Abstract
Microcephaly affects ∼1% of the population and is associated with mental retardation, motor defects and, in some cases, seizures. We analyzed the mechanisms underlying brain size determination in a mouse model of human microcephaly. The Hertwig's anemia (an) mutant shows peripheral blood cytopenias, spontaneous aneuploidy and a predisposition to hematopoietic tumors. We found that the an mutation is a genomic inversion of exon 4 of Cdk5rap2, resulting in an in-frame deletion of exon 4 from the mRNA. The finding that CDK5RAP2 human mutations cause microcephaly prompted further analysis of Cdk5rap2an/an mice and we demonstrated that these mice exhibit microcephaly comparable to that of the human disease, resulting from striking neurogenic defects that include proliferative and survival defects in neuronal progenitors. Cdk5rap2an/an neuronal precursors exit the cell cycle prematurely and many undergo apoptosis. These defects are associated with impaired mitotic progression coupled with abnormal mitotic spindle pole number and mitotic orientation. Our findings suggest that the reduction in brain size observed in humans with mutations in CDK5RAP2 is associated with impaired centrosomal function and with changes in mitotic spindle orientation during progenitor proliferation.
Keywords: CDK5RAP2, Centrosome, Neurogenesis, Mouse
INTRODUCTION
The cerebral cortex that provides humans with our unique cognitive abilities consists of six morphologically and functionally distinct neuronal layers (Rakic, 2009). All cortical neurons are produced from a neuroepithelium lining the ventricular cavity of the developing brain (Noctor et al., 2001). During neurogenesis, postmitotic neurons differentiate and migrate away from the ventricular zone (VZ) towards their final destination in the cerebral cortex (Rakic, 2009). Earlier born neurons localize to deeper layers of the cortex, whereas later born neurons populate progressively more superficial layers (Angevine and Sidman, 1961; Rakic, 1974). Hence, proper development of the cerebral cortex requires the coordination of cell proliferation, fate determination and migration. Defects in these processes can cause neurodevelopmental disorders such as microcephaly, a disease associated with mental retardation (Mochida, 2008). Primary microcephaly is defined as decreased head circumference and a disproportionately reduced cerebral cortex, but no other major developmental malformations (Woods et al., 2005).
The centrosome plays key roles in neuronal progenitor proliferation and fate determination, as well as neuronal differentiation and migration (Higginbotham and Gleeson, 2007). Recent work suggests that asymmetric inheritance of daughter and mother centrosomes regulates cell fate determination in cortical progenitors (Wang et al., 2009). Moreover, mutations in several genes that encode centrosomal proteins cause human primary microcephaly (Woods et al., 2005). Among them is CDK5RAP2 (cyclin-dependent kinase 5 related activator protein 2 or CDK5 regulatory subunit-associated protein 2) (Bond et al., 2005), which was originally identified in mammals by its interaction with p35Nck5a (p35; Cdk5r1), an activator of the neuronal cyclin-dependent kinase 5 (CDK5) (Ching et al., 2000). CDK5RAP2 encodes a 1893 amino acid, 215 kDa centrosomal protein. In somatic cells, CDK5RAP2 promotes centrosomal cohesion (Graser et al., 2007) and recruits the γ-tubulin ring complex (γ-TuRC) – the microtubule nucleator – to the centrosome (Fong et al., 2008). However, the role of CDK5RAP2 in neuronal development remains poorly understood.
Hertwig's anemia (an) arose in the progeny of a heavily irradiated male mouse (Hertwig, 1942). Homozygous mutant animals have a macrocytic, hypoproliferative anemia and leukopenia attributable to intrinsic defects that are progressively more severe in the more mature hematopoietic progenitors (Barker and Bernstein, 1983; Barker et al., 2005; Eppig and Barker, 1989). Male animals are infertile due to severe deficiencies in germ cells, whereas female animals have less severe defects in oocytes, but cannot deliver pups (Russell et al., 1985). There is a high level of spontaneous aneuploidy in primary cultures of hematopoietic fetal liver, bone marrow and kidney epithelial cells derived from mutant animals (Eppig and Barker, 1984). Both the severity of the hematological disease and a coexisting predisposition to hematopoietic tumors are highly dependent upon the genetic background of the animal (Barker et al., 2005). Here, we show by positional cloning that an is due to a mutation in Cdk5rap2, and that an mutants show additional severe neurological defects that were not previously recognized.
Mutations in human CDK5RAP2 cause severely reduced brain size with relatively well-preserved cortical patterning (Bond et al., 2005), but without anemia or tumor predisposition, the hallmarks of an. Although CDK5RAP2 is one of several centrosomal proteins implicated in human microcephaly (Bond et al., 2002; Bond et al., 2005), we present the first analysis of any of these centrosomal proteins in mutant mice. Our analysis reveals important roles for Cdk5rap2 in mitotic progression and mitotic spindle orientation. Our data also suggest that Cdk5rap2 is essential for cortical progenitor proliferation and survival. These results illustrate the fundamental role of centrosome function in disease mechanisms associated with human primary microcephaly.
MATERIALS AND METHODS
Mouse strains and genetic mapping
We maintained the Hertwig's anemia (an) mutation on the WB/ReJ (WB) and C57BL/6J (B6) inbred backgrounds, as homozygous mutant mice are poorly viable, particularly on the C57BL/6J background. WB.Cg an/+ × B6.Cg an/+ F1 (WBB6F1) animals are nearly fully viable and were used for studies in adults. To identify the gene underlying the an phenotype, we performed an intersubspecific intercross between B6.Cg an/+ and the +/+ Mus musculus castaneus (CAST/Ei) inbred strain to generate second filial generation (F2) intercross animals segregating the an allele. F2 animals were phenotyped by complete blood count at 3 weeks of age. We reconfirmed linkage to the brown locus on chromosome 4 in 46 consecutive F2 animals at 3 weeks of age, and further delineated the genetic interval by genotyping an additional 734 F2 animals and correlating the phenotype with crossover events within the originally defined locus. Genotyping and breeding of additional novel crossover events in the B6.Cg an/+ colony further narrowed the critical region.
Genotyping
Subsequent to mutation identification, animals were genotyped using standard PCR reactions specific for the wild-type (1040 bp, primers 5′-TCACTGAGCTGAAGAAGGAGAA-3′ and 5′-GCAATCACTAAAATGTCCGATT-3′) and mutant (507 bp, primers 5′-GCAATCACTAAAATGTCCGATT-3′ and 5′-TGTCTTTCTGCCCTGACAGT-3′) alleles.
Northern blot analysis
Total RNA from Cdkd5rap2an/an and Cdk5rap2+/+ tissue was extracted and purified as described (Baelde et al., 2001). A 32P-labeled Cdk5rap2 probe (500 bp) was generated by PCR amplification of exon 24 from mouse genomic DNA (129S6.Tac) with primers 5′-GCCTTATTACCAGCATGCAA-3′ and 5′-TCACCGAAAAGTTCCAAGTTC-3′. A commercial mouse multi-tissue northern blot (Origene) and the Cdk5rap2an/an and Cdk5rap2+/+ blot were run, transferred, hybridized and processed as previously described (Lee et al., 2007).
Immunoblotting
Cdk5rap2+/+ and Cdk5rap2an/an tissues were homogenized and protein extracted in RIPA buffer (50 mM Tris-HCl pH 7.2, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% Nonidet P40) with Complete Protease Inhibitors (Roche). Protein (30 μg per sample) was loaded onto a 7% Novex Tris-acetate gel (Invitrogen) and run according to the manufacturer's instructions. Gels were transferred to nitrocellulose membranes using a semi-dry transfer system (Hoeffer) and analyzed with rabbit anti-Cdk5rap2 antibody at 1:1500 (Bethyl Laboratories).
Histology and immunostaining
Embryonic and postnatal day 0 (P0) brains were dissected in ice-cold 1×PBS, fixed in 4% paraformaldehyde by immersion or cardiac perfusion, embedded in paraffin and sectioned at 5 μm. Immunohistochemistry was performed as previously described (Feng and Walsh, 2004). Animals were handled according to protocols approved by the IACUC of the Beth Israel Deaconess Medical Center and the Children's Hospital Boston.
