Abstract
S-adenosylmethionine (SAM) minimizes alcohol hepatotoxicity; however, the molecular mechanisms responsible for SAM hepatoprotection remain unknown. Herein, we use proteomics to determine whether the hepatoprotective action of SAM against early-stage alcoholic liver disease is linked to alterations in the mitochondrial proteome. For this, male rats were fed control or ethanol-containing liquid diets ± SAM and liver mitochondria were prepared for proteomic analysis. Two-dimensional isoelectric focusing (2D IEF/SDS-PAGE) and blue native gel electrophoresis (BN-PAGE) were used to determine changes in matrix and oxidative phosphorylation (OxPhos) proteins, respectively. SAM coadministration minimized alcohol-dependent inflammation and preserved mitochondrial respiration. SAM supplementation preserved liver SAM levels in ethanol-fed rats; however, mitochondrial SAM levels were increased by ethanol and SAM treatments. With use of 2D IEF/SDS-PAGE, 30 proteins showed significant changes in abundance in response to ethanol, SAM, or both. Classes of proteins affected by ethanol and SAM treatments were chaperones, beta oxidation proteins, sulfur metabolism proteins, and dehydrogenase enzymes involved in methionine, glycine, and choline metabolism. BN-PAGE revealed novel changes in the levels of 19 OxPhos proteins in response to ethanol, SAM, or both. Ethanol- and SAM-dependent alterations in the proteome were not linked to corresponding changes in gene expression. In conclusion, ethanol and SAM treatment led to multiple changes in the liver mitochondrial proteome. The protective effects of SAM against alcohol toxicity are mediated, in part, through maintenance of proteins involved in key mitochondrial energy conserving and biosynthetic pathways. This study demonstrates that SAM may be a promising candidate for treatment of alcoholic liver disease.
Keywords: alcohol, liver steatosis, mitochondrial dysfunction, proteomics
alcoholism is the third leading cause of preventable death in the US, with alcoholic liver disease continuing to be a significant cause of morbidity and mortality. It is estimated that 12,000 deaths occur each year in the US from alcohol-related chronic liver disease and cirrhosis (41). Chronic alcohol consumption causes liver disease by a combination of pathological insults including inflammation, redox alterations, fibrogenesis, and mitochondrial damage. Damage to mitochondrial DNA (mtDNA), inhibition of mitochondrial protein synthesis, defects in the oxidative phosphorylation (OxPhos) system complexes, depressed ATP synthesis, and enhanced susceptibility of hepatocytes to hypoxic and apoptotic stimuli implicate the mitochondrion as a “central hub” in the pathobiology of alcoholic liver disease (35). Alcohol-dependent defects in mitochondrial function are proposed to contribute to the initiation of liver disease through increased oxidative/nitrative stress and bioenergetic deficiency leading to hepatocyte cell death, which is now considered to be a critical event responsible for the progression of steatosis to steatohepatitis (27).
Even though the events responsible for chronic alcohol-mediated mitochondrial dysfunction have been extensively studied, the impact of alcohol consumption on the global content of liver mitochondrial proteins, “the mitochondrial proteome,” has only recently been investigated. Early studies by Coleman and Cunningham (16) showed that alcohol consumption for 5–6 wk in rats decreased the synthesis of the 13 mitochondrial-encoded proteins that comprise OxPhos complexes I, III, IV, and V. Our laboratories have extended these studies to examine alcohol-dependent changes in nuclear encoded proteins that comprise the OxPhos complexes and other mitochondrial proteins (61). This is an important avenue of investigation because it is estimated that there are at least 600+ proteins that comprise the mitochondrial proteome (44). Furthermore, it is known that toxicant-mediated alterations to the mitochondrial proteome negatively impact organelle function (3, 64). Therefore, studies aimed at determining alcohol-mediated alterations to the mitochondrial proteome represent an appealing approach to interrogate molecular events responsible for alcohol toxicity and will aid in development of new therapies for prevention and treatment of alcoholic liver disease. Clearly, this last point is important because there are no FDA-approved therapies for the treatment of alcoholic liver disease.
Methionine metabolism is disrupted in alcohol- and other toxicant-related liver diseases, resulting in decreased hepatic levels of S-adenosylmethionine (SAM), the primary methyl donor for biological reactions and a key intermediate for both glutathione and polyamine synthesis (38). Depletion of hepatic SAM is associated with oxidative stress, mitochondrial glutathione (GSH) reduction, and DNA damage (40, 48). Consequently, SAM supplementation protects against alcohol hepatotoxicity in animal models (32) and improved outcomes in a clinical trial (39). Emerging evidence from several laboratories indicates that SAM hepatoprotection may be mediated through effects on the mitochondrion. Early studies reported that SAM supplementation during chronic alcohol exposure maintained mitochondrial GSH transport and levels (23). Mitochondria isolated from liver of mice lacking methionine adenosyltransferase 1, the rate-limiting enzyme for SAM synthesis, have decreased cytochrome c oxidase subunits and reduced membrane potential (54). More recently, Cahill and colleagues (59) have shown that SAM prevents alcohol-dependent defects in mitochondrial ribosomes with our laboratory showing (8) that SAM prevents alcohol-dependent increases in mitochondrial superoxide production and mtDNA damage. These findings are also supported by observations that mitochondrial SAM depletion enhances TNF-α-mediated toxicity (58).
Although accumulating evidence demonstrates a protective effect of SAM against alcohol-mediated mitochondrial dysfunction and hepatotoxicity, the underlying molecular mechanisms and targets responsible for these benefits are unknown. Herein, we propose that SAM preserves mitochondrial function during chronic alcohol feeding through impacts on the mitochondrial proteome. We tested this hypothesis by feeding rats an ethanol-containing diet ± SAM for 5 wk and examined mitochondrial bioenergetics and the proteome using complementary proteomics techniques to assess alcohol- and SAM-dependent changes in mitochondrial proteins.
