Abstract
Population approaches, traditionally employed in pharmacokinetic-pharmacodynamic studies, have shown value also in the context of glucose-insulin metabolism models by providing more accurate individual parameters estimates and a compelling statistical framework for the analysis of between-subject variability (BSV). In this work, the advantages of population techniques are further explored by proposing integration of covariates in the intravenous glucose tolerance test (IVGTT) glucose minimal model analysis. A previously published dataset of 204 healthy subjects, who underwent insulin-modified IVGTTs, was analyzed in NONMEM, and relevant demographic information about each subject was employed to explain part of the BSV observed in parameter values. Demographic data included height, weight, sex, and age, but also basal glycemia and insulinemia, and information about amount and distribution of body fat. On the basis of nonlinear mixed-effects modeling, age, visceral abdominal fat, and basal insulinemia were significant predictors for SI (insulin sensitivity), whereas only age and basal insulinemia were significant for P2 (insulin action). The volume of distribution correlated with sex, age, percentage of total body fat, and basal glycemia, whereas no significant covariate was detected to explain variability in SG (glucose effectiveness). The introduction of covariates resulted in a significant shrinking of the unexplained BSV, especially for SI and P2 and considerably improved the model fit. These results offer a starting point for speculation about the physiological meaning of the relationships detected and pave the way for the design of less invasive and less expensive protocols for epidemiological studies of glucose-insulin metabolism.
Keywords: NONMEM, population modeling, nonlinear mixed-effects modeling, insulin sensitivity, glucose metabolism
variability in biological systems is a widespread trait and poses an extremely challenging problem. Issues related to variability impinge on the design and execution of experiments on biological systems, especially in the clinical setting, where measurements are few and far between. The challenge posed by variability is compounded by the fact that, often, the traits to be measured cannot be quantified directly but are amenable only to indirect quantification. It can be argued that, when such quantification is only possible through identification of a mathematical model, new, state-of-the-art tools are required to both quantify and explain sources of in vivo variability. An example of indirect quantification of traits is provided by model-based biomedical investigation, exemplified here by the so-called minimal model of glucose kinetics (6). When fitted to individual responses from an intravenous glucose tolerance test (IVGTT) (16), the model provides useful indexes of an individual subject's metabolic state, including measures of insulin sensitivity and glucose effectiveness. The traditional paradigm to estimate the IVGTT glucose minimal model parameters is by means of weighted least squares (WLS), performed on each single subject experimental data. However, for epidemiological studies, encompassing large numbers of subjects, the use of a population approach (5, 11, 29) has been shown both to improve the quality of the individual parameter estimates and to provide more refined insight on the between-subject variability of the parameters in the population of IVGTTs (12, 13, 15, 23). Conventionally, in fact, the analysis of the population variability is performed only as an a posteriori step, by means of a statistical moment analysis (namely, sample mean and covariance) on the individual results previously obtained by the traditional WLS approach. This method, however, does not take into account the uncertainty in the estimates of the individual parameters and is therefore prone to provide biased values (32).
Population approaches, instead, are based on the assumption that all subjects are random realizations of the population and they share some typical features common to the population itself, called “fixed effects”. The population information is employed to support the individual parameter estimation process, because each subject is characterized by “random effects,” which shape its deviation from the population typical behavior. In particular, parametric population approaches assume a probability distribution for the different subjects and then estimate the parameters characterizing this distribution, i.e., means, variances, and correlations.
A further step in epidemiological studies consists of trying to inspect the causes underlying the variability present in the data in an attempt to identify whether some independently assessed characteristics of the subjects significantly correlate with the model parameter values. These features, normally referred to in population analysis as covariates, can be integrated into the population model itself to improve its predictive power. The coefficients driving the relationships between the individual parameter values and the covariates can, in fact, be introduced in the model as additional parameters and therefore optimized together with the remaining population fixed effects. In this way, a part of the population variability, previously accounted for by the individual random effects, is explained in a deterministic fashion. Moreover, the equations linking the model parameters to the independently measured covariates can be used to predict individual parameter values, thus potentially reducing the need for blood samples and invasive trials.
In this work, methods for covariate selection are applied to a large database of IVGTTs, showing that it is feasible to use independently measured, relatively easy to obtain covariates to explain observed variation in the metabolic parameters provided by the glucose minimal model. This work lays the foundation for further research aimed at investigating reduced protocols, based not only on blood sampling but also on individual-specific covariate values, for the identification of the glucose minimal model metabolic indexes.
MATERIALS AND METHODS
Data.
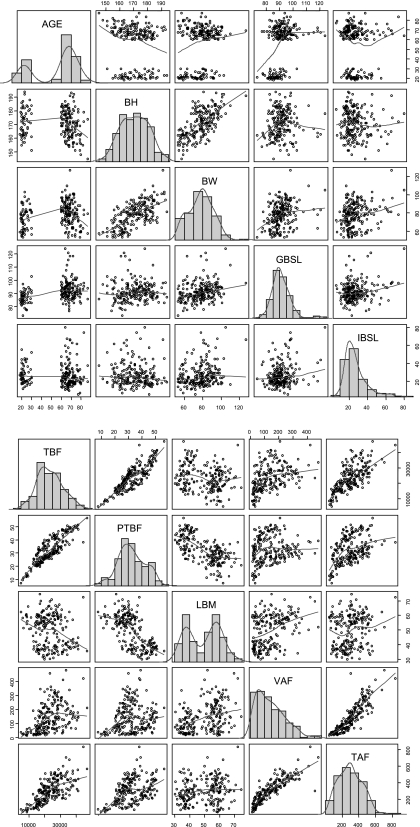
The dataset encompasses 204 healthy subjects [mean age 56 yr (range 18–87); mean BMI 27 kg/m2 (range 20–35)] who underwent an insulin-modified (16), full sampling schedule IVGTT (240 min, 21 samples, insulin infusion at 20 min lasting 5 min) (2, 3). Additional patient information was also collected with the purpose of investigating which physiological characteristics were significant predictors of glucose-insulin metabolism. More in detail, this information included age (AGE), sex (SEX), height (BH), weight (BW), and basal levels of glucose (GBSL) and insulin (IBSL). In addition, as described in Basu et al. (2), body composition was assessed using dual-energy X-ray absorptiometry (DEXA), and the following covariates were obtained: total body fat (TBF), also expressed as a percentage of total body weight (%TBF), and lean body mass (LBM). Data from a single-slice computed tomographic (CT) scan at the level of L2/L3 were employed, as explained in Jensen et al. (20), to obtain estimates of visceral (VAF) and total abdominal body fat (TAF). Last, some derived covariates were calculated from primary measurements: body mass index (BMI) and body surface area (BSA). All these covariates are listed in Table 1 and graphic summaries of their distributions, also showing regressions between the most correlated pairs, can be seen in Fig. 1. Three subjects had some missing values for VAF, TAF, and %TBF; therefore, their values were assumed to be the population averages. This seemingly naïve approach is justified by the fact that, when the model was fitted, regression was carried out on the mean-centered covariate values; so a null covariate effect was imputed for the subjects with missing data.