Antibodies
The following antisera were used at the specified dilutions: rabbit anti-Cdk5rap2 (1:1000) (Bethyl laboratories, BL2320); rabbit anti-Ki67 (1:500) (Vector Labs, VP-RM04); rabbit anti-Dcx (1:500) (Covance); goat anti-Cux1 (1:10) (Santa Cruz, SC6327); rat anti-Ctip2 (1:500) (Abcam, ab18465); mouse anti-aurora kinase A (1:200) (Abcam, ab13824); rabbit anti-Tbr2 (1:300) (Abcam, ab23345); rabbit anti-phospho-histone H3 (1:500) (Upstate, 07-492); rabbit anti-Sox2 (1:200) (Novus-Biologicals, NB110-372335C); rat anti-BrdU (1:300) (abdSerotec, MCA2060); rabbit anti-Tbr1 (1:500) (gift from R. Hevner, University of Washington, Seattle, WA, USA); goat anti-Brn1 (1:200) (Santa Cruz, SC-6028); mouse anti-aurora kinase B (1:200) (BD biosciences, 611083); rabbit anti-Par3 (1:300) (Upstate, 07-330); and rabbit anti-activated cleaved caspase 3 (1:50) (Cell Signaling, 9661). Secondary detection reagents (1:500) included Alexa Fluor 488-, 594- or 546-conjugated anti-rabbit, mouse or goat IgG (Invitrogen); Cy5-conjugated anti-rabbit, mouse or goat IgG, Cy3-conjugated or unconjugated Fab fragments of anti-rabbit IgG (Jackson ImmunoLaboratories); and Cy3-conjugated Streptavidin reagent.
Analysis of cells in S phase and of cell cycle exit
For analysis of S-phase cells, E10.5 and E14.5 pregnant females were injected intra-peritoneally with 70 μg BrdU/g body weight and sacrificed 30 minutes after injection. The percentage of cells in S phase was calculated by counting cells stained with BrdU as a function of total nuclei. For analysis of cell cycle exit, E14.5 pregnant females were injected with BrdU as above, sacrificed 24 hours after injection, and analyzed as previously described (Chenn and Walsh, 2002).
Image acquisition and data analysis
Images were acquired and analyzed as previously described (Sepp et al., 2008). For spindle orientation analysis, images were acquired as 5 μm z-stacks using a Zeiss confocal microscope and angles were measured in three-dimensional (3D) reconstructed projections using ImageJ (NIH). At least three coronal brain serial sections from at least three different animals were analyzed per genotype per experiment. Unless otherwise noted, brain sections were chosen at the mid-anterior level of the brain, and micrographs were taken of the dorsal and dorsal-lateral cortex region. Cell counts were performed as a percentage of total nuclei per high-magnification field per coronal section. P-values were calculated with an ANOVA single-factor test.
RESULTS
Hertwig's anemia is due to a mutation in Cdk5rap2
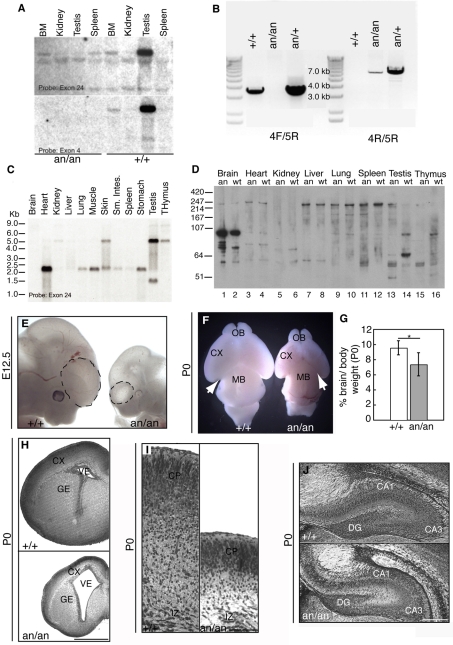
We mapped Hertwig's anemia (an) to a 290 kb genomic interval between 68.91 and 69.94 Mb (genomic assembly NCBI m35, Dec 2005; www.ensembl.org/mus_musculus) on mouse chromosome 4, which includes the 5′ end of Cdk5rap2 and the multiple epidermal growth factor-like domains 9 (Megf9) gene. Sequencing of the entire coding regions of Megf9 and Cdk5rap2 revealed no exonic or splice junction mutations. However, RT-PCR of Cdk5rap2 from WBB6F1 an/an animals demonstrated an in-frame 111 bp deletion resulting from loss of exon 4 from the mRNA (see Fig. S1A in the supplementary material). This change predicts a 37 amino acid deletion of Cdk5rap2, but as the mutant mRNA maintains the reading frame a shortened protein product could be formed (see Fig. S1B in the supplementary material). Northern analysis demonstrated that exon 4 was uniformly omitted from the mutant mRNA (Fig. 1A). Amplification of genomic DNA indicated that the basis of this exon skipping was an inversion of exon 4 (Fig. 1B). Furthermore, B6.Cg an/+ females bred to male animals chimeric for a Cdk5rap2 gene-trap allele yielded F1 compound heterozygous animals with leukopenia and macrocytic anemia, similar to the an mutants (see Table S1 in the supplementary material), confirming Cdk5rap2 as the Hertwig's anemia gene and allowing the designation of the mutation as Cdk5rap2an.
Fig. 1.
Hertwig's anemia (an) is due to an inversion of exon 4 of Cdk5rap2, resulting in exon 4 skipping and abnormal cortical development. (A) Exon 4 is skipped in an/an homozygotes. Cdk5rap2 expression is detected in both +/+ and an/an mice using an exon 24 probe (upper panel). However, reprobing the same blot with an exon 4 probe yields no signal in an/an animals (lower panel). (B) PCR of genomic DNA from +/+, an/+ and an/an WBB6F1 confirmed the presence of an inversion surrounding exon 4. (C) Northern blot of adult mouse tissues demonstrating a 2.3 kb transcript expressed in most tissues, a 5.8 kb transcript expressed in several tissues and particularly strongly in testis, and a 1.4 kb transcript expressed only in testis. (D) Western analysis of Cdk5rap2 protein in +/+ and an/an adults. Most tissues have a large (∼230 kDa) isoform, whereas the testis, thymus and brain also contain a smaller (∼105 kDa) isoform. A testis-specific protein isoform (∼70 kDa) is also observed. (E) B6.Cg an/an embryos have reduced brain and body sizes already at E12.5, as compared with controls. Dashed lines highlight cortical vesicles. (F) Transmitted light micrograph of P0 B6.Cg Cdk5rap2+/+ and Cdk5rap2an/an brains. Mutant brains are smaller with a disproportionately reduced cortex (CX) and smaller olfactory bulbs (OB). Arrows indicate that more of the midbrain (MB) area is exposed in Cdk5rap2an/an than in the control. (G) Analysis of the brain-to-body weight ratio in the B6.Cg background, showing that the mean brain-to-body weight ratio at P0 is decreased in Cdk5rap2an/an animals. Error bars indicate s.d. *, P=7×10–5. (H) Histological analysis of P0 brains from B6.Cg animals. Coronal sections (5 μm) stained with Cresyl Violet show a thinner cortex, reduced ganglionic eminence (GE) and enlarged ventricle (VE) in the mutants. (I) At P0, the cortex of Cdk5rap2an/an animals is smaller but shows a laminar organization that is similar to that of control littermates. A high-magnification micrograph shows the cortical plate (CP) and intermediate zone (IZ) in control (left) and mutant (right) animals. (J) At P0, the hippocampus is smaller in Cdk5rap2an/an (bottom) than in controls (top). DG, dentate gyrus; CA1 and CA3, cornus ammonis 1 and 3. Scale bars: 200 μm in H; 100 μm in I,J.
