Abstract
In this issue of Developmental Cell, Soper and colleagues (2008) report that the mammalian MAELSTROM (MAEL) protein is critical for transposon silencing in the male germ line. Loss of MAEL is associated with meiotic failure and DNA damage, suggesting that efficient transposable element restraining mechanisms must be in place for the preservation of germ line integrity.
Sexually reproducing species face the daunting challenge of transmitting their genetic material to subsequent generations without the accumulation of mutations or major genomic alterations. Since the discovery of transposable elements (TEs) by Barbara McClintock sixty years ago, it has been recognized that these mobile sequences pose a considerable threat to genomic integrity during germ line transmission. It is therefore not surprising that diverse mechanisms have evolved to silence germ line TE mobilization. A paper in this issue of Developmental Cell reveals the importance of the MAELSTROM protein in transposon control and highlights the danger posed by mobile elements when left unchecked in the mammalian germ line.
DNA methylation is one well-understood mechanism of TE silencing. Recently, a novel transposon-silencing pathway reliant on small RNAs has attracted much attention (Aravin et al, 2007). At the center of this pathway is a class of small RNAs (termed piRNAs) that bind the germ line-specific “Piwi” class of Argonaute proteins. Many of these piRNAs are complementary to mobile elements. Mutations in two mouse Piwi members, Mili (Piwil2) and Miwi2 (Piwil4), result in the loss of TE DNA methylation in the testes (Kuramochi-Miyagawa et al 2008). These findings suggest that small RNAs guide the methylation machinery to transposable elements in the germ line to mediate silencing. Recently, the mouse MAELSTROM protein was found to interact with two Piwi proteins, MILI (PIWIL2) and MIWI (PIWIL1), in the germ line-specific structure called nuage (Costa et al, 2006), hinting that MAEL may function in this pathway.
Nuage (cloud in French) is a perinuclear electron-dense structure found in germ line cells of many species. Its function, however, has remained mysterious. Similar to P bodies in somatic cells, the germ line-specific nuage is enriched for multiple components of the small RNA silencing pathway, including the helicases Armitage and Spindle-E, and the Piwi proteins Argonaute3 and Aubergine in flies, ZIWI in zebrafish, and MILI and MIWI in mice (Klattenhoff and Theurkauf, 2008). Recent studies have established a connection between members of the Piwi pathway and transposon control. Moreover, derepression of retrotransposons was observed in two Drosophila nuage component mutants, maelstrom and krimper (Lim and Kai, 2007), suggesting that the nuage coordinates activity of the RNA silencing pathway in TE control in the Drosophila germ line.
In flies, Maelstrom is expressed in the female germ line, yet its biochemical function remains undefined. It was originally identified based on is role in oocyte axis specification (Clegg et al, 1997; Findley et al, 2003). The name refers to the distorted appearance of microtubules in mael mutant oocytes. However, subsequent studies suggested that the patterning phenotype is a secondary consequence of DNA damage checkpoint activation (Klattenhoff et al, 2008). Recent work demonstrated that Mael is required for the accumulation of repeat-associated siRNAs (rasiRNAs) and repression of TEs in flies (Lim and Kai, 2007).
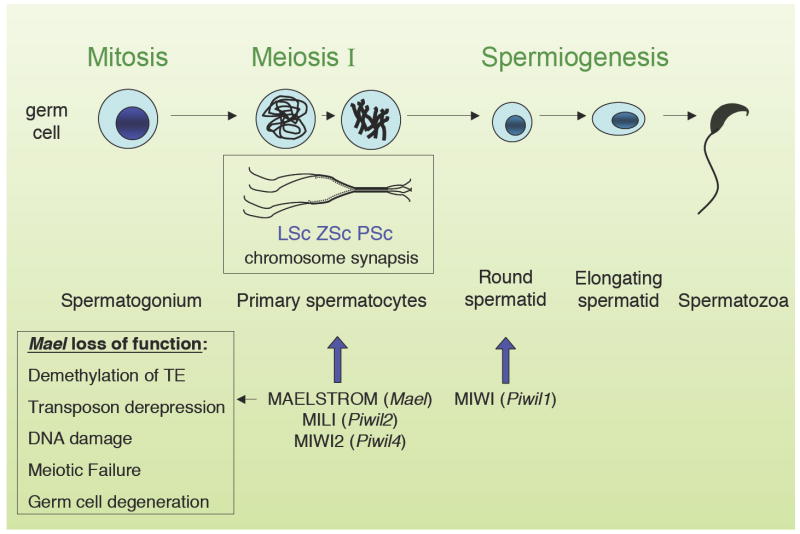
Until now, it remained unknown whether the mammalian counterpart of nuage and MAELSTROM protein(s) function in transposon silencing in the germ line. In mammals, MAEL is expressed in the male germ line. Soper and colleagues (2008) first examined the localization of MAEL during spermatogenesis (Figure 1). They observed a stage-specific recruitment of MAEL to nuage during the late pachytene and diplotene stages.
Figure 1.