MATERIALS AND METHODS
Chronic alcohol feeding protocol.
Male Sprague-Dawley rats (n = 6 animals per treatment group) were individually housed and maintained under a 12-h light-dark cycle. Nutritionally adequate Lieber-DeCarli control and ethanol-containing liquid diets (33) were formulated by Bio-Serv (Frenchtown, NJ). The ethanol diet contains 36% of total daily calories as ethanol, 35% as fat, 18% as protein, and 11% as carbohydrate. The control diet is identical except ethanol calories are substituted with dextrin maltose. Control animals were pair fed to ethanol counterparts so that each pair was isocaloric. A second set of animals were pair fed control and ethanol diets supplemented with SAM (0.8 mg active SAM/ml diet). Animals were maintained on these feeding protocols for at least 31 days before experiments. All animal protocols were approved by the institutional animal care and use committee, and animals received humane care in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (NIH Publication no. 86-23).
Liver histology and blood alcohol measurements.
Formalin-fixed liver tissue was processed for hematoxylin-eosin staining and evaluated for steatosis and inflammatory foci by a pathologist blinded to the experimental design (8). Blood alcohol levels were measured in serum by an alcohol dehydrogenase-linked spectrophotometric assay (Pointe Scientific, Canton, MI).
HPLC measurements.
Standards and samples were analyzed by an HPLC method modified from (17, 29). SAM and S-adenosylhomocysteine (SAH) standards (Sigma Chemical, St. Louis, MO) and SAM used in feeding studies (Orchid Chemicals and Pharmaceuticals, Tamil Nadu, India) were dissolved in cold 0.5 N perchloric acid and filtered through 0.22 μm filters into HPLC vials, and 10 μl was injected onto an HPLC system LC-2010CHT HPLC (Shimadzu Scientific Instruments, Columbia, MD). For liver and mitochondrial SAM and SAH measurements, 0.2 g of fresh liver and 44.4 mg/ml of fresh mitochondria were homogenized in 0.5 N perchloric acid, respectively. The acid extracts were cleared of precipitated proteins by centrifugation and HPLC was performed as described. The system was equipped with an autosampler temperature control set to 4°C using a reverse-phase column [Luna C18(2), 150 × 4.60 mm, Phenomenex, Torrance, CA] provided with a Phenomenex guard column (ODS, 4 mm length and 3 mm ID). The mobile phase was isocratic, was run at a flow rate of 1.0 ml/min, and contained 50 mM NaClO4, 0.1 M NaCH3COO−, 2.4 mM CH3(CH2)6SO3Na·H2O, and 4.2% (vol/vol) acetonitrile, pH 3.5. As shown in the online supplemental data section, the SAM used for these feeding studies was pure and contained no contaminating levels of SAH.
Liver mitochondria isolation.
Mitochondria were isolated by standard differential centrifugation techniques (7). Mitochondria function was assessed by measuring state 3 and 4 respiration rates and calculating the respiratory control ratio (RCR; = state 3/state 4) using glutamate/malate as substrate. Mitochondrial protein yield was unaffected by ethanol and/or SAM treatments (8).
2D IEF/SDS-PAGE.
Mitochondrial protein (100 μg) was added to isoelectric focusing (IEF) gel strip rehydration buffer containing 7 M urea, 2 M thiourea, 2% (wt/vol) CHAPS, 0.5% (wt/vol) n-dodecyl-β-d-maltoside, 0.002% (wt/vol) bromophenol blue, ampholine electrophoresis reagent (Sigma, St. Louis, MO; range pH 3–10), 0.04 M DTT, and 2 mM tributylphosphine. Following protein extraction, samples were applied to IEF gel strips (Invitrogen ZOOM Strips, pH 3–10, Carlsbad, CA) and rehydration of IEF strips was done overnight. For SDS-PAGE, IEF gel strips were placed horizontally on top of a 10% resolving gel with 4% stacking gel, and sealed into place using warm agarose (1%, wt/vol) and gels were run at 100 V for 1 h. After electrophoresis, gels were stained with Sypro Ruby (Invitrogen, Carlsbad, CA) for total protein. Protein stained gels were imaged via a Bio-Rad Fluor-S Imager (Bio-Rad, Hercules, CA).
2D gel image analyses.
Gels were analyzed for differences in protein density by using PDQuest Image Analysis software (Bio-Rad) as described in Refs. 61 and 62. For image analysis of 2D gels, individual protein spots on each gel were identified by the software program and manually verified to generate a match set of gels for control, ethanol, control + SAM-, and ethanol + SAM-treatment gels. The protein spot density was compared across all gels and a reference gel was selected, which served as the master gel image. This reference gel contained the highest abundance of detected protein spots. By use of a built-in algorithm, automatic matching of protein spots in each gel to the corresponding protein spots in the master gel was performed and then manually verified to correct for any proteins that may have been incorrectly matched to proteins in the reference gel. To correct for intergel protein loading differences, the density data for all protein spots in each gel was normalized to the total density in valid protein spots for that particular gel. Statistical analysis was performed as described in Refs. 3 and 61.
2D BN-PAGE.
