did not overproliferate and instead created only small clones, of a size similar to Vha55 clones (Fig. 3J, compare with Fig. 4E), indicating that the production of an active signaling form of Notch is blocked in these cells. Finally, Vha55 mutant cells expressing NICD overproliferated similarly to WT clones expressing either transgene, revealing that the requirement for V-ATPase activity in Notch signaling can be bypassed by providing a cleaved form (Fig. 3K). These data show that V-ATPase activity is required downstream of ligand binding, but upstream of the S3 activating cleavage.
Acidification of the early endosomal lumen is a prerequisite for efficient activation of Notch signaling
Although the above data suggest a role for V-ATPase activity in Notch cleavage, they do not specify in which organelle this activity might be required, as V-ATPases acidify most compartments of the endocytic pathway. Previous work has shown that efficient Notch activation following ligand binding requires entry of Notch into early endosomes, whereas a failure to sort and transport Notch out of the Hrs-positive early endosomal compartment, such as in ESCRT or lgd mutant cells, causes ectopic Notch activation (reviewed by Brou, 2009; Fortini and Bilder, 2009; Fürthauer and González-Gaitán, 2009; Tien et al., 2009). To determine whether V-ATPase activity is required in the early endosome for Notch activation, we generated chromosomes that were double mutant for Vha55 and the ESCRT-III component vps22 (larsen – FlyBase). Similar to vps22 mutant cells, vps22 Vha55 double-mutant cells accumulated ubiquitin and showed extensive colocalization of Notch with Hrs, indicating that V-ATPase activity is not required for Notch to access the early endosomal compartment (Fig. 4A-F). However, in contrast to vps22 mutant cells, expression of mβ-lacZ was significantly reduced in vps22 Vha55 double-mutant cells, to a level that was similar to that of Vha55 mutant cells (Fig. 4A-C,G-I). These data indicate that although Notch is trapped in early endosomes in vps22 Vha55 mutant cells, as in cells mutant for ESCRT components alone, it is nevertheless unable to signal. Consistent with a suppression of ectopic Notch signaling, in contrast to vps22 mutant cells, vps22 Vha55 mutant cells in mosaic eyes were unable to overproliferate (Fig. 4J-L). Thus, whereas ESCRT phenotypes are epistatic to V-ATPase with respect to Notch trafficking, V-ATPase phenotypes are epistatic to ESCRT with respect to Notch signaling. These results indicate that V-ATPase function is required for Notch activation in early endosomes.
DISCUSSION
Recent evidence suggests that the route and rate of Notch traffic through endocytic compartments can be regulated to either potentiate or diminish signaling activity. Our results identify a physiological mechanism to account for this effect on signaling. We identify a set of genes required for endosomal Notch signaling that encode subunits of the V-ATPase, the molecular machine that creates a proton motive force to acidify most compartments of the endosomal system. The data thus establish a novel role of V-ATPase that is relevant to both physiological and pathological Notch signaling. While this work was under review, an article describing a similar role for V-ATPase regulators and a different subunit of the V-ATPase in Notch signaling was published (Yan et al., 2009). Our independent results confirm those findings and expand them by reporting the ultrastructure of V-ATPase mutant cells, extending the role to tumor contexts that depend on excess Notch signaling activity and, importantly, providing evidence for the early endosome as the site of the requirement for V-ATPase activity. Interestingly, the aquaporin Big brain (Bib) was recently shown to promote Notch signaling at a similar step of Notch processing, and bib mutant clones, like V-ATPase mutant clones, show defects in endosomal acidification (Kanwar and Fortini, 2008). It is possible that some other, as yet uncharacterized, V-ATPase function unrelated to its well-established role in proton pumping might be required for Notch signaling activation. Alternatively, the common acidification and Notch signaling phenotypes of V-ATPase and bib mutant cells might indicate that Notch signaling normally requires endosomal acidification.