The three phases of spermatogenesis in mice: mitosis, meiosis, and spermiogenesis. During mitosis, stem cells divide to generate primary spermatocytes. These proceed through meiosis I and II to generate haploid round spermatids. Meiotic prophase I is further divided into several stages: In leptotene spermatocytes (LSc), chromosomes condense and sister chromatids pair. In zygotene spermatocytes (ZSc), synaptonemal complex components are organized for pairing of homologs. Crossover occurs in pachytene spermatocytes (PSc), resulting in the exchange of genetic information. Homologous chromosomes separate in diplotene and resolve in diakinesis. During spermiogenesis, after the second meiotic division, round spermatids mature to elongated forms and spermatozoa. Meiotic arrest points are indicated for mouse Maelstrom and Piwi family mutants.
To investigate the role of the mammalian ortholog of MAEL in germ line development, a null allele of Mael was generated in mice. Elimination of Mael led to germ cell degeneration in the testes and male sterility. Mutant spermatocytes fail to mature beyond zygotene of meiotic prophase I and are eliminated by apoptosis by mid-pachytene. Furthermore, examination of chromosome pairing reveals that MAEL is essential for meiotic chromosome synapsis. In contrast to Drosophila Maelstrom, expression of the mammalian ortholog has not been demonstrated during oogenesis, and no mention is made of a female infertility phenotype in Mael null mice.
Interestingly, the Mael mutants undergo meiotic arrest at the same point in spermatogenesis as Miwi2 and Mili mutants, suggesting a functional relationship between these proteins. This led the authors to ask whether MAEL is required for piRNA biogenesis. Mael mutants are competent for production of pre-pachytene piRNAs, which target repeat elements, indicating that MAEL is dispensable for piRNA expression and accumulation. Subsequent pachytene piRNAs are not expressed, presumably reflecting meiotic crisis and germ cell loss. These data do not support a primary role for MAEL in piRNA processing, although additional studies may shed a brighter light on this issue.
A fascinating consequence of the loss of Mael is a dramatic increase in expression of LINE-1 (L1) retrotransposons in adult mutant testes relative to WT, as measured by RT-PCR. Moreover, activation of L1 correlated with impairment of DNA methylation, reminiscent of the Mili and Miwi2 mutant phenotypes (Kuramochi-Miyagawa et al 2008). A significant increase in L1 ORF1p, which encodes a structural protein required for retrotransposition, was also observed. Expression of L1 in the nucleus led to extensive DNA damage, evidenced by co-localization of ORF1p and γH2AX, a marker of double strand breaks (DSBs). This is consistent with reports demonstrating that overexpression of L1 resulted in the accumulation of γH2AX foci and DSBs in cell culture (Gasior et al, 2006).
The authors then performed an elegant series of experiments to distinguish DSBs that occur normally during meiosis and DNA damage that occurs in Mael -/- mutants. SPO11 is a topoisomerase that generates DSBs during meiosis. In contrast to mice that lack Spo11 alone, double mutant mice lacking both Spo11 and Mael show massive γH2AX signal in meiotic nuclei. These findings provide evidence that the DSBs in Mael -/- mutants are independent of normal breaks that facilitate genetic recombination.
Soper and colleagues (2008) have demonstrated that the mammalian MAELSTROM protein is essential for silencing of retrotransposons and provide evidence that early meiosis is a critical time-point when transposon control is ensured in the male germ line. Many questions remain. Since pre-pachytene piRNAs accumulate to normal levels in Mael mutants, MAEL likely functions downstream of piRNA biogenesis and perhaps piRNA-Piwi complex assembly. Instead, MAEL may function at a later step such as shuttling Piwi complexes from the nuage to sites of TE methylation, as Soper et al. speculate. Indeed, MAEL localization is dynamic, shuttling from nuage to the nucleus (Findley et al, 2003). It will thus be important to examine the localization of Piwi proteins in Mael mutants. The authors also suggest that MAEL may play a more prominent role in MIWI2-dependent processes based on the similarity between their mutant phenotypes. MAELSTROM interacts with MILI and MIWI, however additional studies are necessary to determine whether MAEL associates with MIWI2. Perhaps most importantly, the primary cause of DNA damage and meiotic failure in Mael mutants remains elusive. Is it solely driven by mobilization of TEs or could there be other consequences of Maelstrom loss? If the former, what burden of transposon activity can be tolerated during meiosis? A lesser compromise of TE silencing mechanisms (i.e., hypomorphic alleles of Mael or Piwi family members) or circumvention of the meiotic crisis phenotype, coupled with availing technologies for endogenous transposon mapping, may provide a new means for forward genetics in mice. Finally, conspicuously lacking is an understanding of TE silencing in mammalian oogenesis, which appears resistant to meiotic failure in several mouse models (Hunt and Hassold, 2002). Despite many unanswered questions, the emerging story of nuage function draws our attention to possible threats transposable elements pose when not efficiently silenced. Much like the chaos created by the infamous whirlpool described in Poe’s essay A Descent into the Maelstrom, inappropriate transposon activation may well wreak havoc in the germ line.
References
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Clegg NJ, Frost DM, Larkin MK, Subrahmanyan L, Bryant Z, Ruohola-Baker H. maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: posterior localization of grk mRNA. Development. 1997;124:4661–4671. doi: 10.1242/dev.124.22.4661. [DOI] [PubMed] [Google Scholar]
- Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet. 2006;15:2324–2334. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PA, Hassold TJ. Sex matters in meiosis. Science. 2002;296:2181–2183. doi: 10.1126/science.1071907. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper SFC, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse Maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Developmental Cell. 2008 doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]