Expression analysis of Cdk5rap2 and Cdk5rap2an
Cdk5rap2an/an animals had defects in multiple organs, including brain, thymus and testis. Using a probe to exon 24, we found that the predominant Cdk5rap2 transcript differs from tissue to tissue, with a 5.8 kb major transcript in testis and thymus, and a 2.3 kb transcript most abundant in heart and most other tissues (Fig. 1C). A unique 1.4 kb transcript was also present in testis. Although the mRNA was undetectable in adult brain using a probe to exon 24, the GENSAT in situ hybridization reference database (www.ncbi.nlm.nih.gov/projects/gensat/) and published work (Bond et al., 2005) show that Cdk5rap2 is highly expressed in embryonic neural progenitors.
Immunoblotting of Cdk5rap2 protein in the mutant indicated that deletion of exon 4 does not substantially destabilize the protein. In most tissues, a large (∼230 kDa) species was seen to predominate. A smaller isoform (∼105 kDa) was present in testis and thymus and predominated in adult brain, but was barely detectable in fetal brain or other tissues (Fig. 1D and data not shown). This suggests that the 230 kDa isoform disrupted in Cdk5rap2an/an animals is the predominant functional species in fetal brain. The major protein isoforms might correspond to the two largest of the three mRNAs. Furthermore, as in the northern analysis (Fig. 1C), there was an additional testis-specific protein isoform (∼70 kDa). All of these proteins isoforms are too large to permit the resolution of a shortened an mutant protein by western analysis.
Cdk5rap2 is essential for normal brain development
The identification of Cdk5rap2 as the an gene prompted us to examine the brain of an mutants, which had not previously been evaluated. WBB6F1 hybrid Cdk5rap2an/an adult animals showed moderate cortical hypoplasia visualized by shortening of the forebrain, which did not extend fully over the superior colliculus of the midbrain, as it does in normal animals (see Fig. S1C in the supplementary material). However, inbred B6.Cg Cdk5rap2an/an animals showed a dramatic reduction in brain size and rarely survived beyond 1 week of age. They were runted, with disproportionately smaller brains both prenatally and at birth (Fig. 1E,F). The mutants exhibited a significant decrease in the ratio of brain to body weight as compared with controls (7.5±1.5 versus 9.5±0.9 mg/g, P<1×10–4) (Fig. 1G). The brain phenotype had a variable expressivity, being more severe in some individuals than others (Fig. 1E,F), including variably increased ventricular size and variably decreased cortical thickness (Fig. 1H,I). Other brain structures, including the hippocampus, were also considerably smaller than normal (Fig. 1J). Together, these data demonstrate that Cdk5rap2 is essential for normal brain development.
Cdk5rap2an/an mice show reduced superficial cortical layers
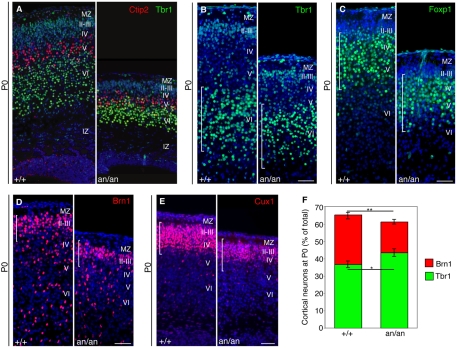
The cortical neuronal layers are formed in an `inside-out' fashion, whereby the deepest layers form first and the most superficial layers form last (Rakic, 2009). In newborn (P0) Cdk5rap2an/an animals, immunostaining for Tbr1 and Ctip2 (Bcl11b – Mouse Genome Informatics) (layers V-VI) (Hevner et al., 2001; Leid et al., 2004), Foxp1 (layers III-V) (Ferland et al., 2003), Brn1 (Pou3f3) and Cux1 (layers II-IV) (McEvilly et al., 2002; Nieto et al., 2004) showed profound defects in neurogenesis but relatively preserved cortical layering (Fig. 2A-E), suggesting that neuronal migration was generally normal. However, Cdk5rap2an/an mice showed consistent thinning of the cerebral cortex (Fig. 2A and see Fig. S2A,B in the supplementary material), accompanied by preferential reduction of the superficial, later-born neuronal layers (Fig. 2D-F), as demonstrated by a significant decrease in Brn1-positive cells compared with controls (17.8±1.4% versus 28.6±2.43%, P<1×10–9). The preferential decrease of superficial neurons in mutant animals was confirmed by Cux1 immunostaining (Fig. 2E). The dramatic decrease in superficial layers resulted in fewer total cells and was associated with correspondingly increased early-born, Tbr1-positive cells in mutants as compared with controls (43.4±2.2% versus 36.7±1.9%, P<0.001) (Fig. 2B,C,F). Thus, Cdk5rap2an/an animals have fewer total neurons and the last-born superficial neurons are particularly reduced.
Fig. 2.
Cdk5rap2an/an mice show reduced neuronal number and thinner superficial layers but preserved cortical layer organization. (A) Cdk5Rap2 mutant mice show preserved cortical layer organization, although each layer is far thinner than normal. P0 wild-type (+/+) and mutant (an/an) littermates are shown. Coronal sections stained with Tbr1 antisera, which labels deep layer VI neurons (green), and with Ctip2 antisera, which labels layer V neurons (red), show the preservation of cortical layer organization in the mutant. The marginal zone (MZ) and IZ are also shown. (B,C) Deeper cortical layer markers show the overall reduction in cortical thickness in the an/an mutant. (B) Tbr1, layer VI; (C) Foxp1, layers III-V. (D,E) Superficial cortical layers are reduced in the Cdk5Rap2an/an brain. P0 wild-type (+/+) and mutant (an/an) littermate coronal sections stained with antisera against (D) Brn1 (layers II-III) and (E) Cux1 (layers II-IV). In B-E the brackets correspond to wild type and highlight the differences between mutant and control animals. (F) The density of superficial and deeper neuronal layers. The density of layers II-III was calculated as the ratio of Brn1-positive cells to total CP nuclei counted per section (the mean percentage is shown). The Brn1 density (red bars) in wild-type (n=6 animals, 486 cells counted/section) and mutant (n=7 animals, 417 cells counted/section) animals is significantly different; **, P=7.4×10–10. The corresponding increase in the density of deep neuronal layer VI is shown. The percentage of Tbr1-positive cells (green bars) among total CP nuclei is moderately increased in the mutant (n=5 animals, 431 cells counted/section) as compared with control littermates (n=6 animals, 544 cells counted/section); *, P=1.3×10–4. Sections were counterstained with Hoechst 33342 (blue) to label nuclei. Error bars indicate s.e.m. Scale bars: 50 μm.
Cdk5rap2an/an mice have defects in neurogenesis during development
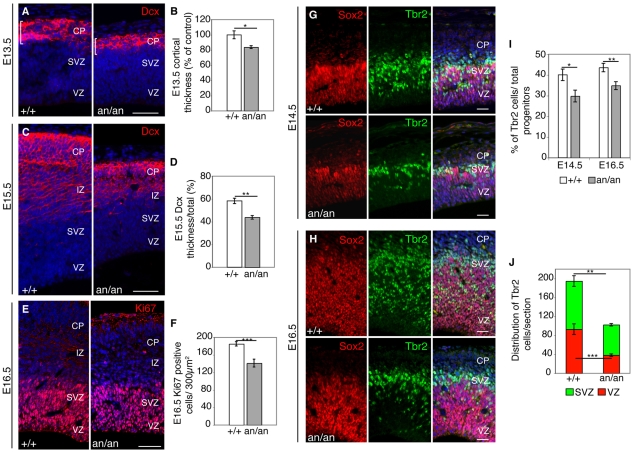
Our observations in newborn animals prompted us to investigate the effect of the Cdk5rap2an/an mutation during embryonic development. We found a significant decrease in overall cortical thickness at E13.5 (Fig. 3A,B), indicating a defect at early stages of neurogenesis. Immunostaining for Dcx, which marks immature neurons (Gleeson et al., 1999), showed that E15.5 Cdk5rap2an/an embryos had a thinner neuronal layer than controls, expressed as the ratio of the thickness of Dcx-immunolabeled neuronal layers over total cortical thickness (43.9±1.6% versus 58.3±2.4%, P<0.001) (Fig. 3C,D). The decrease in cortical neurons was associated with a reduction in the precursor population, as revealed by immunoreactivity for Ki67 (Mki67), a marker of dividing cells, which was diminished in E16.5 Cdk5rap2an/an embryos compared with controls (141.1±9 versus 185±5, P<0.01) (Fig. 3E,F). In addition, short pulses of BrdU at E10.5 (when the developing cortex is primarily composed of neuronal progenitors) and at E14.5 (when most of the deeper layers have already formed), showed that Cdk5rap2an/an embryos have a progressive decrease in the density of S-phase cells (see Fig. S3A-C in the supplementary material), suggesting that progenitor cells decrease as development proceeds.
