Abstract
TBX20 has been shown to be essential for vertebrate heart development. Mutations within the TBX20 coding region are associated with human congenital heart disease, and the loss of Tbx20 in a wide variety of model systems leads to cardiac defects and eventually heart failure. Despite the crucial role of TBX20 in a range of cardiac cellular processes, the signal transduction pathways that act upstream of Tbx20 remain unknown. Here, we have identified and characterized a conserved 334 bp Tbx20 cardiac regulatory element that is directly activated by the BMP/SMAD1 signaling pathway. We demonstrate that this element is both necessary and sufficient to drive cardiac-specific expression of Tbx20 in Xenopus, and that blocking SMAD1 signaling in vivo specifically abolishes transcription of Tbx20, but not that of other cardiac factors, such as Tbx5 and MHC, in the developing heart. We further demonstrate that activation of Tbx20 by SMAD1 is mediated by a set of novel, non-canonical, high-affinity SMAD-binding sites located within this regulatory element and that phospho-SMAD1 directly binds a non-canonical SMAD1 site in vivo. Finally, we show that these non-canonical sites are necessary and sufficient for Tbx20 expression in Xenopus, and that reporter constructs containing these sites are expressed in a cardiac-specific manner in zebrafish and mouse. Collectively, our findings define Tbx20 as a direct transcriptional target of the BMP/SMAD1 signaling pathway during cardiac maturation.
Keywords: Tbx20, Cardiogenesis, Cardiac, Heart, Xenopus laevis, Xenopus tropicalis, SMAD1, SMAD4, T-box, BMP, Zebrafish, Transgenesis
INTRODUCTION
A series of clinical studies has provided direct evidence of a role for T-box genes in heart development and human disease, as mutations in at least three T-box genes, TBX1, TBX5 and TBX20, have been linked to human congenital heart disease (CHD) (Papaioannou and Silver, 1998; Papaioannou, 2001; Prall et al., 2002; Packham and Brook, 2003; Ryan and Chin, 2003; Baldini, 2004; Showell et al., 2004; Mandel et al., 2005; Plageman and Yutzey, 2005; Stennard and Harvey, 2005; Hammer et al., 2008; Kirk et al., 2007; Liu et al., 2008; Qian et al., 2008). Specifically, mutations in TBX20 have been associated with a wide array of congenital abnormalities, including dilated cardiomyopathy (DCM), atrial septal defects (ASD) and mitral valve disease. Moreover, upregulation of TBX20 has been reported in patients with tetralogy of Fallot (Hammer et al., 2008; Kirk et al., 2007; Liu et al., 2008; Qian et al., 2008). These findings are consistent with studies of Tbx20 orthologs in a wide range of model systems, including mouse (Tbx12/20) (Carson et al., 2000; Kraus et al., 2001), zebrafish (Tbx20/HrT) (Ahn et al., 2000; Griffin et al., 2000), chick (Iio et al., 2001) and Xenopus (Brown et al., 2003), which have shown a requirement for Tbx20 in a number of cardiac cellular processes. The effects of Tbx20 loss appear to be in part mediated through its endogenous role in restricting expression of Tbx2, a T-box-containing protein required for the repression of chamber-specific genes (Singh et al., 2009). Despite the essential role of Tbx20 in cardiac development, little is known about the signal transduction pathways that function upstream to regulate Tbx20 expression in the developing heart.
Members of the bone morphogenetic protein (BMP) family and their downstream mediators, the SMADs, have also been shown to be required for many cellular events in early heart development, including cardiac progenitor specification, proliferation and differentiation. The role of BMPs in cardiac development is evidenced by the cardiac-associated defects in mice mutant for components of the BMP pathway and by the observation that SMAD proteins, mediators of BMP signaling, are upregulated in response to cardiac stress or injury. However, identification of a specific cellular role for any single component of the BMP pathway in cardiac development is frequently confounded by genetic redundancy within the BMP and SMAD families, and by temporal and spatial differences in the activities of individual pathway components (for reviews, see Klaus and Birchmeier, 2009; Euler-Taimor and Heger, 2006; Wijk et al., 2007). An alternative means of dissecting the roles of BMPs in early heart development would be to identify the direct transcriptional targets of BMP signaling; however, the cardiac targets of the BMP pathway remain poorly characterized.
In efforts to define the direct targets of growth factor pathways in heart development, we have identified a 334 bp regulatory element that is both necessary and sufficient for Tbx20 expression during cardiac chamber formation in Xenopus. We further show that the Tbx20 cardiac element is a direct transcriptional target of the BMP/SMAD1 arm of the transforming growth factor β (TGFβ) pathway and that its activation is independent of the TGFβ/activin/nodal/SMAD3 pathway. We further demonstrate that Tbx20 is co-expressed with nuclear SMAD1 in cardiomyocytes during cardiac chamber formation and that blocking SMAD1 activity in vivo leads to a specific loss of cardiac Tbx20 but not other markers of cardiac tissue. We go on to demonstrate that the minimal cardiac Tbx20 element contains four crucial non-canonical, high-affinity SMAD-binding sites, which are directly bound by phospho-SMAD1 and which are necessary for the proper combinatorial regulation of Tbx20. Finally, we demonstrate that the ability to recognize the non-canonical SMAD1 sites is not specific to Xenopus by showing that reporter constructs containing these elements are expressed in a cardiac-specific manner in zebrafish and mouse. Collectively, our studies define a direct target of the BMP/SMAD1 signaling pathways in heart development and indicate a role for BMP signaling in cardiac maturation.
MATERIALS AND METHODS
BAC library screen and RLM-RACE
The ISB-1 Xenopus tropicalis bacterial artificial chromosome (BAC) library [Children's Hospital Oakland Research Institute (CHORI)] was screened with the 5′ terminus of the X. laevis Tbx20 coding region, and BAC DNA prepared according to CHORI. DNA was initially characterized by field inversion gel electrophoresis (FIGE) and Southern blot analysis using a panel of Tbx20-specific probes. Following FIGE and Southern blotting, a 4114 bp EcoRI fragment of Tbx20 was isolated and ligated into the pBSII-KS+ vector (Stratagene). From this fragment, the element ranging from base pair position –2464 to +142 was used in the subsequent cloning of Tbx20-EGFP transgenic constructs. The transcriptional start site of X. tropicalis Tbx20 was identified using the First Choice RLM-RACE Kit (Ambion) and 5′ RLM-RACE as described by the manufacturer using whole X. tropicalis embryos (n=25), as well as brain-enriched and heart-enriched tissues (approximately 250 embryos for each), at stage 28. Primer sequences and details are given in Tables S1 and S2 in the supplementary material.
Tbx20-EGFP and Xenopus transgenesis
Tbx20-EGFP reporter constructs were generated by introducing EGFP in-frame into exon 1 of Tbx20 at position +142. A Tbx20-EGFP deletion series was generated by substituting elements of Tbx20 ranging from 471-2106 bp, each containing a 5′ EcoRI linker and a 3′ BamHI linker, for the original 2601 bp of the Tbx20-EGFP construct. Details and primer sequences are given in Table S1 in the supplementary material. All Tbx20 reporter constructs were linearized by KpnI and transgenesis performed according to Kroll and Amaya (Kroll and Amaya, 1996). For each construct, more than 10 EGFP-positive embryos were examined from at least three independent sets of injections.
XTbx20-EGFP transgenic mice
The XTbx20(–2464)-EGFP plasmid was prepared for microinjection by digestion with SacII and KpnI to release the linear transgene. The transgene DNA was purified by agarose gel electrophoresis and injected into the pronuclei of C57BL/6×DBA2 hybrid embryos at the UNC Animal Model core facility. Fertilized ova were subsequently implanted into pseudo-pregnant females and offspring were analyzed for the presence of the transgene. Founders were identified by PCR analysis of tail DNA, using the following primers: 5′-CCCTATTTGATCAGCAAACG-3′ and 5′-CACTTCCATGGGCTGATGCT-3′. Embryos resulting from timed matings between one of the male founders and a wild-type C57BL/6 female were screened for EGFP expression on a Leica MZ16F stereomicroscope. Animal care and animal experiments were in accordance with the Animal Care Committee at the University of North Carolina-Chapel Hill.