Table 1.
Overview of the covariates
| Covariate | Name | Units | Mean | Min | 1st Q | Median | 3rd Q | Max |
|---|---|---|---|---|---|---|---|---|
| AGE | Age | years | 55.53 | 18 | 27 | 65 | 71 | 87 |
| BH | Body height | cm | 170.9 | 145 | 163 | 171 | 178 | 194 |
| BW | Body weight | kg | 77.94 | 53 | 68.9 | 79 | 87 | 127 |
| BMI | Body mass index | kg/m2 | 26.62 | 19.6 | 24.23 | 26.76 | 29.06 | 34.85 |
| BSA | Body surface area | m2 | 1.917 | 1.505 | 1.771 | 1.937 | 2.047 | 2.596 |
| LBM | Lean body mass | kg | 49.53 | 30.1 | 38.5 | 51.84 | 58.68 | 74.58 |
| VAF | Visceral abdominal fat | cm2/CT slice | 141.8 | 11.86 | 62.62 | 127.5 | 204.7 | 478.2 |
| TAF | Total abdominal fat | cm2/CT slice | 301.8 | 43.94 | 195.1 | 294.5 | 404.4 | 837.5 |
| TBF | Total body fat | grams | 23,410 | 4,884 | 17,370 | 22,570 | 28,420 | 46,990 |
| %TBF | Percent total body fat | % | 32.39 | 7.3 | 25.85 | 31.55 | 39.68 | 56.7 |
| GBSL | Basal glucose (fasting, predose) | mg/dl | 91.34 | 72.96 | 86.74 | 90.31 | 94.72 | 123.8 |
| IBSL | Basal insulin (fasting, predose) | pmol/ml | 26.98 | 5.4 | 18.56 | 23.85 | 31.35 | 80.25 |
Continuous covariates for the glucose-insulin system measured in our 204 subject database. Statistics include minimum and maximum value, 1st and 3rd quartiles, and mean and median.
Fig. 1.
Overview of covariate values. Histogram plots and smoothed densities for the covariate set (see also Table 1). Regressions between the most correlated covariates are also shown, and a smoothed tendency line is superimposed to depict the trend of the relation. A: AGE, age; BH, body height; BW, body weight; GBSL, basal glucose level; IBSL basal insulin level. B: TBF, total body fat; PTBF, %TBF; LBM, lean body mass; VAF, visceral abdominal fat; TAF, total abdominal fat.
Minimal model.
The glucose minimal model (6) was used to analyze glucose-insulin IVGTT data. The model can be described by the following equations:
| (1) |
where D denotes the glucose dose, G(t) (in mg/dl) is plasma glucose concentration, I(t) (in pmol/l) is the plasma insulin concentration, Gb and Ib are their basal values, and X(t) is the insulin action (min−1). The model has four uniquely identifiable parameters: glucose effectiveness (SG, min−1), insulin sensitivity (SI, liter·min−1·pmol−1), the insulin action parameter (P2, min−1), and the apparent volume of glucose distribution per unit of body mass (VOL, dl/kg). I(t) is assumed as a known, error-free input (forcing) function. Usually, glucose measurements prior to 8 min are excluded from the parameter estimation, because the one-compartment model is not designed to account for the quickest phase of glucose kinetics (10).
Covariate selection.
The outline of the covariate selection process was the following:
1) a population “base” model (without any covariate effect) was fitted to the data and individual post hoc estimates were obtained for all model parameters;
2) the individual parameter values from the base model were treated as dependent variables in a traditional multivariate linear regression analysis aimed at screening the data for significant correlations;
3) the relations detected in the previous step were further investigated with a nonlinear mixed-effects (NLME) implementation in order to select the optimal covariate set.
The process is explained in greater detail in the following sections.
Base model.
As a first step, a population model without covariates was implemented (base model, BM), similarly to Denti et al. (13), and estimates were obtained for all the population parameters and the individual random effects (individual parameters). Consistently with previous literature (1, 15), the parameters pi were assumed lognormally distributed around the typical values θ, and the covariance matrix of the random effects Ω comprised only the correlation terms between SI-P2 and VOL-SG. In formulae
| (2) |
| (3) |
In addition, a combined error structure (with both proportional and additive components) was used, as explained below:
| (4) |
where y denotes the observed value, g(pi) the model prediction for subject i, and ε1 and ε2 are respectively the proportional and additive errors.
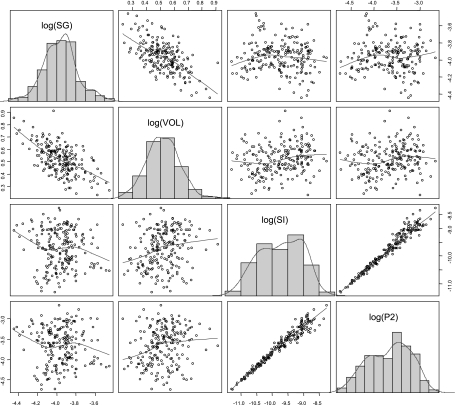
A statistical summary of the individual estimates is shown in Table 2, and a graphic representation of the parameter distributions is shown in Fig. 2.
Table 2.