Changes in the levels of OxPhos system proteins were assessed by 2D blue native polyacrylamide gel electrophoresis (BN-PAGE) (2, 61). In this specialized gel electrophoresis technique, the five OxPhos complexes are first separated intact in the first dimension (i.e., 1D) under nondenaturing (native) conditions. The intact 1D gel “strip” containing all five complexes is then transferred to a denaturing gel to separate the complexes into their individual polypeptides components resulting in a two-dimensional (i.e., 2D) map (see Ref. 2 for a detailed description). OxPhos complexes were extracted from 1 mg of mitochondrial protein using 10% (wt/vol) n-dodecyl-β-d-maltoside in 0.75 M aminocaproic acid, 50 mM Bis-Tris, pH 7.0. Protein extracts were mixed with 6.3 μl 5% (wt/vol) suspension of Coomassie brilliant blue G-250 in 0.5 M aminocaproic acid before they were applied to 5–12% nondenaturing gradient gels. After native gel electrophoresis, the vertical gel lane containing all complexes was cut from the gel and laid on top of a 10% denaturing Tris/Tricine/SDS-PAGE gel to resolve the individual polypeptides of the OxPhos complexes. Gels were stained with Coomassie blue, and images were obtained using Bio-Rad Fluor-S Imager (Bio-Rad). For analysis, the gel images were loaded into Scion Image (Scion, Frederick, MD), the individual protein subunits were “boxed,” densitometry was obtained for each individual protein, and average densities were calculated as described in Refs. 8 and 61.
Protein identification with MALDI-TOF mass spectrometry.
Proteins were excised from gels and were processed via standard methods in the University of Alabama at Birmingham (UAB) mass spectrometry shared facility (http://www.uab.edu/proteomics). For matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) analysis a focused laser beam is used to “evaporate” peptides from a solid matrix into a gas phase as ions. The resulting ions are injected into a tube, accelerated, and allowed to drift toward the detector. Their time of flight is then measured and is proportional to their molecular weight. Samples were destained with three successive 30-min washes in 50% 50 mM NH4HCO3/50% acetonitrile solution. Samples were treated with 10 mM DTT in 50 mM NH4HCO3 for 60 min at 60°C to reduce cysteine residues, which was followed by alkylation of free cysteines with 55 mM iodoacetamide in 50 mM NH4HCO3 for 60 min at room temperature. This was followed by 16-h incubation at 37°C with trypsin (12.5 ng/μl, Promega Gold Trypsin) to digest proteins. The resulting peptide solution was extracted by two successive 30-min washes in a 50/50 solution of 5% formic acid and acetonitrile; supernatants were then collected and dried by using a Savant SpeedVac. Dried peptide samples were resuspended in 0.1% formic acid, desalted (C18 ZipTips, Millipore), and diluted 1:10 with a saturated solution of α-cyano-4-hydroxycinnamic acid matrix before application to MALDI-TOF target plates. After plating, samples were dried and then analyzed with a Voyager De-Pro mass spectrometer in the positive mode. Spectra were analyzed with Voyager Explorer software, and peptide masses were submitted to the MASCOT database (see www.matrixscience.com) for protein identification.
Gene expression.
Total RNA was isolated from liver tissue by using TRIzol (Invitrogen) according to the manufacturer's directions. Reverse transcription of 1 μg of total RNA was performed by using RT2 First Strand Kit (SABiosciences). Real-time PCR was performed by use of an Applied Biosystems 7300 with verified, gene-specific primers purchased from SABiosciences (RT2 qPCR SYBR green-based primers). Relative expression changes were determined by normalizing the relative amount of gene-specific mRNA CT to the Gapdh (housekeeping gene) CT using the comparative cycle threshold (ΔΔCT) method. The following rat-specific RT2 quantitative PCR primer assays were used: heat shock 70-kDa protein 5 (Hspa5, PPR45224A), dimethylglycine dehydrogenase (Dmgdh, PPR43174A), hydroxylacyl-CoA dehydrogenase (Hadh, PPR53155A), ATP synthase subunit d (ATP5h, PPR42259A), and NADH dehydrogenase 1 α subcomplex 8 (Ndufa8, PPR42413A).
Statistical analysis.
Two-way ANOVA was performed with KaleidaGraph software version 4.0 (Synergy Software, Reading, PA). Student's t-tests were performed with Microsoft Excel statistical analysis, with statistical significance set at P < 0.05. The Mann-Whitney rank sum test was used for pathology analyses. Blood alcohol levels, body weights, liver weights, and histopathology were determined with n = 6 animals per treatment group. Six animals per group were used for two-dimensional isoelectric focusing/SDS-PAGE (2D IEF/SDS-PAGE) proteomics, gene expression experiments, and measurement of SAM and SAH levels. Four animals per group were used for BN-PAGE proteomics. Proteins identified from 2D IEF SDS-PAGE gels were classified according to the Universal Protein Resource website, www.uniprot.org, which is maintained by the UniProt Consortium (European Bioinformatics Institute, Swiss Institute of Bioinformatics and Protein Information Resource).
RESULTS
Effect of ethanol and SAM histopathology.