Xenopus embryo and explant culture
Xenopus embryos were obtained and staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1967). For tissue explants, tissue posterior to the cement gland and including the heart field was excised at stage 35/36 and cultured in 1×MBS (Chemicon) at 23°C until stage 40. The cardiac explants included overlying pharyngeal endoderm and some foregut endoderm. Anterior regions of whole embryos were excised and cultured in identical conditions as cardiac explants. Explants were treated at stage 40 with either 7 μM DMSO or 5 μM dorsomorphin (also referred to as Compound C; Calbiochem) in 1×MBS for 6 hours at 23°C (Hao et al., 2008; Yu et al., 2008). Explants were then fixed for 2 hours at room temperature in either Dent's Fix (80% methanol in DMSO) for whole-mount antibody staining, MEMFA for in situ hybridization, or 4% paraformaldehyde for immunohistochemistry.
Zebrafish embryo culture and transgenesis
For transient expression in zebrafish, the XTbx20(–334)-EGFP reporter construct was flanked by Tol2 arms in a pT2 vector for Tol2 transposase-mediated transgenesis (Fisher et al., 2006). Embryos were injected at the 10-cell stage with 100 pg of capped mRNA encoding Tol2 transposase and 50-100 pg of the transgene plasmid. Injected embryos were examined and photographed at 48 hours post-fertilization (hpf) on Zeiss M2Bio and Axioplan microscopes.
Cell culture and luciferase assays
Transient transfections were conducted as previously described (Wang et al., 2001). Each assay was conducted in triplicate at least two times in 12-well plates using the following expression plasmids: myocardin (Wang et al., 2001), SRF (Wang et al., 2001), Mef2c (Wang et al., 2001), SMAD3 (Feng et al., 2000), SMAD4 (Feng et al., 2000), pRK5 N-Flag Smad1 (Liu et al., 1996), pGL3-Nkx2.5 (Lien et al., 2002), Gata4 (Oh et al., 2004) and SM22 (Li et al., 1996). Fold induction was calculated as induction compared with that of reporter alone, and error bars refer to the standard deviation of fold induction.
In situ hybridization and immunohistochemistry
In situ hybridization and immunohistochemistry were conducted as previously described (Goetz et al., 2006), with the following addition: anti-Phospho-Smad1(Ser463/465) / Smad5(Ser463/465) / Smad8(Ser426/428) (1:100; Cell Signaling).
Protein-DNA binding assays
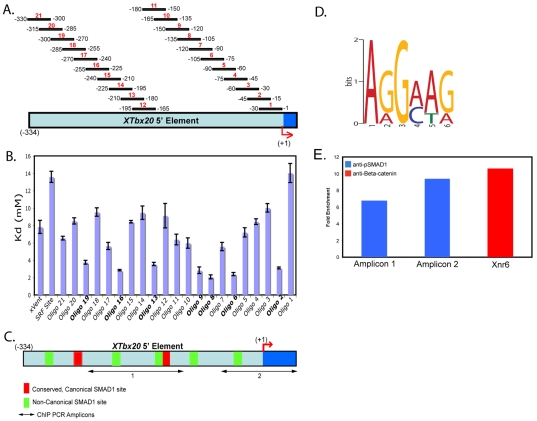
For 2× coverage of the Tbx20(–334) regulatory element, 21 double-stranded, 30 basepair, 5′-FAM oligonucleotides were designed to overlap by 15 bases beginning from base –1 (Fig. 6A), and XVent and SRF binding site oligonucleotides were designed as positive and negative controls, respectively, based on previously published work (Chang et al., 2001; Henningfeld et al., 2000). All fluorescence polarization experiments were performed in a PHERAstar microplate reader (BMG Labtechnologies) with reactions performed in a 50 μl volume containing 250 nM 5′-FAM oligonucleotide and increasing concentrations of GST-SMAD1 (0-7.41667 mM) in 10 mM Tris-HCl (pH 8.0), 100 mM NaCl. Each assay was performed in triplicate at 25°C. Anisotropy was measured by excitation with vertically polarized light, using 490 nm excitation and 520 nm emission filters with the gain optimized for maximum signal and normalized to `no protein' controls. Data analysis was performed using SigmaPlot 8.0 software, and dissociation constants (Kd) determined for each oligonucleotide using the single rectangular I, three parameter equation y=yo+ax/(b+x), where b is equal to Kd.
Fig. 6.
SMAD1 binds to seven regions within the 334 bp Tbx20 regulatory element in vitro and occupies a combination of canonical and non-canonical SMAD1-binding sites in vivo. (A) Double stranded, 5′ carboxyfluorescein-labeled, 30 bp oligonucleotides designed for 2× coverage of the 334 bp Tbx20 cardiac regulatory element for use in fluorescence polarization assays. (B) The dissociation constants (Kd) for each oligonucleotide analyzed in fluorescence polarization studies. Bold type indicates oligonucleotides bound by SMAD1. (C) Schematic of the location of seven putative SMAD1-binding sites located within the 334 bp cardiac regulatory element, including the regions to be amplified by two separate sets of ChIP PCR primers. (D) Position weight matrix generated by MEME software from the sequence analysis of oligonucleotides 19, 13, 9, 6 and 2 reveals a novel non-canonical SMAD1-binding site within the 334 bp cardiac regulatory element. (E) Phospho-SMAD1 occupies a combination of canonical and non-canonical SMAD1-binding sites within the 334 bp cardiac regulatory element. ChIP assay was performed on stage 41 X. tropicalis tadpoles with a phospho-SMAD1/5/8 antibody, and precipitated DNA was probed with primers against either a combination of canonical and non-canonical SMAD1 sites (Amplicon 1) or a single non-canonical SMAD1 site (Amplicon 2). For comparison, ChIP assay was performed on stage 9 X. laevis embryos with a β-catenin antibody, and precipitated DNA was probed with primers against Xnr6. Values are fold enrichment relative to background (no antibody control).
Chromatin immunoprecipitation in X. tropicalis embryos
Stage 41 embryos (n=30) were cross-linked in 1% formaldehyde in PBS for 60 minutes and washed in 0.125 M glycine for 10 minutes and then three times in PBS. Embryos were homogenized in 500 μl cell lysis buffer [50 mM Tris-HCl (pH 8), 2 mM EDTA, 0.1% NP-40, 10% glycerol, and protease/phosphatase inhibitors], centrifuged, and the pellet rinsed twice in cold PBS. Nuclei were lysed in 200 μl nuclei lysis buffer [50 mM Tris-HCl (pH 8), 10 mM EDTA, 1% SDS, and protease/phosphatase inhibitors] and nuclear extracts were diluted in 400 μl IP dilution buffer [20 mM Tris-HCl (pH 8), 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, and protease/phosphatase inhibitors]. ChIP extracts were sonicated three times for 30 seconds on ice, centrifuged, and supernatants were pre-cleared with 50 μl Protein A/G agarose beads (Santa Cruz Biotechnology) at 4°C for 1.5 hours. Pre-cleared ChIP extracts were diluted in 400 μl IP dilution buffer and incubated with 2 μg antibody on a rotating wheel at 4°C overnight. Protein A/G agarose beads (50 μl) were added to the ChIP samples for 2 hours at 4°C, and beads were subsequently washed in IP dilution buffer, ChIP wash buffer [10 mM Tris-HCl (pH 8), 1 mM EDTA, 1% sodium deoxycholate, 1% NP-40, 0.25 M LiCl, and protease/phosphatase inhibitors], IP dilution buffer, and TE buffer [10 mM Tric-HCl (pH 8) and 1 mM EDTA]. The material was eluted in elution buffer [50 mM Tris-HCl (pH 8), 10 mM EDTA, and 1% SDS] at 65°C, digested with RNase A for 3 hours at 37°C, and incubated in 0.3 M NaCl overnight at 65°C to reverse cross links. ChIP samples were subsequently digested with proteinase K for 4 hours at 55°C, phenol extracted, and precipitated, and the recovery of specific DNA sequences was determined by quantitative PCR using SYBR Green PCR reagents (Sigma) and an Applied Biosystems 7900 HT Fast Real-Time PCR machine. Anti-Phospho-Smad1(Ser463/465) / Smad5(Ser463/465) / Smad8(Ser426/428) (Cell Signaling) antibody was used. As a control for this procedure, ChIP was also performed on stage 9 X. laevis embryos (n=50) with a rabbit anti-β-catenin antibody (Cocalico Laboratories; Reamstown, PA, USA), as previously reported (Blythe et al., 2009). Fold enrichment relative to a no antibody control was calculated using the comparative CT method (ΔΔCt). See Table S3 in the supplementary material for ChIP primer sequences.