Overview of individual parameter values from base model
| Name | Units | Mean | Min | 1st Q | Median | 3rd Q | Max | |
|---|---|---|---|---|---|---|---|---|
| SG (log) | Glucose effectiveness | min−1 | 0.0193 (−3.95) | 0.0117 (−4.45) | 0.0170 (−4.08) | 0.0190 (−3.96) | 0.0209 (−3.87) | 0.0316 (−3.45) |
| VOL (log) | Glucose volume of distribution | dl/kg | 1.69 (0.527) | 1.26 (0.229) | 1.56 (0.443) | 1.68 (0.518) | 1.81 (0.592) | 2.50 (0.914) |
| SI (log) | Insulin sensitivity | min−1 per pmol/ml | 7.32E-05 (−9.52) | 1.23E-05 (−11.30) | 3.43E-05 (−10.28) | 6.11E-05 (−9.70) | 0.000107 (−9.14) | 0.000258 (−8.26) |
| P2 (log) | Insulin action | min−1 | 0.0284 (−3.56) | 0.0088 (−4.74) | 0.0180 (−4.02) | 0.0275 (−3.59) | 0.0362 (−3.32) | 0.0701 (−2.66) |
Summary of minimal model individual parameters obtained for 204 subjects with the base model. Along with the values in original unit, log-transformed values (used for regression) are also reported in brackets.
Fig. 2.
Overview of individual parameter values from base model. Scatterplots and histograms of the logarithm of the individual parameter values, as obtained with the base model. SG, glucose effectiveness; VOL, apparent volume of glucose distribution per unit of body mass; SI, insulin sensitivity; P2, insulin action. The correlation is apparent between log(SI) and log(P2) and between log(SG) and log(VOL), as accounted for by the relative terms in the covariance matrix Ω.
Multivariate linear regression.
Given the high number of candidate predictors available and the exponentially related number of potential models, neither the implementation of all the possible combinations directly as NLME models nor a full traditional stepwise approach (36) is feasible due to long computational times. Thus, an off-line multivariate regression of the post hoc estimates of the individual effects from BM was carried out using the statistical software R (28). The results from this exploratory analysis were used to narrow down the pool of potential candidate models to test with a full-fledged NLME model analysis.
Even though the algorithms for multivariate linear regression are fast, the number of possible models grows exponentially with the number of covariates, in our case 14. Thus, branch-and-bound techniques as implemented in the R package leaps (26) were used. This allowed us both to explore more options than a pure stepwise regression and to dramatically reduce the number of models that would have been included in a naïve exhaustive exploration.
The main criterion to rank and select the most significant models was the “adjusted R2” (Ra2), an unbiased version of R2. Ra2 measures the strength of the correlation existing between the dependent variable and a group of predictors (covariates) and is adjusted to take into account the number of predictors. Its mathematical formula is the following:
| (5) |
where VARres and VARtot denote the “unbiased” estimate of the variance of, respectively, the model residuals and the observations, similarly SSres and SStot are their sums of squares, and n is the number of observations (the individual estimates of the minimal model parameters) and p the number of predictors (covariates) used in the model. Intuitively, it can be interpreted as the ratio of variance in the data that is explained by the linear regression model.
NLME implementation.
For each parameter (VOL, SG, SI, and P2), the model that showed the highest Ra2 values was selected for analysis by NLME modeling. When a group of models was characterized by similar values of Ra2, the models were all selected for NLME implementation. In addition, even though Ra2 is already corrected for the degrees of freedom, it was decided to include also for full analysis the best fitting models (largest Ra2) among those with a higher or lower number of predictors. All the selected models were implemented in both SPK (30) and NONMEM (4). The method FOCE with INTERACTION was applied, and the initial values provided by the regression analysis in R were used as initial estimates. NONMEM runs were facilitated by using Perl Speaks NONMEM (25), diagnostic plots were produced with the R package Xpose [Xpose (21)], and the runs were collected and compared with Census (34). Once the mixed-effects optimal parameter values were obtained, the objective function minima were used to assess the actual significance of the covariates in the models, as it is commonly done (35). A p value of 0.01 was used as the threshold of statistical significance. Although both NONMEM and SPK were used and the analyses showed good agreement, NONMEM results are reported here for simplicity.
In the implementation of the NLME models, covariates (with the exception of SEX) were centered on their mean; i.e., the difference of the individual value from the mean was entered in the model, as opposed to the raw value of the covariate. This does not affect the covariate selection process or the minimum of the objective function that is reached, but it was introduced mainly for two reasons. The first, as we have mentioned, is that it allows imputing the values of missing covariates to the mean value for several subjects (in this dataset, only 3); those specific subjects should thus not influence the estimation of the covariate effect. In addition, centering the regressors on their mean makes the interpretation of the value of the fixed effects modeling each parameter's typical values easier, since they can be construed as changes with respect to the population's typical values.
RESULTS
Results from the exploratory covariate ranking using the Ra2 are reported in Table 3, and the covariate incorporation process is summarized in Table 4. For each minimal model parameter (VOL, SI, P2, SG), all the candidate models identified by the explorative regression analysis in R were implemented, ranked according to their OFV χ2 p value with respect to the BM, and the most statistically significant model was selected. In addition, during the covariate selection process, care was taken in analyzing the precision (standard error) of the parameter estimates provided by the mixed-effects modeling software, checking that the asymptotic 95% confidence interval for the regression parameters excluded zero. At the end of the incorporation process, each covariate was tested for elimination with a p value of 0.01.
Table 3.
Candidate predictors: values of Ra2 for the candidate models
| Covariates | No. of Predictors | Adj R2 |
|---|---|---|
| Log(SG) best covariates based on Ra2: BH, BW, BSA, TAF, GBSL | ||
| BH BW BSA VAF GBSL | 5 | 0.038 |
| BH BW BSA VAF | 4 | 0.035 |
| BH BW BSA | 3 | 0.030 |
| BW BMI BSA | 3 | 0.026 |
| GBSL IBSL | 2 | 0.013 |
| GBSL | 1 | 0.006 |
| AGE | 1 | 0.003 |
| Log(VOL) best covariates based on Ra2: SEX, AGE, VAF, %TBF, GBSL | ||
| SEX AGE VAF %TBF GBSL | 5 | 0.324 |
| SEX AGE TAF %TBF GBSL | 5 | 0.323 |
| AGE TAF %TBF GBSL | 4 | 0.317 |
| SEX AGE %TBF GBSL | 4 | 0.316 |
| SEX AGE VAF %TBF | 4 | 0.315 |
| AGE VAF %TBF | 3 | 0.307 |
| AGE TAF %TBF | 3 | 0.302 |
| SEX AGE %TBF | 3 | 0.301 |
| AGE TBF | 2 | 0.284 |
| Log(SI) best covariates based on Ra2: SEX, AGE, BMI, VAF, IBSL | ||
| SEX AGE BMI VAF IBSL | 5 | 0.503 |
| AGE VAF %TBF IBSL | 4 | 0.502 |
| SEX AGE VAF IBSL | 5 | 0.502 |
| AGE VAF IBSL | 3 | 0.491 |
| AGE TAF IBSL | 3 | 0.488 |
| VAF IBSL | 2 | 0.455 |
| TAF IBSL | 2 | 0.453 |
| AGE IBSL | 2 | 0.440 |
| Log(P2) best covariates based on Ra2: SEX, AGE, VAF, TAF, IBSL | ||
| SEX AGE VAF TAF IBSL | 5 | 0.475 |
| SEX AGE BSA VAF IBSL | 5 | 0.475 |
| AGE TAF %TBF IBSL | 4 | 0.475 |
| SEX AGE TAF IBSL | 4 | 0.474 |
| AGE TAF IBSL | 3 | 0.468 |
| AGE VAF IBSL | 3 | 0.461 |
| AGE IBSL | 2 | 0.426 |
| TAF IBSL | 2 | 0.419 |
| VAF IBSL | 2 | 0.400 |
Overall “best” model based on exploratory analysis is suggested to be the one with the highest Ra2 (boldface).