Fig. 3.
Neurons and neuronal progenitors are reduced in Cdk5rap2an/an mice. (A) E13.5 coronal sections immunostained for Dcx (a marker for immature neurons) in wild-type (+/+) and mutant (an/an) littermate pairs. (B) Overall cortical thickness is reduced at E13.5 in Cdk5rap2an/an animals (n=5) compared with controls (n=3). The mean is shown as a percentage of the control mean (+/+, 100±5.2%; an/an, 83.9±1.8%); *, P=0.001. (C) E15.5 coronal sections show a significantly reduced CP in mutants. (D) CP thickness at E15.5 in mutants and controls (n=4) shown as the ratio of Dcx-positive cortical thickness over total cortical thickness. **, P=6.3×10–4. (E) Total proliferating cells are reduced in the mutants (n=7) compared with controls (n=4). Coronal sections of E16.5 littermates were analyzed by Ki67 immunostaining. (F) Quantitation of proliferating cells, showing the total number of Ki67-positive cells counted per 300 μm2. ***, P=0.002. (G,H) Coronal sections of E14.5 and E16.5 littermates were analyzed using Tbr2 (green), a marker for basal progenitors, and Sox2 (red), a marker for neuronal progenitors. (G) E14.5 embryos show some Tbr2-labeled cells in the VZ, SVZ and CP. (H) At E16.5, the reduction in Tbr2-positive cells is also evident across the VZ and SVZ in Cdk5rap2an/an embryos. (I) At E14.5, Cdk5rap2an/an embryos (n=5, 214 cells counted/section) had 11.9% fewer Tbr2-positive cells than Cdk5rap2+/+ (n=5, 366 cells counted/section); *, P=2×10–4. At E16.5, Cdk5rap2an/an embryos (n=5, 420 cells counted/section) had 9.3% fewer Tbr2-positive cells than Cdk5rap2+/+ (n=5, 630 cells counted/section); **, P=1.3×10–6. (J) The distribution of Tbr2-positive cells changes in the Cdk5rap2an/an animals. The mean number of Tbr2-positive cells is shown for the VZ and the SVZ. Control embryos had 2.4-fold more Tbr2-positive cells in the VZ and 1.5-fold more in the SVZ than the mutants. **, P=0.01; ***, P=0.003. Error bars indicate s.e.m. (B,D,F,I) or s.d. (J). Scale bars: 50 μm in A,C,E; 200 μm in G,H.
Neuronal progenitors form two groups that undergo mitosis either at the apical surface of the ventricle or on the basal side away from the VZ (Gotz and Huttner, 2005), and both progenitor types were affected in Cdk5rap2an/an mutants. Apical progenitors give rise to deep layer neurons and basal progenitors, whereas basal progenitors primarily produce superficial layer neurons (Pontious et al., 2008). Radial glial cells, a subset of apical progenitors, produce most CNS neurons and provide a scaffold for newly formed neurons to migrate (Misson et al., 1988). We immunostained E15.5 Cdk5rap2an/an mice for Glast (Slc1a3 – Mouse Genome Informatics), a radial glial marker (Shibata et al., 1997), and found no apparent changes in their morphology (data not shown), although the decreased Ki67 and Sox2 staining (Fig. 3E-J) suggested that their number was reduced. In addition, E14.5 and E16.5 Cdk5rap2an/an embryos showed a marked loss of basal progenitors upon immunostaining for the basal progenitor marker Tbr2 (Eomes – Mouse Genome Informatics) (Englund et al., 2005) and for Sox2, a marker for apical progenitors and a subset of basal progenitors (Bani-Yaghoub et al., 2006) (Fig. 3G-I). The ratio of Tbr2-positive cells to total progenitors was decreased in mutants at both E14.5 (28.9±1.8% versus 40.8±2.2%, P<5×10–4) and E16.5 (34.5±1.2% versus 43.8±1.1%, P<1×10–5) (Fig. 3I), demonstrating that the decrease in superficial layer neurons correlates with a reduction in basal progenitors. Interestingly, at mid-neurogenesis, a fraction of these Tbr2-positive cells normally localizes to the VZ, whereas in mutant animals fewer Tbr2-positive cells were located within the VZ. Whereas E15.5 control embryos had roughly an equal distribution of Tbr2-positive cells in the VZ (93.2±17.8) and subventricular zone (SVZ) (101.7±11.3), Cdk5rap2an/an embryos had 59.8% fewer Tbr2-positive cells within the VZ (38.4±5.9) compared with the SVZ (64.2±2.6) (Fig. 3J). This profound reduction of basal progenitors at later stages in neurogenesis might reflect a more generalized loss of all progenitors over the course of neurogenesis, although a more specific defect in basal progenitors cannot be ruled out.
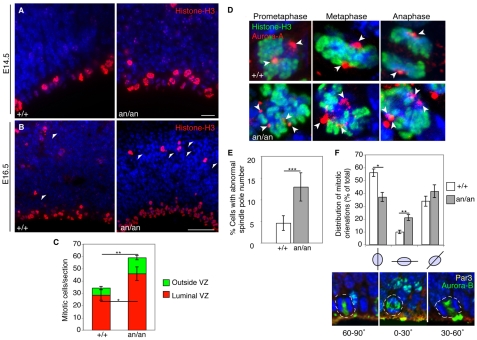
Mitotic abnormalities in Cdk5rap2an/an mice
Since Cdk5rap2 is a centrosomal protein, we hypothesized that the neuronal progenitor deficiency in Cdk5rap2an/an animals reflects underlying mitotic defects. Using the mitotic (M)-phase marker phospho-histone H3, we analyzed E12.5 (see Fig. S4 in the supplementary material), E14.5 and E16.5 embryos (Fig. 4A,B). We found 1.8-fold more M-phase cells in mutants compared with controls at E16.5 (61.75±6.24 versus 35.26±4.8, P<1×10–7). This increase in mitotic cells reflected increases at the luminal VZ (45.95±5.57 versus 28.57±4.23, P<1×10–5) and outside the VZ (13.07±1.81 versus 5.78±1.27, P<1×10–7) (Fig. 4C). Since the overall progenitor number decreases, this surprising increase in M-phase cells suggests that Cdk5rap2 deficiency leads to a delay in mitotic progression. To determine the cause of this delay, we examined aurora kinase A immunoreactivity (Katayama et al., 2003) in neuronal precursors. Aurora kinase A labels the mitotic spindle poles as two distinct dots at the focused ends of a bipolar spindle structure. Cdk5rap2an/an embryos showed a 2.8-fold increase in pro-metaphase and metaphase precursor cells with mono-, tri- and other aneupolar spindle poles labeled with aurora kinase A, as compared with controls (12.95±3.26% versus 4.62±1.69%, P<1×10–5) (Fig. 4D,E). These abnormal spindles confirm previous data indicating that atypical meioses are readily apparent in Cdk5rap2an/an embryonic germ cells and suggests a mechanism for the predisposition to chromosomal aneuploidy seen in Cdk5rap2an/an primary cells (Eppig and Barker, 1984).
Fig. 4.