RESULTS
A Tbx20-EGFP transgene recapitulates endogenous expression of Tbx20 in mid-tadpole stage embryos
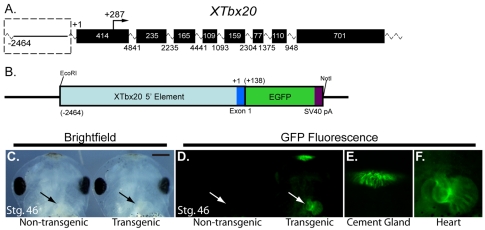
Given the evolutionarily conserved role for Tbx20 in heart development and its role in human congenital heart disease, we sought to determine the regulatory pathways that are required for the proper spatial and temporal expression pattern of Tbx20. To this end, we mapped the cardiac transcriptional start site of Tbx20 and inserted an EGFP reporter cassette in-frame with the TBX20 translational start site (see Materials and methods) in a 4116 bp fragment corresponding to the 5′ end of the Xenopus tropicalis Tbx20 locus (Fig. 1A,B). Based on our observations that Tbx20 is expressed in an identical pattern in X. tropicalis and X. laevis (Brown et al., 2003; Showell et al., 2006), we introduced the Tbx20(–2464)-EGFP reporter into X. laevis embryos by restriction enzyme-mediated integration (REMI) transgenesis. Consistent with endogenous Tbx20 expression, the Tbx20 reporter directed expression of EGFP to the developing heart and cement gland (≥5 rounds of injections; n≥20 EGFP-expressing embryos per experiment; Fig. 1C-F; see also Fig. S1 in the supplementary material). Specifically, EGFP expression was first observed in the cement gland at stage 24 and in the heart at stage 32. Identical to endogenous Tbx20 expression, as the cells of the cement gland began to undergo apoptosis, expression of EGFP decreased and it was completely absent by late tadpole stages (stages 48). By contrast, EGFP expression in the heart was maintained throughout chamber differentiation and heart looping, and continued to be expressed until later stages of cardiac development (>stage 46; Fig. 1C-F; data not shown). We did not observe EGFP expression in the heart prior to early tadpole stages (stage 32) nor in any other tissue types, including those that endogenously express Tbx20, such as the hindbrain and eye.
Fig. 1.
A regulatory element 5′ to the Tbx20 genomic locus is sufficient to drive gene expression in the Xenopus cement gland and heart. (A) Schematic of the X. tropicalis Tbx20 genomic locus. X. tropicalis Tbx20 consists of eight exons spanning approximately 20 kB. The Tbx20 transcriptional start site is located 287 bp upstream of the translation start site in exon 1. A putative cardiac regulatory element is located at the 5′ end of the Tbx20 locus (dashed box). (B) Schematic of the 2464 bp region of the 5′ end of Tbx20 cloned in frame to the EGFP reporter to examine its regulatory capacity in X. laevis transgenics. (C-F) As with endogenous Xenopus Tbx20 expression, the Tbx20 EGFP reporter is expressed in the cement gland and heart of living X. laevis transgenic embryos. (C) Ventral views of the anterior ends of stage 46 sibling non-transgenic (left) and transgenic (right) embryos. (D) Fluorescence views of siblings in C. (E,F) Magnified view of the EGFP expression driven by the Tbx20 regulatory element in the cement gland (E) and heart (F) of the transgenic embryo in D.
A 334 bp regulatory element is sufficient for cardiac Tbx20 expression
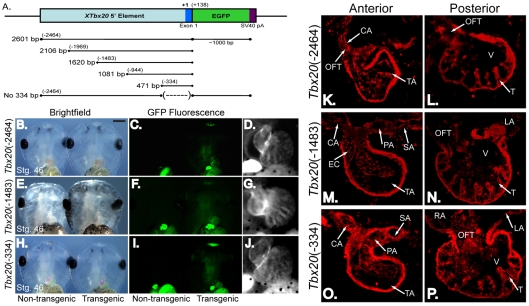
To define the minimal regulatory element necessary to drive Tbx20 cardiac-specific expression, we generated transgenic embryos with a panel of 5′ deletion constructs. Results from these injections showed that –2464 bp, –1483 bp and –334 bp Tbx20 reporters (≥3 rounds of injections; ≥20 EGFP-expressing embryos per experiment; Fig. 2A-J) all recapitulated endogenous Tbx20 expression in cardiac tissue and the cement gland at stage 46 (8/8 EGFP-expressing embryos per construct; Fig. 2K-P). Collectively, these data show that sequences within 334 bp upstream of the Tbx20 transcriptional start site contain elements that function to regulate Tbx20 cardiac expression at this stage of heart development.
Fig. 2.
A 334 bp regulatory element recapitulates the endogenous expression of Tbx20 throughout the X. laevis heart. A deletion series of the 5′ regulatory element was created to determine a reduced element sufficient to drive EGFP transgene expression. (A) Schematic of the deletion series of Tbx20 elements fused to EGFP for X. laevis transgenesis. (B,E,H) Ventral view of the anterior regions of living stage 46 (late tadpole) X. laevis embryos (left) and siblings transgenic for constructs shown in A (right) under white light. (C,F,I) Embryos in B, E and H as viewed under fluorescent light. Green autofluorescence in the gut can be noted in both control and transgenic embryos. (D,G,J) Magnified views of the EGFP-expressing hearts of embryos in C, F and I demonstrating that EGFP expression in the heart is maintained under the control of a Tbx20(–334) element. (K-P) Transverse sections were cut through the embryos expressing Tbx20-EGFP shown in B-J, and expression of the Tbx20(–2464)-EGFP (K,L), Tbx20(–1483)-EGFP (M,N) and Tbx20(–334)-EGFP (O,P) transgenes was demonstrated by antibody staining for EGFP. Anterior (K,M,O) and posterior (L,N,P) sections show EGFP transgene expression throughout the heart. CA, carotid arch; EC, endocardial cushion; LA, left atrium; OFT, outflow tract; PA, pulmocutaneous arch; RA, right atrium; SA, systemic arch; T, trabeculae; TA, truncus arteriosis; V, ventricle.