Table 4.
Covariate selection process
| Model | OFV | ΔOFV | Marginal ΔOFV |
|---|---|---|---|
| BASE | 18,213.3 | 0 | 0 |
| VOL∼SEX+AGE+%TBF+GBSL | 18,071.7 | −141.6 | −141.6 |
| SI∼AGE+VAF+IBSL | 17,977.7 | −235.6 | −94.0 |
| P2∼AGE+IBSL | 17,914.0 | −299.3 | −63.7 |
Outline of covariate selection process. OFV, NONMEM objective function value. ΔOFV and marginal ΔOFV are the differences in OFV with the base model and the previous step, respectively.
The covariate selection proceeded as outlined below and in Table 4; the order for the inclusion (VOL, SI, P2) was chosen according to the improvement in the objective function value. As a first step, the candidate models for VOL were tested, and the lowest p value was obtained with SEX, AGE, %TBF, and GBSL. The improvement in the objective function value was very pronounced (−141.6 OFV, +4 parameters); therefore, the p value was much lower than the selected cutoff of 1%. This provisionally optimal model for VOL was incorporated in the model, and, in the following step, the predictors for SI were evaluated. The most statistically significant OFV decrease was detected (−94.0 OFV, +3 parameters) when AGE, VAF, and IBSL were used as predictors. The next candidates tested were the models for P2, and the best option was including AGE and IBSL (−63.7 OFV, +2 parameters). A correlation was also detected with TAF, but just below the required level of statistical significance and was therefore not included in the model. For SG, the model that resulted in the most significant drop in the OFV was employing BH, BW, and BSA, but it did not explain a significant fraction of the BSV and was arguably only due to the high level of collinearity among the covariates, especially between BW and the derived variable BSA (∼0.98). A very high condition number for the covariance matrix of the estimates confirmed this, so the model was not selected. Alternative models (e.g., BW, BMI, and BSA) had very similar problems in terms of collinearity of the predictors, and no other option proved statistically significant. Therefore, no covariates were used for SG. This is in agreement with the results of the exploratory analysis, which detected only very low Ra2 for SG. It can also be pointed out that SG presented the highest level of shrinkage [as reported also by Krudys et al. (23)], ∼17% (see below for more complete data on shrinkage). This level of shrinkage is not considered alarming, but this result still indicates that SG is the parameter for which the between-subject differences are the weakest to detect.
A 1,000-samples nonparametric bootstrap (14, 24) was also employed to test the robustness of the model and obtain more reliable confidence intervals for the parameter estimates. The bootstrap analysis confirmed the significance of the relations detected, and both the parameter estimates and their precision were in agreement with the values obtained with the asymptotic standard errors returned at the end of the NONMEM run.
The final model was then the following:
| (6) |
where COV (all caps) is a covariate (AGE, %TBF, etc.) and ΔCOV represents the deviation of the covariate COV from its average value. Note that for VOL, two separate typical values were estimated for males and females; therefore, separate averages where also used for the two sexes.
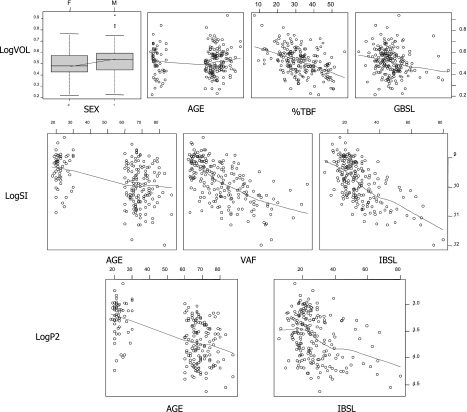
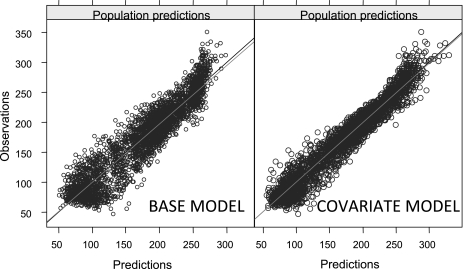
The optimal parameter values for the base and the final covariate model are reported in Table 5, along with their precision, and the estimates obtained with the bootstrap. In our reports, to facilitate interpretation of the results, we reported the values of the square root of the elements on the diagonal of Ω, which can be interpreted in first approximation as %CV values, while the off-diagonal elements were reported as the corresponding correlations rather than covariances. Scatterplots of the individual parameter values obtained from the BM and used for the regression vs. the selected covariates are shown in Fig. 3. Our analysis detects a negative correlation between log(SI) and AGE, VAF, and IBSL, and similarly log(P2) is found to be negatively correlated with AGE and IBSL. All these relationships are apparent also from the scatterplots in Fig. 3, and indeed the population variability appreciably decreases from 72.7 to 47.5% and from 50.7 to 37.9% for SI and P2, respectively. This means that the model has increased its descriptive (and arguably predictive) capabilities and is able to explain a portion of the BSV in a deterministic way rather than attributing it to random differences among subjects. The drop in population variability for VOL is more limited (from an initial 12.2 to 10.4%), but statistically significant. A lower typical value of volume is detected for females; a negative correlation is identified with %TBF and GBSL, and a positive correlation with AGE. However, although the relationships with SEX, %TBF, and GBSL are also apparent in the scatterplots of the initial regression analysis performed on the BM individual estimates, interestingly the correlation is not so manifest for AGE. The portion of variability explained by the covariates is also apparent when one compares plots of the observations vs. the population prediction (PRED) for the base and final models, available in Fig. 4. PRED denotes the model prediction obtained, for each individual, with zero random effects, so that the individual parameters are calculated only with the deterministic part of the model, i.e., the fixed effects and the measured covariates. For the BM, this means that all the subjects share the same typical values, whereas in the final model the covariates modify the expected value of each subject's parameters. It can clearly be seen that the amount of variability decreases significantly, with all the prediction-observation pairs more tightly clustering around the identity line. In both the BM and the final covariate model, shrinkage of the individual effects was calculated and was below the generally accepted upper threshold of 20–30% (22) (∼17% for SG, ∼5% for VOL, ∼7% for SI, and ∼12% for P2).