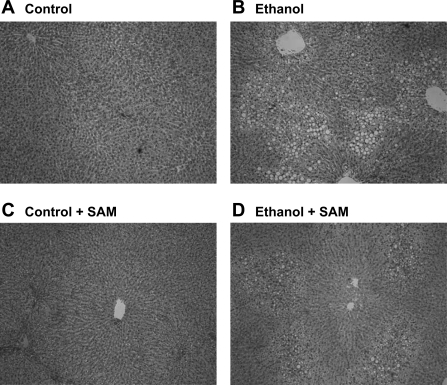
Addition of SAM to the ethanol-containing diet had no effect on diet consumption (data not shown) or blood alcohol concentrations (ethanol, 149 ± 28 and ethanol + SAM, 150 ± 25 mg/dl, P = 0.98). Similarly, there was no difference in body weight among the four groups at the end of the feeding protocol (control, 313 ± 8; ethanol, 304 ± 9; control + SAM, 311 ± 7; ethanol + SAM, 316 ± 7 g, P = 0.73). Ethanol consumption resulted in mild to moderate fat accumulation with ∼20% of hepatocytes being steatotic. In comparison, there was a modest, but not statistically significant, decrease in steatosis in the ethanol + SAM group with only 10–12% of hepatocytes containing fat (P = 0.21) (Fig. 1). Liver weights were elevated in ethanol groups with SAM treatment having no effect (control, 10.4 ± 0.6; ethanol, 12.4 ± 0.7; control + SAM, 9.7 ± 0.5; ethanol + SAM, 12.5 ± 0.5 g, P < 0.001, ethanol effect). Ethanol induced an inflammatory response with 5 of 12 ethanol livers exhibiting at least 1 inflammatory foci per ×10 objective whereas inflammation was absent in all livers in the ethanol + SAM group (P = 0.016).
Fig. 1.
S-adenosylmethionine (SAM) reduces ethanol-induced hepatic steatosis. Representative photomicrographs of rats maintained on control (A), ethanol (B), control + SAM (C), and ethanol + SAM (D) diets. Note that the inclusion of SAM in the ethanol diet (D) reduced the extent of steatosis compared with the ethanol-alone group (B); however, this difference was not statistically significant (P = 0.21). Representative photomicrographs are shown for n = 6 pairs of animals per group.
Effect of ethanol and SAM on levels of tissue and mitochondrial SAM and SAH.
As shown in Table 1, ethanol feeding alone decreased SAM, increased SAH, and decreased the SAM/SAH ratio in liver tissue. Importantly, SAM supplementation prevented the ethanol-dependent decrease in liver SAM (ethanol vs. ethanol + SAM, P = 0.036, t-test), which confirms our previous results (8). SAM supplementation had no effect to normalize liver SAH levels and the SAM/SAH ratio. In contrast to liver measurements, mitochondrial levels of SAM were significantly elevated by chronic ethanol consumption. There was a significant effect of SAM supplementation to increase mitochondrial SAM, SAH, and the SAM/SAH ratio in both control and ethanol-fed groups.
Table 1.
Effect of chronic ethanol consumption and SAM on liver SAM, SAH, and SAM/SAH ratio
| Tissue/Treatment | SAM | SAH | SAM/SAH ratio |
|---|---|---|---|
| Liver* | |||
| Control | 2.09 ± 0.17 | 0.54 ± 0.04 | 3.9 ± 0.22 |
| Ethanol | 1.62 ± 0.13 | 0.73 ± 0.06 | 2.2 ± 0.05 |
| Control + SAM | 2.09 ± 0.10 | 0.58 ± 0.05 | 3.8 ± 0.42 |
| Ethanol + SAM | 1.95 ± 0.09 | 0.88 ± 0.08 | 2.3 ± 0.22 |
| ANOVA (P values)† | |||
| Ethanol effect | 0.029 | 0.0007 | <0.0001 |
| SAM effect | 0.19 | 0.14 | 0.94 |
| Interaction (Ethanol × SAM) | 0.22 | 0.37 | 0.69 |
| Mitochondria‡ | |||
| Control | 0.037 ± 0.002 | 0.010 ± 0.0003 | 3.6 ± 0.17 |
| Ethanol | 0.057 ± 0.005 | 0.014 ± 0.001 | 4.1 ± 0.21 |
| Control + SAM | 0.054 ± 0.010 | 0.009 ± 0.001 | 5.7 ± 0.67 |
| Ethanol + SAM | 0.082 ± 0.004 | 0.013 ± 0.001 | 6.5 ± 0.46 |
| ANOVA (P values)§ | |||
| Ethanol effect | 0.0008 | 0.0027 | 0.138 |
| SAM effect | 0.0022 | 0.31 | <0.0001 |
| Interaction (Ethanol × SAM) | 0.542 | 0.98 | 0.81 |
Data are represented as means ± SE for n = 5 animals per treatment group.
Liver concentrations of S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) are expressed as nmol·liver−1·g body wt−1 (8).
,§Results from 2-factor ANOVA with ethanol effect (control vs. ethanol), SAM effect (absent vs. present), and interaction (Ethanol × SAM). †Liver values and
mitochondria values.
Mitochondrial concentrations of SAM and SAH are expressed as nmol/mg protein.
Effect of ethanol and SAM on mitochondrial respiration.
Chronic alcohol consumption significantly decreased state 3 respiration (ADP-dependent) and the RCR compared with controls (Table 2). In contrast, state 3 respiration and the RCR were preserved in the ethanol + SAM group and did not differ significantly from corresponding controls.
Table 2.
Effect of chronic ethanol consumption and SAM on mitochondrial respiration in liver
| Treatment | State 3* | State 4 | RCR† |
|---|---|---|---|
| Control | 0.085 ± 0.003 | 0.018 ± 0.001 | 4.80 ± 0.15 |
| Ethanol | 0.057 ± 0.003 | 0.019 ± 0.001 | 3.14 ± 0.08 |
| Control + SAM | 0.125 ± 0.004 | 0.024 ± 0.001 | 5.37 ± 0.15 |
| Ethanol + SAM | 0.111 ± 0.007 | 0.024 ± 0.002 | 4.56 ± 0.11 |
| ANOVA (P values)‡ | |||
| Ethanol effect | <0.0001 | 0.682 | <0.0001 |
| SAM effect | <0.0001 | <0.0001 | <0.0001 |
| Interaction (Ethanol × SAM) | 0.128 | 0.994 | 0.00114 |
Mitochondrial respiration was measured by use of the complex I-linked substrate glutamate/malate. Data are represented as means ± SE for n = 8 animals per treatment group.