The best-understood requirement for V-ATPase-dependent acidification is in promoting lysosomal degradation by ensuring the proper targeting, release and activation of lysosomal proteases. Indeed, we find that Notch and other cargoes are trapped in a lysosome-like compartment in V-ATPase-deficient cells, where they accumulate rather than being degraded. The cargo-trapping phenotype resembles that seen in the presence of chemical inhibitors of lysosomal acidification, such as chloroquine, as well as in cells mutant for regulators of lysosomal genesis such as the HOPS components Dor and Car and the PIKFYVE Fab1 (Sevrioukov et al., 1999; Rusten et al., 2006; Akbar et al., 2009). However, in striking contrast to these latter mutants, in which Notch signaling is largely unaffected, a substantial loss of Notch signaling is seen in V-ATPase-deficient cells. This indicates that the requirement for V-ATPase in Notch signaling must precede lysosomal entry.
How could acidification of earlier endosomal compartments promote Notch signaling? One possibility is that the role of acidification reflects the requirement of endosomal transport for Notch activation. In mammalian cells, V-ATPases have been implicated in the recruitment of proteins that modulate traffic between early and late endosomes (Hurtado-Lorenzo et al., 2006). We find a reduced rate of endocytosis in V-ATPase mutant cells, and it is possible that a dampened flux of Notch through the endocytic pathway contributes to reduced Notch signaling. However, we believe this role is unlikely to account for the V-ATPase mutant phenotype for the following reasons. First, although its endocytic rate may be reduced, Notch can clearly reach both early and late endosomal compartments in mutant cells; this mild quantitative reduction in traffic contrasts with the more substantial and qualitative loss of Notch signaling. Second, in mammalian cells, loss of endosomal acidification reduces progression between early and late endosomes. Since the requirement for Notch signaling is in entry into early endosomes, Notch activation would seem to be less sensitive to this step. Moreover, because Notch reaches late endosomal compartments in V-ATPase mutant cells, in Drosophila, at least, redundant mechanisms to energize endocytic traffic must exist. Third, the striking failure of Notch trapped within Hrs-positive early endosomes in V-ATPase ESCRT double mutants to signal suggests that even when endocytic traffic is blocked at the endosomal sorting step, Notch activation still requires V-ATPase activity. The absence of Notch activation in vps22 Vha55 cells is particularly striking because even when most Notch cannot reach endosomes in cells double mutant for the ESCRT component TSG101 (erupted) and the endosomal syntaxin avl (Syx7), the minor fraction of Notch that does reach endosomes is efficiently cleaved and strongly activates ectopic Notch signaling (Vaccari et al., 2008). This suggests that the role of V-ATPase in Notch activation goes well beyond trafficking. The data also reveal that it is not mere access to the early endosome that is required for Notch signaling, but rather that a specific physiological feature of that endosome is central to the signaling activation mechanism.
An attractive alternative is that V-ATPase-dependent acidification creates an endosomal environment that is conducive for productive Notch S3 cleavage and, therefore, signaling. In this scenario, Notch transits through the early endosome in V-ATPase mutant cells, but is not efficiently activated because of altered γ-secretase function. Such a role is consistent with our genetic experiments, which show that a membrane-tethered Notch truncation that is automatically processed to an active form by γ-secretase in WT cells cannot signal in cells that lack V-ATPase function. Although it is appealing to speculate that an acidic pH could promote overall cleavage by γ-secretase, which requires low luminal pH for optimal activity (Pasternak et al., 2003), experiments testing the acid dependence of other γ-secretase substrates have failed to show a reduction in overall S3-type processing (Vingtdeux et al., 2007). Although these assays are limited in their ability to reflect the in vivo situation, an interesting possibility suggested by mammalian studies is that pH might alter the precision of the S3 cleavage site, producing NICD forms that are quickly degraded and cannot effectively signal (Tagami et al., 2008). Additional possibilities exist, such as that rather than creating a cleavage-promoting environment per se, the V-ATPase could serve as a pH sensor that couples the generation of a low pH to the recruitment of γ-secretase regulators in order to restrict cleavage to an endosomal site. Finally, acidification could directly affect the correct maturation and localization of the γ-secretase enzyme. Future work will distinguish between these possibilities.