Tbx20 reporter expression is conserved in mouse and is regulated by SMAD1/SMAD4 but not SMAD3
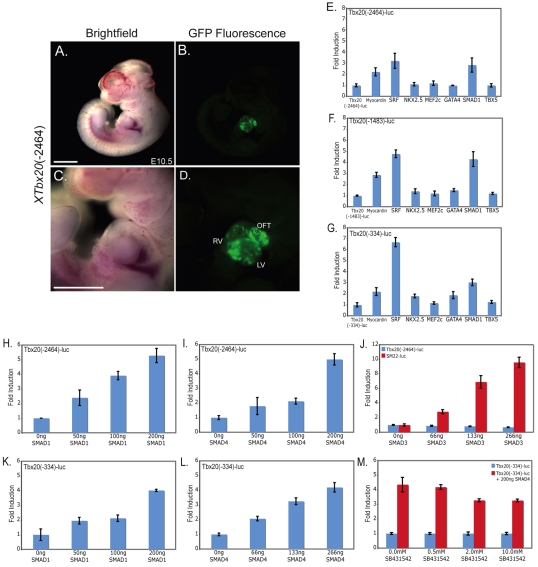
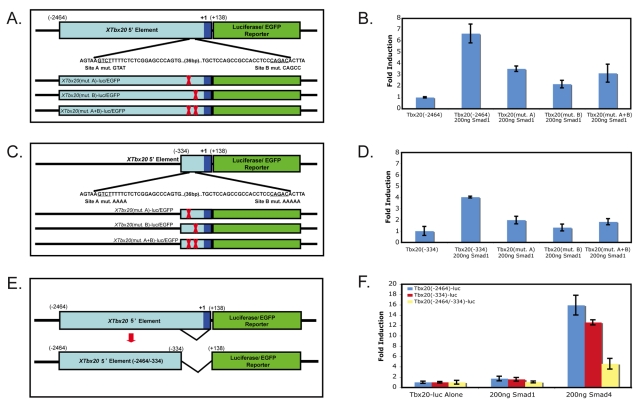
We observed that offspring from a mouse transgenic founder carrying the Xenopus Tbx20(–2464)-EGFP reporter showed an expression pattern analogous to that observed in Xenopus, with strong EGFP expression throughout the developing heart but not in other tissues where Tbx20 is endogenously expressed in the mouse, such as the hindbrain and the lateral plate mesoderm (Carson et al., 2000; Kraus et al., 2001) (Fig. 3A-D). Based on these findings, we determined whether a set of murine cardiac transcription factors for which binding sites were identified in the minimal 334 bp Tbx20 regulatory element by ConSite, Jaspar and Transfac software, including SRF, NKX2.5, MEF2C, GATA4, SMAD1 and TBX5, were capable of activating Xenopus cardiac-specific Tbx20 reporters (Fig. 3E-G). Of the potential transcription factors, SMAD1, SRF, and the SRF co-factor myocardin were found to be capable of activating Tbx20 (Fig. 3E-G). We further note that the 334 bp reporter has a greater response to SMAD1 and SRF than do the 1483 bp or 2464 bp elements, suggesting that sequences upstream of the 334 bp element can attenuate the response to SMAD1 and SRF. Although SRF and myocardin were able to induce Tbx20, upon further analysis we found that the Tbx20 reporters did not respond to myocardin or SRF in a dose-dependent fashion and that mutation of putative SRF-binding sites had no effect on the temporal or spatial expression of the Tbx20 reporter in vivo (data not shown). Although we cannot formally rule out a role for SRF or myocardin in the regulation of Tbx20 expression, we did not analyze the effects of SRF or myocardin on Tbx20 expression in greater detail.
Fig. 3.
XTbx20 5′ regulatory elements are activated by TGFβ/BMP signaling via SMAD1 and SMAD4 but not SMAD3. (A,B) The Xenopus Tbx20 5′ element is expressed in a cardiac-specific manner in E10.5 mouse embryos derived from a transgenic mouse founder expressing the XTbx20(–2464)-EGFP reporter. (C,D) Magnified view of EGFP fluorescence in the heart of a E10.5 XTbx20(–2464)-EGFP+/– mouse embryo. (E-G) Luciferase reporters controlled by three Tbx20 deletion elements were transfected into COS7 cells with a panel of cardiac factor expression plasmids. (H,I,K,L) The Tbx20(–2464) (H,I) and Tbx20(–334) (K,L) reporters are both activated by SMAD1 and SMAD4 in a dose-dependent manner when transfected with increasing amounts of SMAD expression plasmid. (J) SMAD3 transfection does not induce the Tbx20(–2464) reporter, though the control SM22 reporter is dramatically induced. (M) Treatment of COS7 cells with increasing doses of a small molecule inhibitor of activin signaling SB431542 does not affect the activation of the Tbx20(–334) plasmid by SMAD4. Values are the fold increase in luciferase activity relative to that driven by the reporter alone. Error bars represent the standard deviation of fold induction for three trials. LV, left ventricle; OFT, outflow tract; RV, right ventricle. Scale bars: 1 mm in A-D.
To also characterize the activation of Tbx20 in response to the TGFβ family of signaling molecules, we tested the ability of SMAD1, a mediator of BMP signaling, SMAD3, a mediator of TGFβ/activin/nodal signaling, and the common SMAD, SMAD4, to activate Tbx20 luciferase reporters. We observed a dose-dependent activation of both the largest (–2464) and the smallest (–334) Tbx20 regulatory elements with increasing amounts of SMAD1 and SMAD4 (Fig. 3H,I,K,L). Further deletions of the Tbx20 upstream region to –251 and –81 bps greatly decreased the responsiveness to SMAD4 and led to an increase in EGFP in non-specific regions in Xenopus transgenics (see Fig. S2 in the supplementary material).
We further observed that SMAD3 failed to activate any Tbx20 reporter although it did induce expression of the SMAD3 control reporter SM22 (Fig. 3J) (Qiu et al., 2003). Consistent with these findings, treatment of cells with the TGFβ/activin/nodal small molecule inhibitor SB431542 had no effect on the induction of Tbx20 in response to SMAD4 (Fig. 3M). Taken together, these results suggest that Tbx20 cardiac activation occurs in a SMAD1-dependent, SMAD3-independent manner that is, at least in part, mediated by sequences that lie between 81 and 334 bp upstream of the Tbx20 cardiac transcriptional start site.
Tbx20 and SMAD1 are colocalized during cardiac chamber formation
Upon activation of the BMP signaling pathway, SMAD1 is phosphorylated and translocates to the nucleus where it binds DNA and regulates transcription of neighboring genes (for reviews, see Kretzschmar and Massague, 1998; Massague et al., 2005). To determine whether SMAD1 is nuclear localized and co-expressed with Tbx20 in cardiac tissue and, therefore, could function endogenously to regulate Tbx20 expression, we serial sectioned X. laevis hearts (stage 46) and examined Tbx20 expression by in situ hybridization. On adjacent sections, we examined phospho-SMAD1 expression and its cellular localization by immunohistochemistry. Our results demonstrate that at these stages Tbx20 and phospho-SMAD1 are co-expressed throughout the myocardium of the developing heart, including in the ventricle, the atria, the outflow tract and the truncus arteriosus (Fig. 4A-F).
Fig. 4.
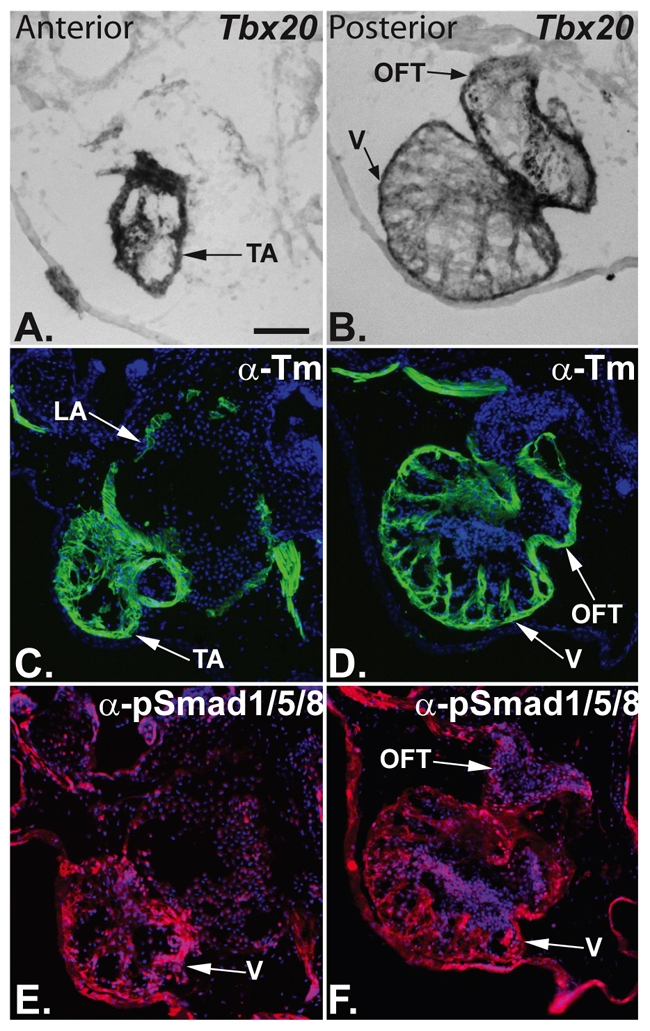
XTbx20 is expressed throughout the myocardium and endocardium of the X. laevis heart. (A,B) Tbx20 is expressed in both the anterior and posterior regions of the X. laevis stage 46 heart. (C-F) Immunohistochemistry of serial sections shows that Tbx20 expression overlaps with that of the myocardial marker tropomyosin (C,D) and with phospho-SMAD1/5/8 expression in the endocardium (E,F); anti-tropomyosin (Tm) staining is labeled green, anti-pSMAD1/5/8 is labeled red, and nuclei are labeled blue with DAPI. LA, left atrium; OFT, outflow tract; TA, truncus arteriosus; V, ventricle.