Table 5.
Final model parameter estimates
| Parameter | Base Model | Base Model (bootstrap) | Covariate Model | Covariate Model (bootstrap) |
|---|---|---|---|---|
| θSG | 0.0193 (2%) | 0.193 (0.0193…0.0200) | 0.0192 (2%) | 0.0192 (0.0184…0.0199) |
| ωSG | 21.2% (8%) | 21.2% (17.6%…24.1%) | 21.0% (8%) | 20.9% (17.9%…24.0%) |
| θVOL | 1.68 (1%) | 1.68 (1.65…1.71) | ||
| θVOL-FEMALE | 1.65 (1%) | 1.65 (1.62…1.68) | ||
| θVOL-MALE | 1.70 (1%) | 1.70 (1.67…1.74) | ||
| θVOL∼AGE | 0.00181 (18%) | 0.00181 (0.00124…0.00238) | ||
| θVOL∼%TBF | −0.0101 (7%) | −0.0102 (−0.0122…−0.00810) | ||
| θVOL∼GBSL | −0.00312 (23%) | −0.00317 (−0.00484…−0.00153) | ||
| ωVOL | 12.2% (6%) | 12.1% (10.8%…13.7%) | 10.4% (7%) | 10.3% (9.1%…11.5%) |
| θSI | 5.74 (7%) | 5.77 (5.10…6.42) | 5.83 (5%) | 5.83 (5.37…6.28) |
| θSI∼AGE | −0.00810 (29%) | −0.00808 (−0.0118…−0.00439) | ||
| θSI∼VAF | −0.00208 (20%) | −0.00210 (−0.00310…−0.00120) | ||
| θSI∼IBSL | −0.0282 (11%) | −0.0281 (−0.0370…−0.0213) | ||
| ωSI | 72.7% (9%) | 72.0% (66.1%…79.5%) | 47.5% (9%) | 46.8% (40.8%…52.2%) |
| θP2 | 0.0253 (6%) | 0.254 (0.0229…0.0278) | 0.0254 (5%) | 0.0254 (0.0233…0.0278) |
| θP2∼AGE | −0.0110 (17%) | −0.0110 (−0.0138…−0.00796) | ||
| θP2∼IBSL | −0.0150 (25%) | −0.0150 (−0.0208…−0.0101) | ||
| ωP2 | 50.7% (11%) | 50.4% (43.5%…58.0%) | 37.9% (13%) | 37.3% (30.1%…44.3%) |
| ρSG-VOL | −0.60 (16%) | −0.603 (20%) | −0.779 (15%) | −0.784 (16%) |
| ρSI-P2 | 0.909 (18%) | 0.909 (11%) | 0.876 (17%) | 0.877 (15%) |
| σprop | 2.27% (3%) | 2.26% (1.95%…2.56%) | 2.27% (3%) | 2.27% (1.96%…2.55%) |
| σadd | 4.27 (2%) | 4.27 (3.91…4.62) | 4.28 (2%) | 4.27 (3.91…4.65) |
Summary of parameter estimates obtained with nonlinear mixed-effects estimation. Values are reported as follows. Typical values for parameters (θ) are in original units. Given that between-subject variability is modeled as log normal, variance measures are reported as approximate coefficients of variation (CV), whereas the covariance terms (off-diagonal element of Ω) are in terms of correlation. Coefficients for the covariates are in logarithmic units; therefore, as a first approximation they can be interpreted as proportional changes in the minimal model parameter per unit change of the covariate. By way of explanation, SI = 5.83·10−5·exp(−0.0081·ΔAGE), where ΔAGE is the deviation of AGE from the mean AGE. This model can be approximated as SI ≈ 5.83·10−5·(1−0.0081·ΔAGE), which predicts an ∼0.8% drop in SI for every year of AGE above mean AGE. Residual Unknown Variability is reported as CV% for the proportional term and in glucose concentration units for the additive component. Precision of estimates (in brackets) is reported as CV% or as confidence interval for bootstrap results.
Fig. 3.
Individual parameters vs. significant covariates. Scatterplots with the logarithm of the parameter values obtained with the base model regressed vs. the respective covariates included in the final model.
Fig. 4.
Observations vs. population predictions. Scatterplot of the observations (DV) vs. population predictions (PRED) for the base model (left) and the final covariate model (right).
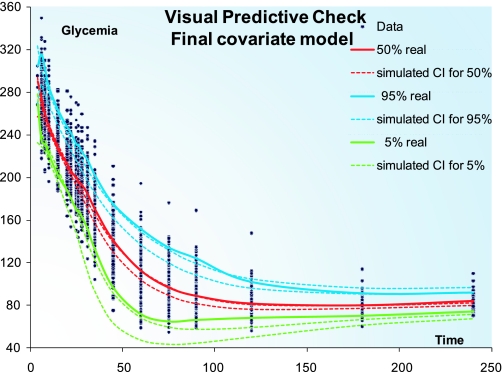
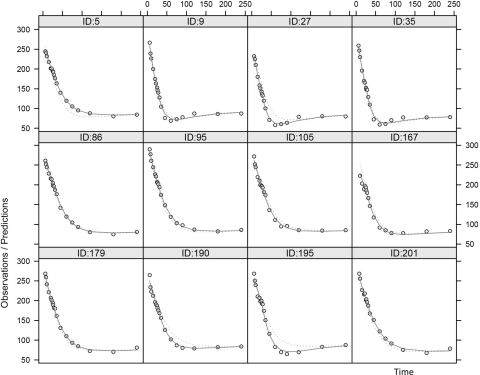
To assess the ability of the model to capture the variability observed in the dataset, a visual predictive check (VPC) (19) was performed with NONMEM and PsN. The final population parameters were used to simulate 3,000 replicates of the dataset, and the observations were stratified by percentiles for each time point. As can be seen in Fig. 5, the model-simulated and the real data superimpose satisfactorily, even though a slight overprediction of the median values for times 50–100 min can be detected. Some sample individual plots are also available in Fig. 6 and show the goodness of fit for the rich individual data provided by the model.