Regardless of the specific mechanism governing the role of V-ATPase in Notch cleavage, overall our data suggest that drugs that impair V-ATPase function and consequently reduce endosomal acidification might be used to curtail pathologic overactivation of Notch, such as that observed in cancers characterized by Notch overexpression (Roy et al., 2007). Considering that such a goal is presently being pursued by the use of γ-secretase inhibitors (Rasul et al., 2009; Tanaka et al., 2009), the use of already established V-ATPase inhibitors, such as Bafilomycin A1, either alone or in combination with γ-secretase inhibitors, could represent a promising therapeutic avenue.
Supplementary Material
Acknowledgments
We thank Tor Erik Rusten, Hugo Bellen and Jim Posakony for reagents; Luis Chia and Emily Crane for technical help; and Takeshi Sasamura, Mark E. Fortini, Marisa Oppizzi and Salvatore Pece for helpful discussions. T.V. is supported by American Heart Association Award #0825176F and by a new unit start-up grant from Associazione Italiana Ricerca contro il Cancro; D.B. is supported by National Institutes of Health grant R01GM068675 and American Cancer Society grant RSG-07-040-01. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.045484/-/DC1
References
- Akbar M. A., Ray S., Krämer H. (2009). The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol. Biol. Cell 20, 1705-1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan A. K., Du J., Davies S. A., Dow J. A. (2005). Genome-wide survey of V-ATPase genes in Drosophila reveals a conserved renal phenotype for lethal alleles. Physiol. Genomics 22, 128-138 [DOI] [PubMed] [Google Scholar]
- Bilder D., Li M., Perrimon N. (2000). Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113-116 [DOI] [PubMed] [Google Scholar]
- Brou C. (2009). Intracellular trafficking of Notch receptors and ligands. Exp. Cell Res. 315, 1549-1555 [DOI] [PubMed] [Google Scholar]
- Davies S. A., Goodwin S. F., Kelly D. C., Wang Z., Sözen M. A., Kaiser K., Dow J. A. (1996). Analysis and inactivation of vha55, the gene encoding the vacuolar ATPase B-subunit in Drosophila melanogaster reveals a larval lethal phenotype. J. Biol. Chem. 271, 30677-30684 [DOI] [PubMed] [Google Scholar]
- Deng W. M., Althauser C., Ruohola-Baker H. (2001). Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128, 4737-4746 [DOI] [PubMed] [Google Scholar]
- Fortini M., Bilder D. (2009). Endocytic regulation of Notch signaling. Curr. Opin. Genet. Dev. 19, 323-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürthauer M., González-Gaitán M. (2009). Endocytic regulation of notch signalling during development. Traffic 10, 792-802 [DOI] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G. H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D. A., et al. (2006). V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8, 124-136 [DOI] [PubMed] [Google Scholar]
- Kanwar R., Fortini M. (2008). The big brain aquaporin is required for endosome maturation and notch receptor trafficking. Cell 133, 852-863 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lloyd T. E., Atkinson R., Wu M., Zhou Y., Pennetta G., Bellen H. J. (2002). Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108, 261-269 [DOI] [PubMed] [Google Scholar]
- Lopez-Schier H., St Johnston M. A. (2001). Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 15, 1393-1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Bilder D. (2005). Endocytic control of epithelial polarity and proliferation in Drosophila. Nat. Cell Biol. 7, 1132-1139 [DOI] [PubMed] [Google Scholar]
- Maegawa Y., Morita H., Iyaguchi D., Yao M., Watanabe N., Tanaka I. (2006). Structure of the catalytic nucleotide-binding subunit A of A-type ATP synthase from Pyrococcus horikoshii reveals a novel domain related to the peripheral stalk. Acta Crystallogr. 62, 483-488 [DOI] [PubMed] [Google Scholar]