Oxygen utilization for state 3 and 4 respiration expressed as μg atom oxygen consumed·min−1·mg protein−1.
Respiratory control ratio (RCR) = state 3 respiration/state 4 respiration.
Results from 2-factor ANOVA with ethanol effect (control vs. ethanol), SAM effect (absent vs. present), and interaction (ethanol × SAM).
Effect of ethanol and SAM on the mitochondrial proteome.
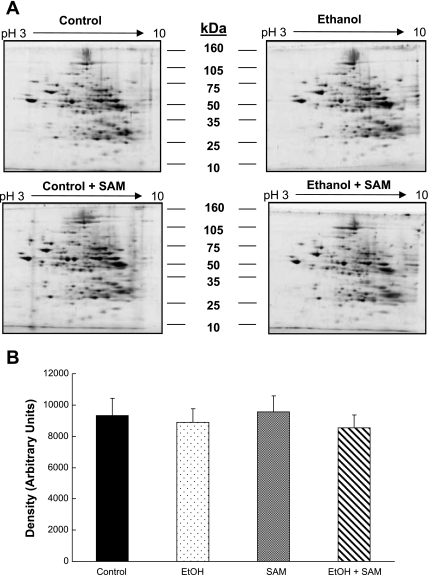
Representative 2D IEF/SDS-PAGE gels from each treatment group are shown in Fig. 2A. Densitometry revealed that there were no significant changes in total (i.e., global) mitochondrial protein content across the four treatment groups indicating equal loading and high reproducibility of gels (Fig. 2B). This result is important because although changes in individual proteins were observed and will be discussed, a lack of change in total protein density among groups demonstrates that there is no massive alteration in global mitochondrial protein content or protein pattern due to ethanol, SAM, or both. It should also be emphasized that mitochondrial preparations used for these proteomic studies were functionally validated, i.e., had high respiratory capacity and were coupled even in the ethanol groups (Table 2), which as suggested by Ping and colleagues (66) is an absolute criteria for valid proteomic analyses of mitochondria.
Fig. 2.
Representative 2D gel images of liver mitochondrial proteins. A: mitochondrial protein from livers of control, ethanol, control + SAM-, and ethanol + SAM-treated groups were separated by use of pH 3–10 isoelectric focusing (IEF) gel strips and 10% SDS-PAGE gels. B demonstrates that there were no significant differences in total protein density on gels prepared from the 4 treatment groups. Data represent the means ± SE, n = 6 animals per each group. 2-factor ANOVA; ethanol P = 0.44, SAM P = 0.96, and interaction (ethanol × SAM) P = 0.75.
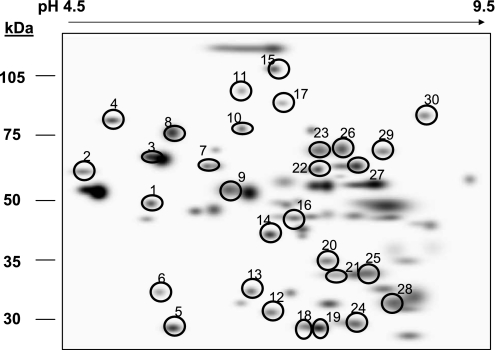
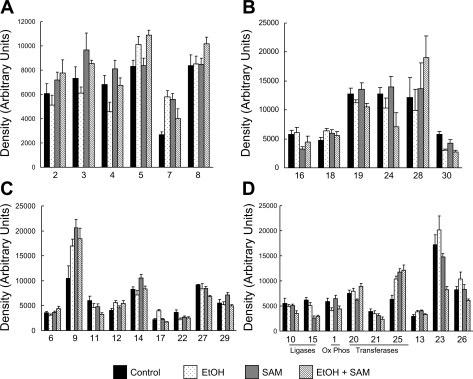
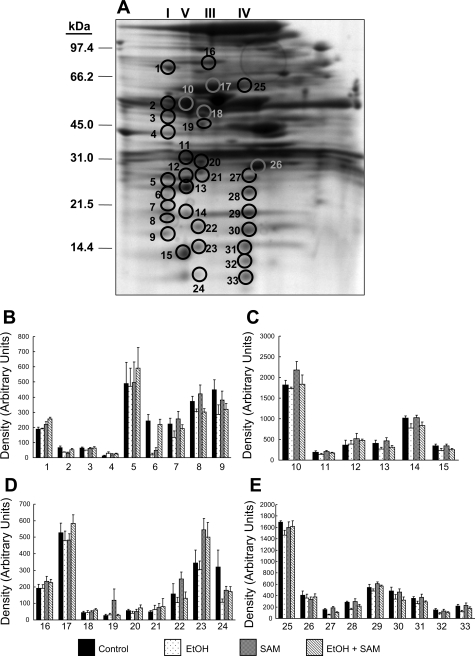
Of the 150–175 proteins detected, 76 proteins were found in common across all gels from the four treatment groups. A master map of these 76 proteins is provided in the supplemental data provided online. Of these 76 proteins, 30 were found to be significantly altered in abundance in response to ethanol, SAM, or both. A master map of these specific 30 proteins is provided in Fig. 3 with a listing of their unique proteins IDs and statistical analysis results provided in Table 3. Moreover, the bar graphs in Fig. 4 illustrate the abundances (i.e., densities) for each of the 30 proteins shown in Fig. 3 that were significantly changed by ethanol, SAM, or both. To simplify presentation and discussion, we have loosely grouped these 30 proteins into the following broad functional groups in Fig. 4: 1) chaperones in A; 2) fatty acid metabolism enzymes in B; 3) oxidoreductases in C; and 4) miscellaneous proteins in D. Note that the numerical labels used on the x-axis to identify the proteins in the bar graphs (Fig. 4) correspond to the protein ID numbers used in Fig. 3 (master map) and Table 3. Thus for the following sections please refer to Fig. 3, Fig. 4, and Table 3 for proteins ID numbers studied by 2D IEF/SDS-PAGE. In contrast, the OxPhos proteins identified by BN-PAGE are referenced within Fig. 6 and Table 4. A brief overview of the results from the proteomics analyses are provided in results. The potential physiological or functional significance of ethanol- and SAM-mediated alterations for select proteins are presented separately in the discussion.