SMAD signaling is required for the maintenance of Tbx20 expression in vivo
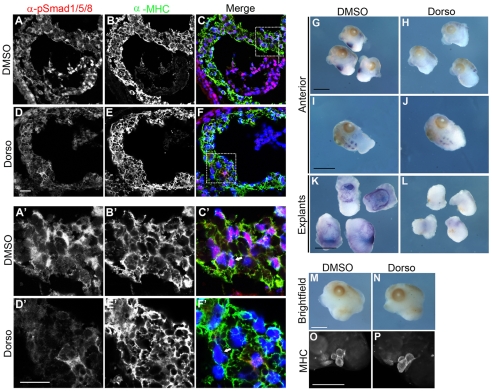
To verify that BMP signaling regulates Tbx20 expression in vivo, we determined the effects of inhibiting SMAD1 activation on Tbx20 expression. In order to bypass the requirements for SMAD1 during the early stages of embryogenesis, we made use of the SMAD1 inhibitor Dorsomorphin and a tissue explant assay (Fukuda et al., 2009; Hao et al., 2008; Langdon et al., 2007; Yu et al., 2008). As reported for tissue culture cells, treatment of stage 40 anterior explants with Dorsomorphin clearly showed that Dorsomorphin blocks the nuclear localization of SMAD1 (Fig. 5A-F,A′-F′) and completely inhibits the expression of Tbx20 in the developing heart, as compared with controls (Fig. 5G,H,K,L). Moreover, the effects of blocking SMAD1 are specific to the cardiac expression of Tbx20, as we saw little change in expression of Tbx20 in the hindbrain (Fig. 5I,J). Furthermore, SMAD1 inhibition had little to no effect on other cardiac markers, including MHC, tropomyosin and Tbx5 (Fig. 5M-P; see also Fig. S3 in the supplementary material). Taken together, these results strongly imply that the BMP pathway signals through SMAD1 to directly regulate the cardiac expression of Tbx20 in vivo.
Fig. 5.
SMAD1 activation is required for cardiac-specific expression of Tbx20 in X. laevis. (A-F′) Immunohistochemistry of transverse sections through the heart of stage 40 anterior explants shows loss of nuclear phospho-SMAD1/5/8 (arrows) in the myocardium of dorsomorphin-treated explants (D-F,D′-F′) compared with DMSO-treated controls (A-C,A′-C′). In the merged images, anti-phospho-SMAD1/5/8 (pSMAD1/5/8) staining is labeled red, anti-myosin heavy chain (MHC) is labeled green, and nuclei are labeled blue with DAPI. (G-L) In situ hybridization for Tbx20 performed on stage 40 anterior and cardiac explants shows complete loss of Tbx20 expression in the heart (H,L) but not the hindbrain (J) of dorsomorphin-treated anterior and cardiac explants compared with DMSO-treated controls (G,I,K). (M-P) Whole-mount antibody staining of stage 40 anterior explants shows normal expression of the myocardial marker MHC in dorsomorphin-treated explants (N,P) compared to DMSO-treated controls (M,O). Dorso, dorsomorphin. Scale bars: 20 μm in A-F′; 1 mm in G-P.
SMAD activation of Tbx20 occurs through direct binding of SMAD1
To identify the specific endogenous SMAD1-binding sites within the minimal 334 bp Tbx20 cardiac regulatory element, and to accurately determine the binding affinity of SMAD1 to the respective elements, we used fluorescence polarization assays. Double-stranded, 30 bp, 5′ carboxyfluorescein-labeled oligonucleotides overlapping by 15 bp were designed across the 334 bp Tbx20 cardiac regulatory element for full 2× coverage (Fig. 6A). Based on the premise that oligonucleotides tumble more slowly in solution when bound by protein as compared with unbound oligonucleotides, we combined fluorescent oligonucleotides with increasing concentrations of GST-SMAD1 fusion protein to evaluate the changes in light depolarization as anisotropy. From the anisotropy data, we plotted binding curves and calculated dissociation constants [Kd(mM)] for each oligonucleotide interaction with SMAD1, and, as controls, the binding of GST-SMAD1 to oligonucleotides containing known SMAD1-binding sequences (XVent) or known SRF-binding sequences (Fig. 6B; see also Table S4 in the supplementary material) (Chang et al., 2001; Henningfeld et al., 2000). SMAD1 bound oligonucleotide 8, covering bases –105 to –135, with the highest affinity (Kd=2.078 mM). However, SMAD1 bound six additional oligonucleotides (2, 6, 9, 13, 16 and 19) with affinities that were equal to or greater than twice that of XVent (Kd=2.078 mM to Kd=3.758 mM; Fig. 6B; see Table S4 in the supplementary material). Results from these studies show that SMAD1 binds the Tbx20 cardiac regulatory element at seven individual sites with an affinity at least twice that of the XVent control oligonucleotide (Kd=7.829 mM), suggesting that these seven sites are high-affinity SMAD1-binding sites (Fig. 6C). Sequence analysis of the oligonucleotides bound by SMAD1 revealed two conserved, consensus SMAD-binding sites with the sequences GTCT and CAGAC in oligos 16 and 8, respectively. We also observed that one region containing a putative binding site failed to bind SMAD1 protein in vitro. From this, we propose the presence of non-traditional SMAD-binding elements between bases –15 and –45 (oligonucleotide 2), –75 and –105 (oligonucleotide 6), –120 and –150 (oligonucleotides 8/9), –180 and –210 (oligonucleotide 13), and –270 and –300 (oligonucleotide 19). We further note that SMAD1 is capable of binding non-canonical sites with affinities equal to those of canonical sites.
The ability of SMAD1 protein to bind non-canonical sites within the Tbx20 334 bp cardiac regulatory element suggests the presence of a common SMAD1 motif within these oligonucleotides. Accordingly, sequence analysis by MEME software revealed a novel SMAD1-binding site with the sequence AGGA/CA/TG within oligonucleotides 19, 13, 9, 6 and 2 (Fig. 6D). Of the oligonucleotides containing the non-canonical SMAD site, SMAD1 bound oligo 6, containing the binding site AGGCAG, with the highest affinity (Fig. 6B; see also Table S4 in the supplementary material). To determine whether SMAD1 directly binds a portion of the Tbx20 cardiac element containing both canonical and non-canonical SMAD1 sites in vivo, we performed ChIP on stage 41 X. tropicalis tadpoles with a phospho-SMAD1/5/8 antibody (Fig. 6C). In parallel and as a positive control, we tested the occupancy of β-catenin on the Xnr6 locus in stage 9 X. laevis embryos, as Xnr6 has been demonstrated by others to be a direct target of β-catenin by ChIP in Xenopus (Blythe et al., 2009). A 6.8-fold enrichment above background of phospho-SMAD1 was observed on the endogenous SMAD1 sites within the Tbx20 cardiac element (Fig. 6E, Amplicon 1). We next tested the occupancy of endogenous phospho-SMAD1 on a single non-canonical SMAD1 site within the Tbx20 cardiac element (Fig. 6C, Amplicon 2). Strikingly, phospho-SMAD1 was enriched 9.4-fold above background on the non-canonical SMAD1 site, which was comparable to the enrichment of β-catenin we saw at the Xnr6 locus (10.6-fold; Fig. 6E). These data are consistent with SMAD1 directly binding a novel non-canonical site within the Tbx20 cardiac element in vivo.