- Menut L., Vaccari T., Dionne H., Hill J., Wu G., Bilder D. (2007). A mosaic genetic screen for Drosophila neoplastic tumor suppressor genes based on defective pupation. Genetics 177, 1667-1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L., Golde T., Osborne B. (2006). Notch signaling in cancer. Curr. Mol. Med. 6, 905-918 [DOI] [PubMed] [Google Scholar]
- Nellesen D. T., Lai E. C., Posakony J. W. (1999). Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev. Biol. 213, 33-53 [DOI] [PubMed] [Google Scholar]
- Newsome T. P., Asling B., Dickson B. J. (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127, 851-860 [DOI] [PubMed] [Google Scholar]
- Nishi T., Forgac M. (2002). The vacuolar (H+)-ATPases-nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94-103 [DOI] [PubMed] [Google Scholar]
- Pasternak S. H., Bagshaw R. D., Guiral M., Zhang S., Ackerley C. A., Pak B. J., Callahan J. W., Mahuran D. J. (2003). Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J. Biol. Chem. 278, 26687-26694 [DOI] [PubMed] [Google Scholar]
- Rasul S., Balasubramanian R., Filipovic A., Slade M. J., Yagüe E., Coombes R. C. (2009). Inhibition of gamma-secretase induces G2/M arrest and triggers apoptosis in breast cancer cells. Br. J. Cancer 100, 1879-1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M., Pear W. S., Aster J. C. (2007). The multifaceted role of Notch in cancer. Curr. Opin. Genet. Dev. 17, 52-59 [DOI] [PubMed] [Google Scholar]
- Rusten T. E., Rodahl L. M., Pattni K., Englund C., Samakovlis C., Dove S., Brech A., Stenmark H. (2006). Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol. Biol. Cell 17, 3989-4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F. (2004). Regulation of notch signaling activity. Curr. Biol. 14, R129-R138 [PubMed] [Google Scholar]
- Sevrioukov E. A., He J. P., Moghrabi N., Sunio A., Kramer H. (1999). A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell 4, 479-486 [DOI] [PubMed] [Google Scholar]
- Soyombo A. A., Tjon-Kon-Sang S., Rbaibi Y., Bashllari E., Bisceglia J., Muallem S., Kiselyov K. (2006). TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 281, 7294-7301 [DOI] [PubMed] [Google Scholar]
- Sun J., Deng W. M. (2005). Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development 132, 4299-4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Deng W. (2007). Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev. Cell 12, 431-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami S., Okochi M., Yanagida K., Ikuta A., Fukumori A., Matsumoto N., Ishizuka-Katsura Y., Nakayama T., Itoh N., Jiang J., et al. (2008). Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol. Cell. Biol. 28, 165-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Setoguchi T., Hirotsu M., Gao H., Sasaki H., Matsunoshita Y., Komiya S. (2009). Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br. J. Cancer 100, 1957-1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Ito N., Dickson B. J., Treisman J. E., Hariharan I. K. (2001). The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105, 345-355 [DOI] [PubMed] [Google Scholar]
- Tien A. C., Rajan A., Bellen H. J. (2009). A Notch updated. J. Cell Biol. 184, 621-629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Bilder D. (2005). The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev. Cell 9, 687-698 [DOI] [PubMed] [Google Scholar]
- Vaccari T., Lu H., Kanwar R., Fortini M. E., Bilder D. (2008). Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol. 180, 755-762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Rusten T., Menut L., Nezis I., Brech A., Stenmark H., Bilder D. (2009). Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J. Cell Sci. 122, 2413-2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingtdeux V., Hamdane M., Bégard S., Loyens A., Delacourte A., Beauvillain J. C., Buée L., Marambaud P., Sergeant N. (2007). Intracellular pH regulates amyloid precursor protein intracellular domain accumulation. Neurobiol. Dis. 25, 686-696 [DOI] [PubMed] [Google Scholar]
- Yan Y., Denef N., Schupbach T. (2009). The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell 17, 387-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.