Fig. 3.
Master map of mitochondrial proteins differentially altered by ethanol and SAM. Analysis of gels shown in Fig. 2 revealed 30 proteins differentially altered in abundance in response to ethanol and SAM treatment alone or in combination (circled proteins). The mass spectrometry identification and statistical analyses for these proteins is presented in Table 3. Please note that a separate and different numbering system is used for the global proteome map and table provided in the supplemental data section.
Table 3.
Mitochondrial proteins shown in Fig. 3 that are differentially altered in abundance by chronic ethanol and/or SAM treatments: results from 2D IEF/SDS-PAGE
| Spot No.* | Protein | Mass, kDa | Ethanol Effect† | SAM Effect† | Interaction (E × SAM)† | C vs. E‡ | C vs. SAM‡ | SAM vs. E+SAM‡ | E vs. E+SAM‡ |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Ubiquinol-cyt c reductase core protein I | 52.8 | P = 0.004 | ||||||
| 2 | Protein disulfide isomerase/ T3 binding protein | 54.0 | P = 0.04 | ||||||
| 3 | Hspd1 protein | 59.4 | P = 0.02 | P = 0.004 | |||||
| 4 | Heat shock 70 kDa protein 5 (GRP78) | 72.3 | P = 0.02 | P = 0.03 | P = 0.002 | ||||
| 5 | Prohibitin | 29.8 | P = 0.001 | P = 0.03 | |||||
| 6 | Pyruvate deHase β subunit | 39.0 | P = 0.04 | P = 0.07 | P = 0.03 | ||||
| 7 | Protein disulfide isomerase A3 | 56.6 | P = 0.0006 | P = 0.0007 | |||||
| 8 | Stress-70 protein, mitochondrial (GRP75) | 73.8 | P = 0.05 | ||||||
| 9 | Mitochondrial aldehyde deHase | 53.3 | P = 0.009 | P = 0.04 | P = 0.01 | ||||
| 10 | Propionyl-CoA carboxylase α | 79.8 | P = 0.05 | ||||||
| 11 | Sarcosine deHase | 101.4 | P = 0.04 | ||||||
| 12 | 3-Hydroxyisobutyrate deHase | 35.3 | P = 0.01 | P = 0.01 | P = 0.03 | ||||
| 13 | Agmatinase | 37.8 | P = 0.01 | P = 0.03 | P = 0.05 | ||||
| 14 | Isovaleryl CoA deHase | 46.4 | P = 0.03 | P = 0.03 | P = 0.04 | ||||
| 15 | Pyruvate carboxylase | 129.7 | P < 0.0001 | P = 0.001 | P = 0.007 | ||||
| 16 | Acetyl CoA deHase, long chain | 47.8 | P = 0.02 | P = 0.02 | |||||
| 17 | Dimethylglycine deHase | 96.0 | P = 0.02 | P = 0.0001 | P < 0.0001 | P = 0.00006 | |||
| 18 | Enoyl-CoA hydratase, hexadienoyl-CoA complex | 28.2 | P = 0.002 | ||||||
| 19 | Enoyl-CoA hydratase, hexadienoyl-CoA complex | 28.2 | P = 0.02 | ||||||
| 20 | Ornithine carbamoyltransferase | 39.9 | P = 0.01 | P = 0.05 | |||||
| 21 | Thiosulfate sulfurtransferase | 33.4 | P = 0.05 | ||||||
| 22 | Dihydrolipoamide deHase | 54.0 | P = 0.04 | ||||||
| 23 | Catalase | 59.7 | P = 0.0008 | P = 0.02 | P = 0.003 | ||||
| 24 | Electron transfer flavoprotein β | 27.7 | P = 0.02 | ||||||
| 25 | Thiosulfate sulfurtransferase | 33.4 | P = 0.01 | P = 0.0004 | P = 0.03 | P = 0.001 | |||
| 26 | Catalase | 59.7 | P = 0.04 | P = 0.04 | P = 0.02 | ||||
| 27 | ALDH6a1 | 57.8 | P = 0.05 | ||||||
| 28 | L-3-Hydroxyacyl-CoA deHase | 34.4 | P = 0.03 | ||||||
| 29 | Choline deHase | 66.2 | P = 0.03 | P = 0.004 | |||||
| 30 | Hadh (trifunctional protein) | 82.6 | P = 0.0003 |
Mitochondrial proteins listed above were matched on all gels from the control (C), ethanol (E), control + SAM (SAM), and ethanol + SAM (E + SAM) groups.
IEF, isoelectric focusing.
Spot no. is the same as those used to identify circled proteins in Fig. 3.
2-factor ANOVA P value.
Student's t-test P value. MOWSE scores and Accession numbers are listed in table provided in supplemental data online.
Fig. 4.