Canonical SMAD sites alone are not sufficient for Tbx20 activation by SMAD1
To determine which SMAD1-binding sites are crucial for cardiac-specific expression of Tbx20, we mutated the two consensus SMAD1-binding sites, alone or in combination, in the context of the Tbx20(–2464)-EGFP or –luc reporter constructs (Fig. 7A,B), and the Tbx20(–334)-EGFP or –luc constructs (Fig. 7C,D). Results from these assays show that constructs lacking SMAD1 consensus sites are still SMAD1 responsive. Thus, these data strongly imply that SMAD1 responsiveness is mediated by non-canonical SMAD1-binding sites. Finally, when we deleted the 334 bp minimal element in the context of the original Tbx20(–2464) cardiac reporter, both the response to SMAD1/4 in tissue culture assays and EGFP expression were greatly reduced (Fig. 7E,F; data not shown). Together, these data show that the 334 bp region directly upstream of the Tbx20 start site is necessary and sufficient for cardiac expression of Tbx20, and that Tbx20 cardiac expression is dependent on SMAD1/4 activity.
Fig. 7.
SMAD1 activation is mediated through non-canonical SMAD1-binding sites. (A-D) Mutation of the two consensus SMAD1-binding sites alone or in combination, in the context of the Tbx20 (–2464)-EGFP or -luc reporter constructs (A,B), and the Tbx20(–334)-EGFP or -luc constructs (C,D), led to a decrease but not loss of activation in response to SMAD1. (E,F) Deletion of the 334 bp regulatory element from the 2464 bp reporters, Tbx20(–2464/–334)-luc and Tbx20(–2464/–334)-EGFP, led to a substantial decrease in response to SMAD1. Fold induction reflects changes in induction relative to induction of the reporter alone; error bars represent standard deviation of three replicates.
Xenopus Tbx20 reporter constructs are expressed in a cardiac-specific fashion in zebrafish
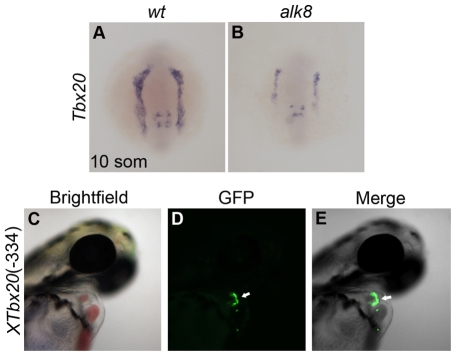
As a further test of whether Tbx20 is a general target of the BMP/SMAD1 pathway, we analyzed expression of Tbx20 in zebrafish mutant for the BMP receptor Alk8. Consistent with our findings in Xenopus, we observed a significant reduction in Tbx20 expression in zebrafish alk8 (lost-a-fin) mutants (Fig. 8A,B). Zygotic alk8 mutants are weakly dorsalized, exhibiting the effects of a mild disruption of BMP signaling (Bauer et al., 2001; Mintzer et al., 2001). Correspondingly, Tbx20 expression was diminished in the bilateral heart fields (Fig. 8B).
Fig. 8.
The Xenopus Tbx20 334 bp cardiac regulatory element drives EGFP expression in a cardiac-specific manner in zebrafish. (A,B) In situ hybridization depicts expression of Tbx20 in wild-type zebrafish embryos and alk8sk42 (Marques and Yelon, 2009) mutant siblings at the 10-somite stage; dorsal views, anterior to the top. Tbx20 expression is reduced in both the anterior lateral plate mesoderm, including the bilateral cardiac primordia, and the midline mesenchyme of zygotic alk8 mutants. (C-E) Lateral views of a live zebrafish embryo at 48 hpf, following injection with the XTbx20(–334)-EGFP transgene. Injected embryos express EGFP in the myocardium (arrows).
The observation that the BMP/SMAD1 pathway directly regulates the cardiac expression of Tbx20 through a set of non-canonical SMAD1-binding sites, and the observation that Tbx20 is downregulated in alk8 mutants, led us to question whether recognition of the canonical and non-canonical SMAD1-binding sites is specific to Xenopus or whether these sites can serve as a general response element to BMP/SMAD1 signaling in other vertebrates. To address these issues, we generated a Tbx20(–334)-EGFP fusion construct flanked by Tol2 transposase sites and injected this transgene together with Tol2 transposase RNA into zebrafish. Injection of reporter constructs in this fashion yields relatively efficient, yet highly mosaic, transgene expression (Fisher et al., 2006). The Tbx20(–334)-EGFP transgene was highly efficient at driving EGFP expression in the zebrafish heart (Fig. 8C-E). Ninety-three percent (92/99) of the injected embryos expressed the transgene, and 100% (92/92) of the expressing embryos displayed mosaic EGFP expression in the heart. As with Xenopus, expression outside the heart was inconsistent and appeared as irreproducible ectopic expression of EGFP. In summary, our data demonstrate that the 334 bp element from the Xenopus Tbx20 locus is sufficient for cardiac-specific expression in Xenopus and zebrafish.
DISCUSSION
Studies of cardiac gene regulation have suggested that heart-specific transcription is regulated temporally and spatially via a set of distinct modular cis-acting elements (Schwartz and Olson, 1999). Here, we report that SMAD signaling is required in vivo for expression of Tbx20 during cardiac chamber formation. We have also identified a 334 bp element in Xenopus that contains a series of seven high-affinity SMAD1/4-binding sites that are necessary and sufficient for the evolutionarily conserved cardiac expression of Tbx20 in mouse and zebrafish. Complementary to this finding, we have identified additional sequences that attenuate the BMP/SMAD1 response. Collectively, our studies demonstrate two distinct temporal requirements for BMP signaling during heart development: BMP signaling is required during the early phases of vertebrate heart development for the establishment of the cardiac lineage and is required later during cardiac chamber formation and maturation through the direct transcriptional regulation of Tbx20.
Tbx20 cardiac expression requires canonical and non-canonical SMAD1-binding sites
Our data demonstrate a requirement for a set of high-affinity, canonical and non-canonical SMAD-binding sites in the regulation of Tbx20 expression. Sequence analysis of regions within the 334 bp Tbx20 cardiac element that were demonstrated to bind SMAD1 reveals two conserved consensus SMAD-binding sites containing the sequences GTCT and CAGAC, as well as a novel non-canonical SMAD-binding motif containing the sequence AGGA/CA/TG. We observed that SMAD1 occupies a combination of these sites in vivo during cardiac maturation. We have demonstrated that mutation of SMAD1/4 canonical-binding sites, either singly or in combination, has little effect on the expression of Tbx20 either in vitro or in vivo, implying that it is the complement of canonical and non-canonical SMAD1-binding sites that is required for cardiac expression of Tbx20. Furthermore, our results suggest that the ability of SMAD1 to bind to DNA is not based on sequence alone. This hypothesis is supported by our observation that a region of the Tbx20 minimal element containing a putative SMAD1/4-binding site failed to bind to SMAD1 in vitro. Taken together, our data support a model in which Tbx20 expression is regulated during the later stages of heart development by BMP signaling. In this model, the BMP pathway acts through a combinatorial set of unique SMAD-binding elements, the individual elements of which differ in their contributions to the response to growth factor signaling, and therefore, to transcriptional output.