Fig. 5.
Visual predictive check. Visual predictive check (3,000 simulations) obtained with the final covariate model. The confidence interval (CI) for the 5, 50, and 95% percentiles for simulated (dashed lines) and real data (solid lines) are displayed.
Fig. 6.
Individual profiles. Individual predictions vs. time for some randomly selected subjects. ID, subject ID number. Dots represent observed concentrations; dashed line is the population prediction; solid line is the individual prediction.
The population covariance matrix (Ω) used in our analysis included, in addition to the parameter variance terms, also the correlations SI-P2 and SG-VOL, as previously proposed (13). Both these off-diagonal terms remained statistically significant throughout the covariate selection process; i.e., their estimation confidence intervals never included zero. The same can be said about the estimate of the RUV, which was not overly changed by the incorporation of the covariates. More in general, it can be pointed out that the precision of all the population parameters included in the BM (θSG, θVOL, θSI, θP2, and the Ω terms) did not change significantly with the introduction of the covariates.
DISCUSSION
First of all, it is interesting to discuss the methodology used for the covariate selection. The models suggested by the Ra2 analysis normally included more predictors than the ones selected by the population ML approach, but overall, the degree of accord between the two regressions (the exploratory one in R and the ML in NONMEM and SPK) has proved to be fairly satisfactory. Even if the multivariate linear regression did not single out the most parsimonious model, it still provided a very good guess, allowing dramatic reduction in duration and improving the efficiency of the covariate selection process. Specifically, not only was the number of NLME runs decreased with respect to a traditional stepwise approach, but models could also be evaluated in which two or more covariates were jointly included, a possibility that is normally not explored with a stepwise approach (36). With specific regard to the optimal model, it can be said that, even if the dataset comprised only healthy patients (no previous diagnosis of glucose metabolic disorders), a fairly large span of age and BMI was covered and some interesting results were obtained. The covariates included in the final model as predictors of SI have a robust physiological explanation, and they have been reported previously in the literature. It is widely known that insulin sensitivity decreases with aging, and high levels of basal insulin are normally used as a marker for insulin resistance and incipient impaired glucose tolerance. Already in a pioneering publication by Bergman et al. (Ref. 7 and Fig. 7 therein), an inverse relationship among basal insulin and the minimal model SI is detected and reported. Godsland et al. (18) have also detected a similar correlation, even though in this case the log-transformation was not employed. In addition, many studies in the literature have pointed out a correlation between VAF and insulin resistance (17, 27, 33). Basu and coworkers (2, 3) have already recognized the role of VAF when performing a regression analysis on part of the same dataset that we used here. Besides SI, a parallel model was selected for P2, a parameter measuring the inverse of the delay in insulin action with respect to plasma insulin; the behavior of the predictors AGE and IBSL in this model is similar. In addition, it should be mentioned that, although on the borderline of statistical significance, TAF also appeared as a good predictor for P2. It is not clear whether indeed the same physiological factors alter the two different phenomena (namely insulin sensitivity and insulin's activation latency) or if this is due to a limitation of the model, which may not be able to efficiently separate these two aspects of the insulin metabolic system. In any case, the term in the model expressing the correlation of the random effects of SI and P2 does not decrease in value or statistical significance with the introduction of the covariates, meaning that the remaining BSV of the two parameters is also highly correlated. All the relationships contained in the final model for SI and P2 are also graphically visible in the scatterplots, with the individual values from the base model displayed in Fig. 3. The model predicts for SI a drop of ∼0.8% per year of age above average age, ∼2.8% per pmol/ml of basal insulinemia above average, and ∼0.2% per square centimeter of VAF in a CT scan slice. Similar changes are predicted for P2, due to age (−1.1%) and basal insulin (−1.5%). Some of the relationships found for VOL are also evident in the scatterplots in Fig. 3, i.e., for %TBF and GBSL, and so is the difference in the values for males and females; however, it is worth noting that the relationship with AGE is not qualitatively detectable from the plots. This probably means that this correlation is apparent only after the individual volume has been corrected for SEX, %TBF, and GBSL. We have no knowledge of studies in the literature regarding regression analysis of the apparent volume of distribution for the glucose minimal model, so no comparison with other data is possible. However, it should be borne in mind that that the apparent volume of distribution provided by the minimal model parameterization, and therefore used for the regression analysis, is intended per kilogram of the subject's body mass. If this is taken into account, the negative correlation with %TBF might find an explanation in the fact that the volume can be considered proportional to the lean body mass of the subject, and therefore, subjects with a greater percentage of body fat have a relatively lower volume per kilogram of body mass (the model predicted an ∼1% drop in volume for each percentage increase in total body fat above the population mean). In formulae:
| (7) |
Slightly different comments can be made about SG. From the scatterplots, no noteworthy relationship is graphically detectable between log(SG) and any of the covariates, taken separately. However, a statistically significant correlation is detected when the sets BH-BW-BSA, or BW-BMI-BSA are used for the regression. It was chosen not to incorporate this component into the model, because the amount of BSV explained with the introduction of these covariates was negligible (less than 1% of a total BSV of 21%). Moreover, a very high condition number of the covariance matrix of the parameter estimates pointed out the computational issues linked to the strong collinearity of these predictors, a phenomenon previously reported by Bonate (8). Indeed, BMI and BSA are derived variables, calculated from BH and BW. In any case, it should also be borne in mind that SG is affected by model simplification from a two-compartmental into a one-compartment glucose kinetics (9, 10, 31), and this can have a confounding effect on this composite parameter that hinders the proper identification of physiologically plausible predictors.