Quantification of mitochondrial proteins differentially altered by ethanol and SAM. This series of bar graphs illustrates the change in abundance (increase or decrease) of mitochondrial proteins found to be altered by ethanol, SAM, or both. The numbers below each set of bars correspond to the protein spot number given in Table 3 and the master map shown in Fig. 3. For ease in graphical presentation, proteins were separated into 4 broad functional classifications: proteins associated with chaperone functions (A), fatty acid metabolism proteins (B), oxidoreductase (dehydrogenase) proteins of key metabolism pathways (C), and miscellaneous ligases, transferases, and other proteins (D). Statistical analysis was performed as stated in materials and methods, and P values are listed in Table 3. Data represent means ± SE, n = 6 animals per group.
Fig. 6.
BN-PAGE master map of oxidative phosphorylation system proteins. A: BN-PAGE master protein map of the oxidative phosphorylation (OxPhos) proteins that were found in common in all gels across the 4 treatment groups. The circled proteins and adjacent number labels correspond to those proteins listed in Table 4. B: relative abundances of those proteins (1–9) present in the first lane (I), which corresponds to complex I and associated proteins. C: relative abundances of those proteins (10–15) present in the second lane (V), which corresponds to complex V and associated proteins. D: relative abundances of those proteins (16–24) present in the third lane (III), which corresponds to complex III and associated proteins. E shows the relative abundances of those proteins (25–33) present in the fourth lane (IV), which corresponds to complex IV and associated proteins. Numbers listed under each correspond to circled proteins located on the master map (A) and in Table 4. Data represent the means ± SE, n = 4 animals per group. The mass spectrometry identification and statistical analyses for these proteins are presented in Table 4. Note that not all OxPhos proteins were identified by this “miniaturized” high-throughput version of BN-PAGE gel electrophoresis.
Table 4.
Oxidative phosphorylation and associated proteins shown in Fig. 6 that are differentially altered in abundance by chronic ethanol and/or SAM treatments: results from BN-PAGE
| Spot No.* | Protein | Mass, kDa | Ethanol Effect† | SAM Effect† | Interaction (E×SAM)† | C vs. E‡ | C vs. SAM‡ | SAM vs. E+SAM‡ | E vs. E+SAM‡ |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NADH dehydrogenase Fe-S Protein 1 | 79.7 | P = 0.003 | P = 0.04 | P = 0.002 | ||||
| 2 | NADH dehydrogenase flavoprotein 1 | 50.8 | P = 0.01 | P = 0.05 | |||||
| 3 | NADH dehydrogenase Fe-S protein 2 | 53.3 | P = 0.04 | P = 0.05 | |||||
| 4 | NADH dehydrogenase 1α sub- complex 10 | 40.6 | |||||||
| 5 | NADH dehydrogenase flavoprotein 2 | 27.3 | |||||||
| 6 | NADH dehydrogeanse 1 α sub-complex 8 | 20.0 | P = 0.0002 | P = 0.05 | P = 0.01 | P = 0.005 | |||
| 7 | NADH dehydrogenase 1 α sub-complex subunit 12 | 17.1 | P = 0.005 | ||||||
| 8 | NADH dehydrogenase 1 β sub-complex 4 | 15.1 | P = 0.04 | ||||||
| 9 | NADH dehydrogenase Fe-S protein 6 | 13.0 | P = 0.04 | ||||||
| 10 | ATP synthase, H+ transporting F1 complex α subunit | 59.7 | |||||||
| ATP synthase, H+ transporting F1 complex β subunit | 48.1 | ||||||||
| 11 | ATP synthase, γ chain | 32.9 | P = 0.05 | ||||||
| 12 | ATP synthase, H+ transporting F0 complex subunit b isoform 1 | 28.9 | |||||||
| 13 | ATP synthase, H+ transporting F0 complex subunit d | 18.7 | P = 0.03 | P = 0.04 | |||||
| 14 | Cytochrome c oxidase, subunit IV isoform1 | 19.5 | P = 0.02 | P = 0.05 | |||||
| 15 | ATP synthase, H+ transporting F0 complex subunit g | 11.4 | P = 0.04 | P = 0.009 | |||||
| 16 | Mitochondrial trifunctional protein α subunit | 82.6 | |||||||
| 17 | 60 kDa heat shock protein, mitochondrial | 61.0 | |||||||
| 18 | Ubiquinol-cytochrome c reductase, core protein 1 | 52.7 | |||||||
| 19 | Ubiquinol-cytochrome c reductase, core protein 2 | 48.2 | |||||||
| 20 | Cytochrome c1 | 29.5 | |||||||
| 21 | Ubiquinol-cytochrome c reductase, Rieske Fe-S polypeptide 1 | 29.3 | |||||||
| 22 | Ubiquinol-cytochrome c reductase, subunit 7 | 13.5 | P = 0.05 | ||||||
| 23 | Ubiquinol-cytochrome c reductase, subunit 8 | 9.8 | P = 0.02 | P = 0.05 | P = 0.05 | ||||
| 24 | Ubiquinol-cytochrome c reductase, subunit 9 | 7.4 | |||||||
| 25 | 2-hydroxyacyl-CoA lyase | 63.6 | |||||||
| 26 | Electron transferring flavoprotein β | 27.6 | |||||||
| 27 | Cytochrome c oxidase, subunit II | 25.8 | P = 0.001 | P = 0.004 | P = 0.02 | P = 0.04 | |||
| 28 | Unidentified§ | P = 0.008 | P = 0.002 | ||||||
| 29 | Cytochrome c oxidase, subunit IV isoform 1 | 19.5 | P = 0.04 | ||||||
| 30 | Cytochrome c oxidase, subunit Va | 16.0 | P = 0.05 | ||||||
| 31 | Cytochrome c oxidase, subunit Vla | 12.3 | P = 0.03 | P = 0.02 | |||||
| 32 | Cytochrome c oxidase, subunit VIb polypeptide 1 | 10.1 | P = 0.009 | ||||||
| 33 | Unidentified‡ | 6.0 | P = 0.05 | P = 0.007 |
Oxidative phosphorylation and associated proteins listed above were matched on all 2D blue native gel electrophoresis (BN-PAGE) gels from the control, ethanol, control + SAM, and ethanol + SAM groups.