Our finding that a complement of SMAD1/4-binding sites is required for Tbx20 cardiac expression is broadly consistent with the results of studies on two other BMP-responsive genes, XVent and Nkx2.5. Early mesodermal expression of XVent is dependent on five putative SMAD1/4-binding sites, whereas cardiac expression of Nkx2.5 is dependent on twelve individual SMAD1/4-binding sites (Henningfeld et al., 2000; Liberatore et al., 2002; Lien et al., 2002). Similar to our findings, point mutations or deletions in multiple SMAD1/4-binding sites in the XVent promoter have no effect on SMAD responsiveness (Henningfeld et al., 2000). However, the regulation of cardiac-specific expression of Nkx2.5 appears to be unique in that it is mediated by a direct interaction of a SMAD1/SMAD4 complex and a member of the GATA transcription factor family (Caban et al., 2004; Liberatore et al., 2002). Although we have identified a GATA consensus site within the minimal Tbx20 cardiac element, none of the Tbx20 reporters respond to GATA4 (Fig. 3) and deletion of the GATA site in the 5′ deletion series has no effect on the cardiac-specific expression of Tbx20 (Fig. 7D). Thus, tissue-specific expression of Tbx20, unlike that of Nkx2.5, appears to occur through a GATA-independent mechanism. The activation of cardiac gene expression via BMP signaling has also been shown to be dependent on additional cardiac transcription factors. For example, the myocardin-dependent expression of cardiac genes is synergistically activated by the direct interaction of SMAD1 with myocardin (Callis et al., 2005). However, data we obtained using a large panel of cardiac transcription factors demonstrate that, with the exception of SRF, none of these factors significantly induce Tbx20 expression in transient transcriptional assays (Fig. 3). Although we cannot formally rule out a potential role of SRF in Tbx20 expression, mutation of the SRF-binding site had no effect on the temporal or spatial expression of Tbx20 reporter constructs in vivo (data not shown).
Cardiac-specific Tbx20 expression
What, then, is the mechanism underlying the cardiac-specific expression of Tbx20? We note that nuclear localization of SMAD1 during heart development is temporally regulated. Based on this observation and the results of reporter analyses, we favor a model in which the complement of SMAD1/4-binding sites directs a pattern of broad temporal expression of Tbx20 in the embryo that is further spatially refined by restriction of expression to the developing cardiac tissue as a result of the action of as yet unidentified transcriptional repressors. This idea is supported by our observations that: (1) the response of Tbx20 reporters to SMAD1 and SMAD4 is enhanced by deletion of regions both outside and within the 334 bp element; and (2) upon reduction of the 334 bp Tbx20 regulatory element to 81 bp, reporter expression substantially increased in non-cardiac tissues in X. laevis transgenic animals (see Fig. S2 in the supplementary material). These findings are consistent with studies that have demonstrated that the BMP arm of the SMAD signaling pathway is associated with the regulation of genes involved in early heart development, whereas the TGFβ/activin/nodal arm of the SMAD pathway appears to drive cardiac regulation of factors associated with fibrotic, apoptotic and anti-hypertrophic events related to progression to heart failure (for a review, see Euler-Taimor and Heger, 2006). Furthermore, it has been suggested that BMP signals act as long-range diffusible morphogens originating from multiple locations in the embryo, including the endoderm, ectoderm, or the cardiac cells themselves (Schlange et al., 2000; Schultheiss et al., 1997). It is therefore interesting to speculate that the regulation of the novel 334 bp Tbx20 cardiac element during late cardiogenesis is a result of a continued or second wave of BMP signaling from the underlying endoderm or from the myocardial cells, mediated by SMAD1/4.
Although the SMAD1/4-binding sites are crucial for expression of Tbx20 during cardiac chamber formation, the regulatory element described here does not activate endogenous expression of Tbx20 in other regions of the embryo. Thus, our minimal Tbx20 element does not comprise all of the information necessary for the complete expression of Tbx20, and elements regulating early cardiac and neural expression of Tbx20 remain to be identified. Based on this observation and on the modular nature of the BMP/SMAD response elements described here, it appears that, as for regulation of Nkx2.5 expression, regulation of Tbx20 occurs in a modular manner. Finally, taking into consideration (1) that the minimal element we have identified is required for expression of Tbx20 during cardiac chamber formation, (2) the established correlation between mutations in Tbx20 and human congenital heart disease, and (3) that not all human mutations that map to Tbx20 occur in the coding region of the gene (Hammer et al., 2008; Kirk et al., 2007; Liu et al., 2008; Qian et al., 2008), it will be interesting to determine if an association exists between mutations in the Tbx20 minimal element and congenital heart disease and/or cardiac hypertrophy.
Supplementary Material
Acknowledgments
We thank Dr Shoko Ishibashi, Dr Enrique Amaya, Scott A. Lujan, Dr Laura M. Guogas and Dr Matthew R. Redinbo for technical assistance, and Dr Christopher Showell for critical reading of the manuscript and helpful suggestions. We also thank Dr Yonqin Wu and the UNC In Situ Hybridization Core Facility for valuable assistance. We thank N. A. Thomas for her contributions to the zebrafish work and Lauren Waldron for her technical assistance with Xenopus transgenesis. The tropomyosin antibody developed by J.-C. Lin was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242, USA. This work is supported by grants to F.L.C. from the NIH/NHLBI and the American Heart Association, and by an award from the UNC Medical Alumni Association. E.M.M. was supported by a National Science Foundation Graduate Research Fellowship and a UNC Graduate School Dissertation Completion Fellowship. E.K. was supported by the UNC Developmental Biology Training Grant and an award from the American Heart Association. Work in the Yelon lab was supported by grants from the National Institutes of Health, the American Heart Association and the March of Dimes. Work in the Wang lab was supported by the NIH/NHLBI and the American Heart Association. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.043588/-/DC1
References
- Ahn D. G., Ruvinsky I., Oates A. C., Silver L. M., Ho R. K. (2000). tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech. Dev. 95, 253-258 [DOI] [PubMed] [Google Scholar]
- Baldini A. (2004). DiGeorge syndrome: an update. Curr. Opin. Cardiol. 19, 201-204 [DOI] [PubMed] [Google Scholar]
- Bauer H., Lele Z., Rauch G. J., Geisler R., Hammerschmidt M. (2001). The type I serine/threonine kinase receptor Alk8/Lost-a-fin is required for Bmp2b/7 signal transduction during dorsoventral patterning of the zebrafish embryo. Development 128, 849-858 [DOI] [PubMed] [Google Scholar]
- Blythe S. A., Reid C. D., Kessler D. S., Klein P. S. (2009). Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev. Dyn. 238, 1422-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. D., Binder O., Pagratis M., Parr B. A., Conlon F. L. (2003). Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev. Genes Evol. 212, 604-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caban A. J., Hama A. T., Lee J. W., Sagen J. (2004). Enhanced antinociception by nicotinic receptor agonist epibatidine and adrenal medullary transplants in the spinal subarachnoid space. Neuropharmacology 47, 106-116 [DOI] [PubMed] [Google Scholar]
- Callis T. E., Cao D., Wang D. Z. (2005). Bone morphogenetic protein signaling modulates myocardin transactivation of cardiac genes. Circ. Res. 97, 992-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson C. T., Kinzler E. R., Parr B. A. (2000). Tbx12, a novel T-box gene, is expressed during early stages of heart and retinal development. Mech. Dev. 96, 137-140 [DOI] [PubMed] [Google Scholar]
- Chang P. S., Li L., McAnally J., Olson E. N. (2001). Muscle specificity encoded by specific serum response factor-binding sites. J. Biol. Chem. 276, 17206-17212 [DOI] [PubMed] [Google Scholar]
- Euler-Taimor G., Heger J. (2006). The complex pattern of SMAD signaling in the cardiovascular system. Cardiovasc. Res. 69, 15-25 [DOI] [PubMed] [Google Scholar]