Cdk5rap2an/an neuronal progenitors exhibit mitotic defects and changes in mitotic spindle orientation. (A,B) Cdk5rap2 mutants have increased numbers of phospho-histone H3-labeled cells. Fluorescent micrographs of Cdk5rap2+/+ and Cdk5rap2an/an coronal sections at (A) E14.5 and (B) E16.5. Phospho-histone H3 (red) immunostaining shows cells in M phase lining the VZ, and away from the VZ (arrowheads). Nuclei are stained with Hoechst (blue). (C) Analysis of mitotic index in E16.5 embryos. M-phase cells counted along the luminal VZ from the medial-dorsal to the dorsal-lateral junction. Dorsal mitotic cells away from the VZ were also measured. The mean total mitotic cells per coronal section and their localization at the luminal VZ, or away from the VZ, are shown. At E16.5, Cdk5rap2an/an mice (n=5, average of 330 mitotic cells counted/animal) had 1.8-fold more M-phase cells than controls (n=5, average of 128 mitotic cells counted/animal); P=1.12×10–8. M-phase cells increased at the luminal VZ (*, P=5.64×10–6) and outside of the VZ (**, P=2.22×10–8). (D) Cdk5rap2an/an animals have abnormal numbers of spindle poles. Aurora kinase A labels spindle poles (red), whereas phospho-histone H3 labels M-phase cells (green). Arrowheads indicate aurora kinase A-labeled spindle poles. Representative confocal images of Cdk5rap2+/+ precursor cells (top) in prometaphase (left), metaphase (center) and anaphase (right) with bipolar spindles, and Cdk5rap2an/an precursor cells (bottom) in prometaphase with a tetrapolar spindle (left), in metaphase with a tripolar spindle (center), and in anaphase with a tetrapolar spindle (right). (E) Cdk5rap2an/an animals show a 2.8-fold increase in the percentage of abnormal mitotic figures per total number of M-phase cells at the luminal VZ. ***, P=3.26×10–6. (F) Analysis of spindle orientation at E16.5. Bar chart shows the distribution of horizontal (0-30°), oblique (30-60°) and vertical (60-90°) cleavage planes. Cdk5rap2an/an embryos have increased horizontal and decreased vertical cleavage planes (n=4, average of 88 cells counted/animal; **, P=0.005), compared with Cdk5rap2+/+ embryos (n=4, average of 89 cells counted/animal; *, P=0.006). Beneath are shown representative three-dimensional reconstructed confocal images of horizontal, oblique and vertical cleavage planes. Nuclei are stained with Hoechst (blue), the central spindle and midbody are stained for aurora kinase B (green), and the apical membrane is stained for Par3 (yellow). Error bars indicate s.d. Scale bars: 10 μm in A; 50 μm in B.
Cdk5rap2an/an cortical progenitors have defective mitotic spindle orientation
During neurogenesis, the orientation of the mitotic division plane relative to the ventricular surface (VS) has been proposed to influence cell fate determination (Zhong and Chia, 2008). The disruption of proteins that regulate mitotic spindle orientation at the VZ correlates with premature and excessive generation of neurons and early depletion of cortical progenitors (Feng and Walsh, 2004; Fish et al., 2006; Sanada and Tsai, 2005), although the relevance of this mechanism to human microcephaly has been uncertain. To investigate mitotic orientation during neurogenesis in Cdk5rap2an/an mice we focused on anaphase and telophase spindles, as metaphase spindles undergo dynamic rotations (Haydar et al., 2003). We defined cleavage plane orientations in relation to the VS as horizontal (0-30°, i.e. mitotic spindle aligned parallel to the VS), oblique (30-60°) or vertical (60-90°, i.e. mitotic spindle aligned along the apical-basal axis of the VS) (Chenn and McConnell, 1995). During cortical development, most cleavage planes are oriented perpendicular to the VS, producing side-by-side daughter cells (Zhong and Chia, 2008). By co-immunostaining for Par3 (Pard3 – Mouse Genome Informatics) to label the VS (Bultje et al., 2009) and aurora kinase B to label the central spindle (anaphase) and the midbody (telophase) (Vader and Lens, 2008), we found increased horizontal cleavage planes in Cdk5rap2an/an mice compared with controls at E14.5 (15.6±3.2% versus 7.7±4.0%, P<0.05) (see Fig. S5 in the supplementary material). E16.5 Cdk5rap2an/an embryos had increased horizontal cleavage planes (21.2±4.5% versus 10.0±2.6%, P<0.01) and reduced vertical cleavage planes (37.2±7.1% versus 56.1±5.8 %, P<0.01) (Fig. 4F). Thus, Cdk5rap2 is important for mitotic spindle orientation in cortical progenitors.
Cdk5rap2an/an neuronal progenitors show premature cell cycle exit and increased cell death
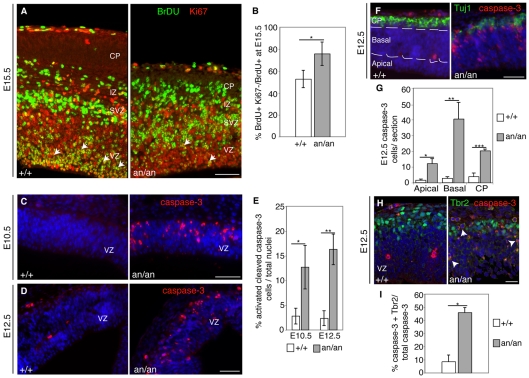
Mutation of Nde1, which encodes another spindle pole protein, causes premature cell cycle exit of neuronal progenitors, depleting the progenitor pool prematurely and reducing brain size (Feng and Walsh, 2004). To investigate whether similar processes cause progenitor cell deficits in Cdk5rap2an/an animals, we labeled progenitors with BrdU at E14.5 and examined the fraction remaining in the cell cycle at E15.5 by double immunostaining for BrdU and the proliferation marker Ki67 (Chenn and Walsh, 2002). The `exit fraction' (Caviness et al., 2003), representing the ratio of BrdU-positive Ki67-negative cells over total BrdU-positive cells, was increased by 25% in mutants compared with controls (P<1×10–4) (Fig. 5A,B), and this may contribute to the premature decrease in progenitors seen in Cdk5rap2an/an embryos.
Fig. 5.
Cdk5Rap2an/an embryos exhibit increased cell death and premature cell cycle exit. (A) Increased cell cycle exit in Cdk5Rap2an/an mice. An E14.5 pregnant female was injected with BrdU and sacrificed after 24 hours. BrdU-labeled progenitors (green) that remain in the cell cycle at E15.5 were identified by co-immunostaining for Ki67 (red). E15.5 Cdk5Rap2+/+ (n=5) and Cdk5Rap2an/an (n=6) embryos were analyzed to determine the fraction of cells leaving the cell cycle by counting BrdU-positive Ki67-negative cells as compared with the total number of BrdU-positive cells. More cells labeled exclusively with BrdU are seen in the IZ and CP of Cdk5Rap2an/an. Greater numbers of cycling cells labeled with both BrdU and Ki67 are found in the VZ of Cdk5Rap2+/+ (arrows). However, Cdk5Rap2an/an had fewer cells that remained in the cell cycle (arrows). (B) Analysis of the `exit fraction' in E15.5 littermates. The mean percentage of BrdU-positive Ki67-negative among total cells is shown for Cdk5Rap2+/+ (336 cells counted/animal) and Cdk5Rap2an/an (420 cells counted/animal). *, P=4.2×10–5. (C,D) Cdk5Rap2an/an animals show increased apoptosis. Coronal sections of (C) E10.5 and (D) E12.5 control and mutant embryos labeled with caspase 3 antibodies (red) and stained with Hoechst (blue) for nuclei. (E) The mean percentage of caspase 3-labeled cells in E10.5 and E12.5 embryos. Analysis of E10.5 littermates showed that Cdk5Rap2an/an embryos (n=3 animals, 293 cells counted/section) had a 4.5-fold increase in the percentage of apoptotic cells as compared with controls (n=3 animals, 354 cells counted/section); *, P=1.7×10–6. Cdk5Rap2an/an E12.5 embryos (n=4 animals, 419 cells counted/section) had 6.8-fold more cell death than controls (n=4 animals, 503 cells counted/section); **, P=1.7×10–13. (F) Cdk5Rap2an/an E12.5 embryos show a higher index of apoptotic neuronal precursor and neuronal cells. Caspase 3-positive cells (red) are present in the apical, basal and CP regions. CP is shown by Tuj1 (green) immunostaining. (G) The average number of caspase 3-labeled cells in each region in E12.5 embryos. Cdk5Rap2an/an embryos (n=3 animals, 73 dying cells counted/section) had 7.7-fold more dying cells in the apical region (*, P=5.8×10–6), 14.4-fold more apoptosis in the basal region (**, P=2.9×10–6), and 5.3-fold more cell death in the CP (***, P=5.7×10–6) than controls (n=3 animals, 8.2 dying cells counted/section). (H) Cdk5Rap2an/an E12.5 embryos have a higher percentage of Tbr2 (green) and caspase 3 (red) double-positive cells (arrows) than controls. (I) The percentage of Tbr2 and caspase 3 double-labeled cells among the total caspase 3-positive cells in Cdk5Rap2an/an (n=3 animals, 370 cells counted/section) and in controls (n=4 animals, 466 cells counted/section); *, P=2.6×10–10. Error bars indicate s.d. (B,E) or s.e.m. (G,I). Scale bars: 50 μm in A; 25 μm in C,D,F; 20 μm in H.