Finally, some considerations can be made about the discrepancies detected in the VPC. These can likely be ascribed to the simulations being unable to capture all the components of observed variability, among which are the time course of insulin and the glucose baseline. Although the glucose parameters are sampled from the mixed-effects model, these structural components are left fixed. This unavoidable situation can potentially lead to a mismatch between the insulin time course and the glucose kinetic parameters and, thus, incorrect predictions. It is, however, reassuring that, by inspection of Fig. 4, the overall agreement of the VPC appears reasonable.
CONCLUSIONS
In this work, we propose a population model for the glucose minimal model, incorporating physiological information such as sex and age, directly measurable anthropometric data such as height and weight, body fat amount and distribution, and fasting levels of glucose and insulin. It is important to mention that many studies encompassing a correlation analysis of glucose-insulin model parameters are present in the literature, but we are not aware of any other study actually implementing a “dynamic” regression model integrating physiological variables and dynamic glucose-insulin measurements in model building such as the one we are proposing in this work. Our model does not simply verify correlations on already calculated parameter values; the regression coefficients are instead integrated in the model. Physiological information thus plays an active role in the parameter estimation process. The introduction of the covariates in the model, in fact, helps to explain a substantial fraction of the population variability for the parameters SI and P2. This is despite the fact that BSV is not considerable in the dataset under test, which is encompassing only healthy subjects, rather than individuals with different degrees of glucose intolerance disorders. The predictive power of the model has been considerably increased with the incorporation of independent physiological information; a significant portion of the variability among individuals is explained in a deterministic fashion, while only a smaller part involves a stochastic component, the random effects. This not only offers a starting point for speculation about the significance of the relationships detected, but also provides a tool to allow the design of less invasive and expensive protocols for epidemiological studies of the glucose disposal metabolic system. The advantages of a population approach to parameter estimates have been shown extensively in the literature. Although so far this has been employed mostly in drug development studies of pharmacokinetics and pharmacodynamics, the modeling of metabolic systems can greatly benefit from the use of these techniques. NLME modeling, in particular, allows not only a more accurate evaluation of the population features but also enhances the individual parameter accuracy and precision, in that the population information is used as a prior that supports the individual parameter estimation process. In addition, thanks to the introduction of the individual values of the significant covariates, this population prior is tailored to each one of the subjects, further improving the estimate precision. Our covariate model, being a first proposal, suffers from several limitations. The existence of more complex and possibly nonlinear relationships should be investigated, as our analysis was limited to linear models. Moreover, alternative parameterizations could be tested to investigate, for example, whether aggregating some parameters (e.g., using SG·VOL or SI·VOL) allows the unveiling of additional or stronger relationships. Finally, since the amount of variability in the data is limited, and therefore the possibility of statistical artifacts and model misspecification cannot be excluded, it would be important to validate and corroborate our results by repeating the analysis on different datasets, possibly encompassing subjects with a broader range of variation concerning glucose disposal.
GRANTS
This work was supported in part by the Fondazione Aldo Gini, Padua, Italy, under the “Borsa Gini” Scholarship (PD). The development of SPK was partially supported by NIH/NIBIB Grant P41 EB-001975. Parts of the results of this paper were presented at the 2009 PAGE meeting in St. Petersburg, Russia (PAGE 18, 2009, Abstract 1642).
DISCLOSURES
No conflicts of interest are reported by the author(s).
ACKNOWLEDGMENTS
We acknowledge Dr. Rita Basu and Dr. Robert Rizza from the Mayo Clinic, Rochester, MN, for providing the dataset used in the analysis.
Current Address of P. Denti: Division of Clinical Pharmacology, University of Cape Town, Cape Town, South Africa.
Current Address of P. Vicini: Pfizer Global Research and Development, San Diego, CA.
REFERENCES
- 1. Agbaje OF, Luzio SD, Albarrak AI, Lunn DJ, Owens DR, Hovorka R. Bayesian hierarchical approach to estimate insulin sensitivity by minimal model. Clin Sci (London) 105: 551–560, 2003. [DOI] [PubMed] [Google Scholar]
- 2. Basu R, Breda E, Oberg AL, Powell CC, Dalla Man C, Basu A, Vittone JL, Klee GG, Arora P, Jensen MD, Toffolo G, Cobelli C, Rizza RA. Mechanisms of the age-associated deterioration in glucose tolerance: contribution of alterations in insulin secretion, action, and clearance. Diabetes 52: 1738–48, 2003 [DOI] [PubMed] [Google Scholar]
- 3. Basu R, Dalla Man C, Campioni M, Basu A, Klee G, Toffolo G, Cobelli C, Rizza RA. Effects of age and sex on postprandial glucose metabolism: differences in glucose turnover, insulin secretion, insulin action, and hepatic insulin extraction. Diabetes 55: 2001–2014, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Beal S, Sheiner L, Boeckmann A. NONMEM Users Guides (1989–2006). Ellicott City, MD: ICON Development Solutions [Google Scholar]
- 5. Beal SL, Sheiner LB. Estimating population kinetics. Crit Rev Biomed Eng 8: 195–222, 1982 [PubMed] [Google Scholar]
- 6. Bergman RN, Ider YZ, Bowden CR, Cobelli C. Quantitative estimation of insulin sensitivity. Am J Physiol Endocrinol Metab Gastrointest Physiol 236: E667–E677, 1979 [DOI] [PubMed] [Google Scholar]