- Feng X. H., Lin X., Derynck R. (2000). Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J. 19, 5178-5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S., Grice E. A., Vinton R. M., Bessling S. L., Urasaki A., Kawakami K., McCallion A. S. (2006). Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 1, 1297-1305 [DOI] [PubMed] [Google Scholar]
- Fukuda T., Kohda M., Kanomata K., Nojima J., Nakamura A., Kamizono J., Noguchi Y., Iwakiri K., Kondo T., Kurose J., et al. (2009). Constitutively activated ALK2 and increased SMAD1/5 cooperatively induce bone morphogenetic protein signaling in fibrodysplasia ossificans progressiva. J. Biol. Chem. 284, 7149-7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S. C., Brown D. D., Conlon F. L. (2006). TBX5 is required for embryonic cardiac cell cycle progression. Development 133, 2575-2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin K. J., Stoller J., Gibson M., Chen S., Yelon D., Stainier D. Y., Kimelman D. (2000). A conserved role for H15-related T-box transcription factors in zebrafish and Drosophila heart formation. Dev. Biol. 218, 235-247 [DOI] [PubMed] [Google Scholar]
- Hammer S., Toenjes M., Lange M., Fischer J. J., Dunkel I., Mebus S., Grimm C. H., Hetzer R., Berger F., Sperling S. (2008). Characterization of TBX20 in human hearts and its regulation by TFAP2. J. Cell. Biochem. 104, 1022-1033 [DOI] [PubMed] [Google Scholar]
- Hao J., Daleo M. A., Murphy C. K., Yu P. B., Ho J. N., Hu J., Peterson R. T., Hatzopoulos A. K., Hong C. C. (2008). Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One 3, e2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningfeld K. A., Rastegar S., Adler G., Knochel W. (2000). Smad1 and Smad4 are components of the bone morphogenetic protein-4 (BMP-4)-induced transcription complex of the Xvent-2B promoter. J. Biol. Chem. 275, 21827-21835 [DOI] [PubMed] [Google Scholar]
- Iio A., Koide M., Hidaka K., Morisaki T. (2001). Expression pattern of novel chick T-box gene, Tbx20. Dev. Genes Evol. 211, 559-562 [DOI] [PubMed] [Google Scholar]
- Kirk E. P., Sunde M., Costa M. W., Rankin S. A., Wolstein O., Castro M. L., Butler T. L., Hyun C., Guo G., Otway R., et al. (2007). Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am. J. Hum. Genet. 81, 280-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A., Birchmeier W. (2009). Developmental signaling in myocardial progenitor cells: a comprehensive view of Bmp- and Wnt/beta-catenin signaling. Pediatr. Cardiol. 30, 609-616 [DOI] [PubMed] [Google Scholar]
- Kraus F., Haenig B., Kispert A. (2001). Cloning and expression analysis of the mouse T-box gene tbx20. Mech. Dev. 100, 87-91 [DOI] [PubMed] [Google Scholar]
- Kretzschmar M., Massague J. (1998). SMADs: mediators and regulators of TGF-beta signaling. Curr. Opin. Genet. Dev. 8, 103-111 [DOI] [PubMed] [Google Scholar]
- Kroll K. L., Amaya E. (1996). Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signalling requirements during gastrulation. Development 122, 3173-3183 [DOI] [PubMed] [Google Scholar]
- Langdon Y. G., Goetz S. C., Berg A. E., Swanik J. T., Conlon F. L. (2007). SHP-2 is required for the maintenance of cardiac progenitors. Development 134, 4119-4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Miano J. M., Cserjesi P., Olson E. N. (1996). SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ. Res. 78, 188-195 [DOI] [PubMed] [Google Scholar]
- Liberatore C. M., Searcy-Schrick R. D., Vincent E. B., Yutzey K. E. (2002). Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev. Biol. 244, 243-256 [DOI] [PubMed] [Google Scholar]
- Lien C. L., McAnally J., Richardson J. A., Olson E. N. (2002). Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev. Biol. 244, 257-266 [DOI] [PubMed] [Google Scholar]
- Liu C., Shen A., Li X., Jiao W., Zhang X., Li Z. (2008). T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur. J. Med. Genet. 51, 580-587 [DOI] [PubMed] [Google Scholar]
- Liu F., Hata A., Baker J. C., Doody J., Carcamo J., Harland R. M., Massague J. (1996). A human Mad protein acting as a BMP-regulated transcriptional activator [see comments]. Nature 381, 620-623 [DOI] [PubMed] [Google Scholar]
- Mandel E. M., Callis T. E., Wang D. Z., Conlon F. L. (2005). Transcriptional mechanisms of congenital heart disease. Drug Disc. Today: Dis. Mech. 2, 33-38 [Google Scholar]
- Marques S. R., Yelon D. (2009). Differential requirement for BMP signaling in atrial and ventricular lineages establishes cardiac chamber proportionality. Dev. Biol. 328, 472-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J., Seoane J., Wotton D. (2005). Smad transcription factors. Genes Dev. 19, 2783-2810 [DOI] [PubMed] [Google Scholar]
- Mintzer K. A., Lee M. A., Runke G., Trout J., Whitman M., Mullins M. C. (2001). Lost-a-fin encodes a type I BMP receptor, Alk8, acting maternally and zygotically in dorsoventral pattern formation. Development 128, 859-869 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J. (1967). Normal Table of Xenopus laevis (Daudin) Amsterdam: North Holland; [Google Scholar]
- Oh J., Wang Z., Wang D. Z., Lien C. L., Xing W., Olson E. N. (2004). Target gene-specific modulation of myocardin activity by GATA transcription factors. Mol. Cell. Biol. 24, 8519-8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packham E. A., Brook J. D. (2003). T-box genes in human disorders. Hum. Mol. Genet. 12, R37-R44 [DOI] [PubMed] [Google Scholar]
- Papaioannou V. E. (2001). T-box genes in human disorders. Hum. Mol. Genet. 12, R37-R44 [DOI] [PubMed] [Google Scholar]
- Papaioannou V. E., Silver L. M. (1998). The T-box gene family. BioEssays 20, 9-19 [DOI] [PubMed] [Google Scholar]
- Plageman T. F., Jr, Yutzey K. E. (2005). T-box genes and heart development: putting the `T' in hearT. Dev. Dyn. 232, 11-20 [DOI] [PubMed] [Google Scholar]
- Prall O. W., Elliot D. A., Harvey R. P. (2002). Developmental paradigms in heart disease: insights from tinman. Ann. Med. 34, 148-156 [PubMed] [Google Scholar]
- Qian L., Mohapatra B., Akasaka T., Liu J., Ocorr K., Towbin J. A., Bodmer R. (2008). Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc. Natl. Acad. Sci. USA 105, 19833-19838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P., Feng X. H., Li L. (2003). Interaction of Smad3 and SRF-associated complex mediates TGF-beta1 signals to regulate SM22 transcription during myofibroblast differentiation. J. Mol. Cell. Cardiol. 35, 1407-1420 [DOI] [PubMed] [Google Scholar]
- Ryan K., Chin A. J. (2003). T-box genes and cardiac development. Birth Defects Res. C Embryo Today 69, 25-37 [DOI] [PubMed] [Google Scholar]
- Schlange T., Andree B., Arnold H. H., Brand T. (2000). BMP2 is required for early heart development during a distinct time period. Mech. Dev. 91, 259-270 [DOI] [PubMed] [Google Scholar]
- Schultheiss T. M., Burch J. B., Lassar A. B. (1997). A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 11, 451-462 [DOI] [PubMed] [Google Scholar]
- Schwartz R. J., Olson E. N. (1999). Building the heart piece by piece: modularity of cis-elements regulating Nkx2-5 transcription. Development 126, 4187-4192 [DOI] [PubMed] [Google Scholar]
- Showell C., Binder O., Conlon F. L. (2004). T-box genes in early embryogenesis. Dev. Dyn. 229, 201-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell C., Christine K. S., Mandel E. M., Conlon F. L. (2006). Developmental expression patterns of Tbx1, Tbx2, Tbx5, and Tbx20 in Xenopus tropicalis. Dev. Dyn. 235, 1623-1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Horsthuis T., Farin H. F., Grieskamp T., Norden J., Petry M., Wakker V., Moorman A. F., Christoffels V. M., Kispert A. (2009). Tbx20 interacts with smads to confine Tbx2 expression to the atrioventricular canal. Circ. Res. 105, 442-452 [DOI] [PubMed] [Google Scholar]
- Stennard F. A., Harvey R. P. (2005). T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development 132, 4897-4910 [DOI] [PubMed] [Google Scholar]
- Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001). Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105, 851-862 [DOI] [PubMed] [Google Scholar]
- Wijk B. V., Moorman A. F. M., Hoff M. J. B. (2007). Role of bone morphogenetic proteins in cardiac differentiation. Cardiovasc. Res. 74, 244-255 [DOI] [PubMed] [Google Scholar]
- Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., Peterson R. T. (2008). Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 4, 33-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.