Defects in spindle orientation also correlate with increased cell death during neuroepithelial stem cell divisions (Yingling et al., 2008). We analyzed cells undergoing apoptosis by immunostaining for activated cleaved caspase 3 (caspase 3). Prior to the onset of neurogenesis (E10.5) we found 4.5-fold more apoptotic cells in mutants compared with control littermates (12.7±4.4% versus 2.8±1.6%, P<1×10–5), suggesting that these increased dying cells represent apical progenitors, the predominant cell type at this stage. At E12.5 we found a 6.8-fold increase in cell death in Cdk5rap2an/an animals compared with controls (16.3±3.1% versus 2.4±1.5%, P<1×10–9) (Fig. 5C-E). To investigate whether a particular cell type was targeted for cell death, we first analyzed the distribution of apoptotic cells in apical (alongside the ventricle), basal (away from the ventricle) and cortical plate (CP) regions at E12.5 (Fig. 5F,G). Cdk5rap2an/an animals had more dying cells than controls in both apical (12.3±3.1 versus 1.6±0.7, P<1×10–5) and basal (40.5±10.9 versus 2.8±1.1, P<1×10–5) regions, as well as in the CP (20.4±1.0 versus 3.8±2.4, P<1×10–5) (Fig. 5G). Double immunostaining for Tuj1 (Tubb3 – Mouse Genome Informatics) and caspase 3 showed increased neuronal cell death in the mutants (Fig. 5F,G). Similarly, immunoreactivity of Tbr2 and caspase 3 showed increased cell death of basal progenitors in Cdk5rap2an/an animals compared with controls (46.0±3.7% versus 8.7±4.9%, P<1×10–9) (Fig. 5H,I). Together, these data suggest that both the precursor and neuronal populations are probably susceptible to cell death in the Cdk5rap2 mutants.
DISCUSSION
Here, we show that Cdk5rap2 is mutated in the Hertwig's anemia mouse, and we demonstrate that Cdk5rap2 is essential for normal progenitor proliferation and survival in the cerebral cortex. Cdk5rap2an/an animals have smaller cerebral cortices that result from an overall reduction of the neuronal layers caused by a decrease in progenitor numbers. Increases in both cell death and premature cell cycle exit reduce the cortical precursor population. Moreover, the decrease of neuronal progenitors in Cdk5rap2an/an animals correlates, paradoxically, with an increased mitotic index, suggesting delays in mitotic progression. We found altered mitotic orientation as well as increased abnormal mitotic figures, with aneupolar spindles, implicating essential roles for Cdk5rap2 in spindle and centrosome function during neurogenesis. These data suggest that CDK5RAP2 mutations in humans could cause microcephaly by mechanisms that include not only mitotic arrest and cell death, but also may include defects in cell fate determination. It is puzzling that anemia has not been reported in the few known human patients with CDK5RAP2 mutations, as it is the defining feature of Cdk5rap2an/an mutants. However, both the microcephaly and anemia in Cdk5rap2an/an mice are variable and strain dependent, and so similar modifiers might affect penetrance in humans.
Mitotic defects have been previously associated with cell fate changes (Buchman and Tsai, 2007). It has been proposed that dividing VZ progenitors with vertical cleavage planes could generate daughter cells with similar cell fates, and that small deviations from the vertical cleavage plane might be enough to disrupt these `symmetric' cell divisions and reduce the progenitor population (Zhong and Chia, 2008). The 2-fold increase in cleavage planes parallel to the VS in Cdk5rap2an/an mice would reduce such symmetric cell divisions and might correlate with reduced progenitor numbers. RNAi knockdown experiments have also provided evidence that Aspm, another gene associated with microcephaly in humans, regulates mitotic spindle orientation and cell fate in early neurogenesis in mice (Fish et al., 2006). Furthermore, Nde1 mutants have increased cell divisions with horizontal cleavage planes that correlate with premature depletion of the progenitor pool (Feng and Walsh, 2004). In these mutants, the decreased progenitor population correlates with premature and excessive generation of deep layer neurons, resulting in a larger fraction of cells leaving the cell cycle (Caviness et al., 2003). Although failure to maintain mitotic fidelity also increases the `exit fraction' in Cdk5rap2an/an animals, unlike Nde1 mutants, Cdk5rap2an/an mice show only modestly increased ratios of deeper versus superficial layer neurons. Lack of apparent overgeneration of deeper layer neurons in Cdk5rap2an/an embryos could result from increased cell death early in development, in contrast to Nde1 mutants that show only modestly increased numbers of apoptotic cells (Feng and Walsh, 2004). Increased cell death in E10.5 Cdk5rap2an/an embryos correlates with a decrease in S-phase cells, suggesting that before neurogenic divisions start, Cdk5rap2 regulates the expansion of the progenitor pool. Moreover, the distribution of caspase 3-labeled cells and their co-immunoreactivity with either neuronal or progenitor markers in E12.5 Cdk5rap2an/an animals suggest that an overall increase in apoptosis affects both neurons and progenitors. In summary, changes in mitotic spindle orientation and premature cell cycle exit in Cdk5rap2an/an animals seem to correlate with cell fate determination defects; however, the dramatic increase in apoptosis might mask the premature and excessive generation of neurons, an expected outcome of faulty cell fate determination.
Cdk5rap2an/an mutants showed preferential thinning of the superficial neuronal layers. In evolutionary terms, these later-born neuronal layers only appeared after mammals diverged from reptiles (Nieuwenhuys, 1994), and the elaboration of the superficial layers has been proposed to be primarily responsible for the disproportionate expansion of the cerebral cortex relative to body size in mammals (Cheung et al., 2007). The evolutionary increase of superficial layers in mammals correlates with expansion of the SVZ, caused by increased basal progenitor numbers (Martinez-Cerdeno et al., 2006; Striedter and Charvet, 2009). In Cdk5rap2an/an animals, the reduction of Tbr2-immunoreactive progenitors could account, in part, for the diminished superficial layers. This is interesting because the CDK5RAP2 gene shows a modest excess of non-conservative amino acid changes in the primate lineage leading to humans, suggesting that it might have been a target of evolution in primates (Evans et al., 2006). Thus, the essential roles of Cdk5rap2 in progenitor proliferation might reflect its relevance to the evolutionary expansion of the cerebral cortex (Kriegstein et al., 2006).