- 7. Bergman RN, Phillips LS, Cobelli C. Physiologic evaluation of factors controlling glucose tolerance in man: measurement of insulin sensitivity and beta-cell glucose sensitivity from the response to intravenous glucose. J Clin Invest 68: 1456–1467, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonate PL. The effect of collinearity on parameter estimates in nonlinear mixed effect models. Pharmaceut Res 16: 709–717, 1999 [DOI] [PubMed] [Google Scholar]
- 9. Caumo A, Vicini P, Cobelli C. Is the minimal model too minimal? Diabetologia 39: 997–1000, 1996 [DOI] [PubMed] [Google Scholar]
- 10. Caumo A, Vicini P, Zachwieja JJ, Avogaro A, Yarasheski K, Bier DM, Cobelli C. Undermodeling affects minimal model indexes: insights from a two-compartment model. Am J Physiol Endocrinol Metab 276: E1171–E1193, 1999 [DOI] [PubMed] [Google Scholar]
- 12. De Gaetano A, Mingrone G, Castageneto M. NONMEM improves group parameter estimation for the minimal model of glucose kinetics. Am J Physiol Endocrinol Metab 271: E932–E937, 1996 [DOI] [PubMed] [Google Scholar]
- 13. Denti P, Bertoldo A, Vicini P, Cobelli C. Nonlinear mixed effects to improve glucose minimal model parameter estimation: a simulation study in intensive and sparse sampling. IEEE Trans Biomed Eng (2009). doi:10.1109/TBME.2009.2020171 [DOI] [PubMed] [Google Scholar]
- 14. Efron B. Nonparametric estimates of standard error—the jackknife, the bootstrap and other methods. Biometrika 68: 589–599, 1981 [Google Scholar]
- 15. Erichsen L, Agbaje OF, Luzio SD, Owens DR, Hovorka R. Population and individual minimal modeling of the frequently sampled insulin-modified intravenous glucose tolerance test. Metab Clin Exper 53: 1349–1354, 2004 [DOI] [PubMed] [Google Scholar]
- 16. Finegood DT, Hramiak IM, Dupre J. A modified protocol for estimation of insulin sensitivity with the minimal model of glucose kinetics in patients with insulin-dependent diabetes. J Clin Endocrinol Metab 70: 1538–1449, 1990 [DOI] [PubMed] [Google Scholar]
- 17. Gastaldelli A, Miyazaki Y, Pettiti M, Matsuda M, Mahankali S, Santini E, DeFronzo RA, Ferrannini E. Metabolic effects of visceral fat accumulation in type 2 diabetes. J Clin Endocrinol Metab 87: 5098–5103, 2002 [DOI] [PubMed] [Google Scholar]
- 18. Godsland IF, Agbaje OF, Hovorka R. Evaluation of nonlinear regression approaches to estimation of insulin sensitivity by the minimal model with reference to Bayesian hierarchical analysis. Am J Physiol Endocrinol Metab 291: E167–E174, 2006 [DOI] [PubMed] [Google Scholar]
- 19. Holford NG. The visual predictive check superiority to standard diagnostic (Rorschach) plots. Abstracts of the Annual Meeting of the Population Approach Group in Europe. ISSN 1871-6032 PAGE 14, 2005, Abstr738, http://www.page-meeting.org/?abstract=738
- 20. Jensen MD, Kanaley JA, Reed JE, Sheedy PF. Measurement of abdominal and visceral fat with computed tomography and dual-energy x-ray absorptiometry. Am J Clin Nutr 61: 274–278, 1995 [DOI] [PubMed] [Google Scholar]
- 21. Jonsson EN, Karlsson MO. Xpose—an S-PLUS based population pharmacokinetic/pharmacodynamic model building aid for NONMEM. Computer Methods Programs Biomed 58: 51–64, 1999 [DOI] [PubMed] [Google Scholar]
- 22. Karlsson MO, Savic RM. Diagnosing model diagnostics. Clin Pharmacol Therapeut 82: 17–20, 2007 [DOI] [PubMed] [Google Scholar]
- 23. Krudys KM, Kahn SE, Vicini P. Population approaches to estimate minimal model indexes of insulin sensitivity and glucose effectiveness using full and reduced sampling schedules. Am J Physiol Endocrinol Metab 291: E716–E723, 2006 [DOI] [PubMed] [Google Scholar]
- 24. Lindbom L, Pihlgren P, Jonsson EN. PsN-Toolkit—a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Computer Methods Programs Biomed 79: 241–57, 2005 [DOI] [PubMed] [Google Scholar]
- 25. Lindbom L, Ribbing J, Jonsson EN. Perl-speaks-NONMEM (PsN)—a Perl module for NONMEM related programming. Computer Methods Programs Biomed 75: 85–94, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Lumley T. Leaps Package for R, v. 2.9 [Online]. http://cran.r-project.org/web/packages/leaps/index.html.
- 27. Miyazaki Y, Glass L, Triplitt C, Wajcberg E, Mandarino LJ, DeFronzo RA. Abdominal fat distribution and peripheral and hepatic insulin resistance in type 2 diabetes mellitus. Am J Physiol Endocrinol Metab 283: E1135–E1143, 2002 [DOI] [PubMed] [Google Scholar]
- 28. R Development Core Team R: A Language and Environment for Statistical Computing, v. 2.8 [Online]. R Foundation for Statistical Computing. 2008. http://www.r-project.org/
- 29. Steimer JL, Mallet A, Golmard JL, Boisvieux JF. Alternative approaches to estimation of population pharmacokinetic parameters: comparison with the nonlinear mixed-effect model. Drug Metab Rev 15: 265–292, 1984 [DOI] [PubMed] [Google Scholar]
- 30. The Team RFPK System for Population Kinetics (SPK). [Online]. Univ. of Washington, 2008. http://spk.rfpk.washington. [Google Scholar]
- 31. Vicini P, Caumo A, Cobelli C. Glucose effectiveness and insulin sensitivity from the minimal models: consequences of undermodeling assessed by Monte Carlo simulation. IEEE Trans Biomed Eng 46: 130–137, 1999 [DOI] [PubMed] [Google Scholar]
- 32. Vicini P, Cobelli C. The iterative two-stage population approach to IVGTT minimal modeling: improved precision with reduced sampling. Intravenous glucose tolerance test. Am J Physiol Endocrinol Metab 280: E179–E186, 2001 [DOI] [PubMed] [Google Scholar]
- 33. Wagenknecht LE, Langefeld CD, Scherzinger AL, Norris JM, Haffner SM, Saad MF, Bergman RN. Insulin sensitivity, insulin secretion, and abdominal fat: the Insulin Resistance Atherosclerosis Study (IRAS) Family Study. Diabetes 52: 2490–2496, 2003 [DOI] [PubMed] [Google Scholar]
- 34. Wilkins JJ. NONMEMory: a run management tool for NONMEM. Computer Methods Programs Biomed 78: 259–267, 2005 [DOI] [PubMed] [Google Scholar]
- 35. Wählby U, Jonsson EN, Karlsson MO. Assessment of actual significance levels for covariate effects in NONMEM. J Pharmacokinet Pharmacodynam 28: 231–252, 2001 [DOI] [PubMed] [Google Scholar]
- 36. Wählby U, Jonsson EN, Karlsson MO. Comparison of stepwise covariate model building strategies in population pharmacokinetic-pharmacodynamic analysis. AAPS Pharm Sci 4: E27, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]