Cdk5rap2 contains a γ-tubulin-association domain that is found in other centrosomal proteins, including several in which mutations alter cell polarity, nuclear positioning, chromosomal alignment, spindle orientation and/or cleavage site specification (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999; Venkatram et al., 2004; Verde et al., 2001). Cdk5rap2 and its fly ortholog Cnn (Centrosomin) recruit the γ-TuRC to the centrosome (Fong et al., 2008; Heuer et al., 1995; Zhang and Megraw, 2007; Zheng et al., 1998). The primary role of the γ-TuRC is the nucleation of microtubules (Zheng et al., 1998). However, mounting evidence suggests additional roles during cell cycle progression for γ-tubulin and other γ-TuRC subunits. When γ-tubulin fails to localize to the centrosome, some cells arrest at G2–M, followed by apoptosis, whereas cells that bypass the G2–M arrest show highly defective mitoses with increased monopolar spindles (Zimmerman et al., 2004). Furthermore, in Aspergillius nidulans, the absence of γ-tubulin arrests cells at the metaphase-to-anaphase transition (Prigozhina et al., 2004). Interestingly, in-frame deletion of exon 4 in the Cdk5rap2an allele disrupts its γ-tubulin-association domain (Fong et al., 2008), and γ-tubulin localization to the centrosome appears decreased in Cdk5rap2an/an mouse embryonic fibroblasts (S.P.M. and M.D.F., unpublished). Therefore, interaction of Cdk5rap2 and γ-tubulin might be required to maintain mitotic fidelity in neuronal progenitors, as has been described in other tissues (Eppig and Barker, 1984).
The abnormal number of spindle poles and consequent aneuploidy in Cdk5rap2an/an mice might reflect defects in centrosome maturation or duplication. Centrosomes that fail to mature are less stable and prone to undergo splitting during mitosis (Fukasawa, 2007). Knockdown of CDK5RAP2 in interphase somatic cells causes abnormal centrosome splitting (Graser et al., 2007), suggesting defective centrosome maturation. Thus, failure of centrosome maturation followed by abnormal centrosome splitting would be an appealing mechanism to explain the aneupolar spindle poles observed in Cdk5rap2an/an neuronal progenitors. Alternatively, defects in centrosome duplication could also cause abnormal numbers of spindle poles (Fukasawa, 2007). In both cases, the abnormal number of centrosomes might produce abnormal mitosis that would either result in cell death or cell cycle arrest, as we have shown in Cdk5rap2an/an cortical progenitors. Alternatively, failure to arrest or exit the cell cycle might produce aneuploidy in Cdk5rap2an/an embryonic germ cells (Eppig and Barker, 1984) and, potentially, in neuronal precursors.
In mammals, an emerging hypothesis suggests that the asymmetric inheritance of the mother and daughter centrosomes (by the progenitor and neuronal daughter cells, respectively) is important for stem cell renewal during neurogenesis (Wang et al., 2009). In mice, the removal of ninein, a component of the mother centriole appendages, disrupts centrosome maturation and impairs centrosome asymmetric inheritance, resulting in neuronal overproduction due to altered cell fate (Wang et al., 2009). Interestingly, mutations in Drosophila cnn randomize centrosome asymmetric inheritance and affect germ cell fate determination (Yamashita et al., 2007). We hypothesize that because Cdk5rap2an/an animals have defects in mitotic orientation, the normal localization of centrosomes might be disrupted and could cause abnormal inheritance of the `maternal' centrosome. Alternatively, disruption of Cdk5rap2-dependent steps of centrosome maturation could impair asymmetric centrosome inheritance in Cdk5rap2an/an mice. Either of these two mechanisms could then contribute to the premature depletion of the neuronal progenitor pool. A similar exhaustion of self-renewing progenitors might contribute to the hematological defects that originally defined the Hertwig's anemia mutation. Further analysis of centrosome maturation, duplication and inheritance in the absence of Cdk5rap2 might provide important additional insight into the relationship of the centrosome to cell fate determination and the pathology of microcephaly.
Supplementary Material
Acknowledgments
We thank S. Tzakas for technical assistance; M. Mahendroo and J. Eppig for contributing to early genetic mapping studies; members of the M.D.F. and C.A.W. laboratories for insightful discussions, especially C. Manzini, E. Morrow, N. Dwyer, E. C. Gilmore, M. Lehtinen and J. Liu; S. White (BIDMC Histology Core) for sample preparation; L. H. An and Y. Zu (BIDMC Imaging Core) and the HMS Genetics Department Imaging Core for confocal microscopy; and R. Hevner for the Tbr1 antibody. Transgenic core services were supported by NIH P30 DK049216 through the Center of Excellence in Molecular Hematology at Children's Hospital Boston. S.B.L was supported by NIH T32AG0222-13, S.P.M. by NIH K08 HL077157, M.H.H. by NIH T32 HL 076115-03 and C.A.W. by NINDS R01 NS32456. C.A.W. is an Investigator of the Howard Hughes Medical Institute. M.D.F. is supported by the Children's Hospital Pathology Foundation Wilkes Tumor Fund. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.040410/-/DC1
References
- Angevine J. B., Jr, Sidman R. L. (1961). Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192, 766-768 [DOI] [PubMed] [Google Scholar]
- Baelde H. J., Cleton-Jansen A. M., van Beerendonk H., Namba M., Bovee J. V., Hogendoorn P. C. (2001). High quality RNA isolation from tumours with low cellularity and high extracellular matrix component for cDNA microarrays: application to chondrosarcoma. J. Clin. Pathol. 54, 778-782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bani-Yaghoub M., Tremblay R. G., Lei J. X., Zhang D., Zurakowski B., Sandhu J. K., Smith B., Ribecco-Lutkiewicz M., Kennedy J., Walker P. R., et al. (2006). Role of Sox2 in the development of the mouse neocortex. Dev. Biol. 295, 52-66 [DOI] [PubMed] [Google Scholar]
- Barker J. E., Bernstein S. E. (1983). Hertwig's anemia: characterization of the stem cell defect. Blood 61, 765-769 [PubMed] [Google Scholar]
- Barker J. E., Deveau S. A., Compton S. T., Fancher K., Eppig J. T. (2005). High incidence, early onset of histiocytic sarcomas in mice with Hertwig's anemia. Exp. Hematol. 33, 1118-1129 [DOI] [PubMed] [Google Scholar]
- Bond J., Roberts E., Mochida G. H., Hampshire D. J., Scott S., Askham J. M., Springell K., Mahadevan M., Crow Y. J., Markham A. F., et al. (2002). ASPM is a major determinant of cerebral cortical size. Nat. Genet. 32, 316-320 [DOI] [PubMed] [Google Scholar]
- Bond J., Roberts E., Springell K., Lizarraga S. B., Scott S., Higgins J., Hampshire D. J., Morrison E. E., Leal G. F., Silva E. O., et al. (2005). A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37, 353-355 [DOI] [PubMed] [Google Scholar]
- Buchman J. J., Tsai L. H. (2007). Spindle regulation in neural precursors of flies and mammals. Nat. Rev. Neurosci. 8, 89-100 [DOI] [PubMed] [Google Scholar]
- Bultje R. S., Castaneda-Castellanos D. R., Jan L. Y., Jan Y. N., Kriegstein A. R., Shi S. H. (2009). Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron 63, 189-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness V. S., Jr, Goto T., Tarui T., Takahashi T., Bhide P. G., Nowakowski R. S. (2003). Cell output, cell cycle duration and neuronal specification: a model of integrated mechanisms of the neocortical proliferative process. Cereb. Cortex 13, 592-598 [DOI] [PubMed] [Google Scholar]
- Chenn A., McConnell S. K. (1995). Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell 82, 631-641 [DOI] [PubMed] [Google Scholar]
- Chenn A., Walsh C. A. (2002). Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365-369 [DOI] [PubMed] [Google Scholar]
- Cheung A. F., Pollen A. A., Tavare A., DeProto J., Molnar Z. (2007). Comparative aspects of cortical neurogenesis in vertebrates. J. Anat. 211, 164-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching Y. P., Qi Z., Wang J. H. (2000). Cloning of three novel neuronal Cdk5 activator binding proteins. Gene 242, 285-294 [DOI] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R. A., Bulfone A., Kowalczyk T., Hevner R. F. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig J. T., Barker J. E. (1984). Chromosome abnormalities in mice with Hertwig's anemia. Blood 64, 727-732 [PubMed] [Google Scholar]
- Eppig J. T., Barker J. E. (1989). Deleterious effects of irradiation and bone marrow transplantation therapy in the genetically anemic an/an mouse. Blood 73, 1373-1379 [PubMed] [Google Scholar]
- Evans P. D., Vallender E. J., Lahn B. T. (2006). Molecular evolution of the brain size regulator genes CDK5RAP2 and CENPJ. Gene 375, 75-79 [DOI] [PubMed] [Google Scholar]
- Feng Y., Walsh C. A. (2004). Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 44, 279-293 [DOI] [PubMed] [Google Scholar]
- Ferland R. J., Cherry T. J., Preware P. O., Morrisey E. E., Walsh C. A. (2003). Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J. Comp. Neurol. 460, 266-279 [DOI] [PubMed] [Google Scholar]
- Fish J. L., Kosodo Y., Enard W., Paabo S., Huttner W. B. (2006). Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA 103, 10438-10443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K. W., Choi Y. K., Rattner J. B., Qi R. Z. (2008). CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the {gamma}-tubulin ring complex. Mol. Biol. Cell 19, 115-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K. (2007). Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer 7, 911-924 [DOI] [PubMed] [Google Scholar]
- Gleeson J. G., Lin P. T., Flanagan L. A., Walsh C. A. (1999). Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron 23, 257-271 [DOI] [PubMed] [Google Scholar]
- Gotz M., Huttner W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777-788 [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Nigg E. A. (2007). Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 120, 4321-4331 [DOI] [PubMed] [Google Scholar]
- Haydar T. F., Ang E., Jr, Rakic P. (2003). Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc. Natl. Acad. Sci. USA 100, 2890-2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig P. (1942). Neue Mutationen und Koppelungsgruppen bei der Hausmaus. Z. Indukt. Abstamm. Vererbungsl. 80, 220-246 [Google Scholar]
- Heuer J. G., Li K., Kaufman T. C. (1995). The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development 121, 3861-3876 [DOI] [PubMed] [Google Scholar]
- Hevner R. F., Shi L., Justice N., Hsueh Y., Sheng M., Smiga S., Bulfone A., Goffinet A. M., Campagnoni A. T., Rubenstein J. L. (2001). Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353-366 [DOI] [PubMed] [Google Scholar]
- Higginbotham H. R., Gleeson J. G. (2007). The centrosome in neuronal development. Trends Neurosci. 30, 276-283 [DOI] [PubMed] [Google Scholar]
- Katayama H., Brinkley W. R., Sen S. (2003). The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 22, 451-464 [DOI] [PubMed] [Google Scholar]
- Kriegstein A., Noctor S., Martinez-Cerdeno V. (2006). Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 7, 883-890 [DOI] [PubMed] [Google Scholar]
- Lee L., DeBono C. A., Campagna D. R., Young D. C., Moody D. B., Fleming M. D. (2007). Loss of the acyl-CoA binding protein (Acbp) results in fatty acid metabolism abnormalities in mouse hair and skin. J. Invest. Dermatol. 127, 16-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid M., Ishmael J. E., Avram D., Shepherd D., Fraulob V., Dolle P. (2004). CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr. Patterns 4, 733-739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cerdeno V., Noctor S. C., Kriegstein A. R. (2006). The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cereb. Cortex 16Suppl. 1, i152-i161 [DOI] [PubMed] [Google Scholar]
- McEvilly R. J., de Diaz M. O., Schonemann M. D., Hooshmand F., Rosenfeld M. G. (2002). Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295, 1528-1532 [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R., Kaufman T. C. (1999). The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829-2839 [DOI] [PubMed] [Google Scholar]
- Misson J. P., Edwards M. A., Yamamoto M., Caviness V. S., Jr (1988). Identification of radial glial cells within the developing murine central nervous system: studies based upon a new immunohistochemical marker. Brain Res. Dev. Brain Res. 44, 95-108 [DOI] [PubMed] [Google Scholar]
- Mochida G. H. (2008). Molecular genetics of lissencephaly and microcephaly. Brain Nerve 60, 437-444 [PubMed] [Google Scholar]
- Nieto M., Monuki E. S., Tang H., Imitola J., Haubst N., Khoury S. J., Cunningham J., Gotz M., Walsh C. A. (2004). Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J. Comp. Neurol. 479, 168-180 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuys R. (1994). The neocortex. An overview of its evolutionary development, structural organization and synaptology. Anat. Embryol. (Berl.) 190, 307-337 [DOI] [PubMed] [Google Scholar]
- Noctor S. C., Flint A. C., Weissman T. A., Dammerman R. S., Kriegstein A. R. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714-720 [DOI] [PubMed] [Google Scholar]
- Pontious A., Kowalczyk T., Englund C., Hevner R. F. (2008). Role of intermediate progenitor cells in cerebral cortex development. Dev. Neurosci. 30, 24-32 [DOI] [PubMed] [Google Scholar]
- Prigozhina N. L., Oakley C. E., Lewis A. M., Nayak T., Osmani S. A., Oakley B. R. (2004). gamma-tubulin plays an essential role in the coordination of mitotic events. Mol. Biol. Cell 15, 1374-1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. (1974). Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science 183, 425-427 [DOI] [PubMed] [Google Scholar]
- Rakic P. (2009). Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10, 724-735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell E. S., McFarland E. C., Peters H. (1985). Gametic and pleiotropic defects in mouse fetuses with Hertwig's macrocytic anemia. Dev. Biol. 110, 331-337 [DOI] [PubMed] [Google Scholar]
- Sanada K., Tsai L. H. (2005). G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell 122, 119-131 [DOI] [PubMed] [Google Scholar]
- Sepp K. J., Hong P., Lizarraga S. B., Liu J. S., Mejia L. A., Walsh C. A., Perrimon N. (2008). Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 4, e1000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Yamada K., Watanabe M., Ikenaka K., Wada K., Tanaka K., Inoue Y. (1997). Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J. Neurosci. 17, 9212-9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striedter G. F., Charvet C. J. (2009). Telencephalon enlargement by the convergent evolution of expanded subventricular zones. Biol. Lett. 5, 134-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G., Lens S. M. (2008). The Aurora kinase family in cell division and cancer. Biochim. Biophys. Acta 1786, 60-72 [DOI] [PubMed] [Google Scholar]
- Vaizel-Ohayon D., Schejter E. D. (1999). Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 9, 889-898 [DOI] [PubMed] [Google Scholar]
- Venkatram S., Tasto J. J., Feoktistova A., Jennings J. L., Link A. J., Gould K. L. (2004). Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell 15, 2287-2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I., Pahlke G., Salanova M., Zhang G., Wang S., Coletti D., Onuffer J., Jin S. L., Conti M. (2001). Myomegalin is a novel protein of the golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem. 276, 11189-11198 [DOI] [PubMed] [Google Scholar]
- Wang X., Tsai J. W., Imai J. H., Lian W. N., Vallee R. B., Shi S. H. (2009). Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461, 947-955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C. G., Bond J., Enard W. (2005). Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am. J. Hum. Genet. 76, 717-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y. M., Mahowald A. P., Perlin J. R., Fuller M. T. (2007). Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315, 518-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J., Youn Y. H., Darling D., Toyo-Oka K., Pramparo T., Hirotsune S., Wynshaw-Boris A. (2008). Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell 132, 474-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Megraw T. L. (2007). Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol. Biol. Cell 18, 4037-4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wong M. L., Alberts B., Mitchison T. (1998). Purification and assay of gamma tubulin ring complex. Methods Enzymol. 298, 218-228 [DOI] [PubMed] [Google Scholar]
- Zhong W., Chia W. (2008). Neurogenesis and asymmetric cell division. Curr. Opin. Neurobiol. 18, 4-11 [DOI] [PubMed] [Google Scholar]
- Zimmerman W. C., Sillibourne J., Rosa J., Doxsey S. J. (2004). Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642-3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.