Spot no. is the same as those used to identify protein spots in Fig. 6.
Two-factor ANOVA P value.
Student's t-test P value.
Unable to identify probable cytochrome c oxidase subunits. Note that not all proteins listed in this table were altered in abundance by ethanol, SAM, or in combination but are included for protein identification. Molecular weight search (MOWSE) scores and accession numbers are listed in the table provided in supplemental data online.
Chaperone proteins from 2D IEF gels.
Figure 4A shows ethanol- and SAM-dependent alterations in those proteins classified as having chaperone functions. These include protein disulfide isomerase/thyroid hormone (T3) binding protein (PDI/T3BP, no. 2), Hspd1 protein (no. 3), heat shock 70-kDa protein 5 (GRP78, no. 4), prohibitin (no. 5), protein disulfide isomerase A3 (PDIA3, no. 7), and mitochondrial stress protein 70 (GRP75, no. 8). Chronic alcohol consumption decreased the levels of PDI/T3BP, Hspd1, and GRP78, whereas prohibitin and PDIA3 were increased by ethanol. Coadministration of SAM prevented the alcohol-dependent decrease in GRP78, Hspd1, and PDI/T3BP.
Fatty acid metabolism protein from 2D IEF gels.
As illustrated in Fig. 4B, there are differential changes in fatty acid metabolism proteins in response to ethanol and SAM. The proteins identified as being altered are long-chain acetyl CoA dehydrogenase (LCAD, no. 16), the enoyl-CoA hydratase and hexadienoyl-CoA complex (nos. 18 and 19), electron transfer flavoprotein β subunit (ETFβ, no. 24), l-3-hydroxyacyl CoA dehydrogenase (LCHAD, no. 28), and the Hadh protein (no. 30). Long-chain l-3-hydroxyacyl CoA dehydrogenase (LCHAD, no. 28), which catalyzes the third step in β-oxidation by converting l-3-hydroxyacyl CoA to 3-ketoacyl CoA (50), was decreased in the ethanol group. Similarly, we observed an ethanol-dependent decrease in the trifunctional protein (Hadh, no. 30) that catalyzes the concerted hydratase, dehydrogenase, and thiolase steps of β-oxidation.
Miscellaneous oxidoreductase proteins from 2D IEF gels.
The ability of ethanol and SAM to modulate key mitochondrial pathways is further supported by effects on the following diverse group of proteins classified as having oxidoreductase (i.e., dehydrogenase) activities (Fig. 4C). These proteins include pyruvate dehydrogenase (PDH) E1 subunit beta (no. 6), mitochondrial aldehyde dehydrogenase (ALDH, no. 9), sarcosine dehydrogenase (SARDH, no. 11), 3-hydroxyisobutyrate dehydrogenase (HIBDH, no. 12), isovaleryl CoA dehydrogenase (IVDH, no. 14), dimethylgylcine dehydrogenase (DMGDH, no. 17), dihydrolipoamide dehydrogenase (no. 22), ALDH 6a1 (no. 27), and choline dehydrogenase (CDH, no. 29) (Fig. 4C). The PDH complex catalyzes the conversion of pyruvate to acetyl CoA and CO2. Proteomic analyses reveal that the PDH E1 beta subunit is increased by SAM treatment under both control and ethanol conditions. This may have the effect of enhancing the supply of acetyl CoA for the TCA cycle, thereby enhancing energy metabolism, which supports data showing increased mitochondrial respiration in the SAM groups (Table 2). In addition, we observed ethanol- and SAM-dependent effects on enzymes that link methionine, choline, and glycine metabolism. There was an ethanol-dependent increase in DMGDH protein (no. 17), which was normalized by SAM treatment. Coupled to this, we found a SAM-dependent increase in CDH (no. 29) whereas SAM treatment significantly decreased SARDH (no. 11) in both control and ethanol groups (Fig. 4C). Ethanol and SAM also had differential effects on enzymes involved in the oxidation of the branch chain amino acids (BCAA) valine and leucine. Ethanol consumption increased HIBDH (no. 12, valine catabolism) and decreased IVDH (no. 14, leucine catabolism), with SAM normalizing IVDH levels in liver mitochondria. The effect of SAM to normalize IVDH levels in ethanol-fed rats is significant since studies show that diets supplemented with BCAA are protective against alcoholic liver diseases (14). Other ethanol- and SAM-dependent effects were found in pathways of amino acid metabolism. We found an ethanol-dependent increase in the protein agmatinase (no. 13) that was normalized by SAM treatment.
Within this broad grouping of enzymes, we observed a SAM-dependent increase in the classic mitochondrial aldehyde dehydrogenase 2 (ALDH2, no. 9) and an ethanol-dependent decrease in ALDH6a1 (no. 27), the only CoA-dependent ALDH (1). The SAM-dependent increase in ALDH2 protein is predicted to be of benefit through increased acetaldehyde detoxification since ALDH2 overexpression inhibits chronic alcohol-induced apoptosis (24). Therefore, the hepatoprotective effects of SAM may extend beyond methionine and lipid metabolism but may include important impacts on liver amino acid and aldehyde metabolism.
Other diverse mitochondrial proteins from 2D IEF gels.
Figure 4D shows those remaining proteins that are altered by ethanol, SAM, or both. First, there was an alcohol-dependent increase in the antioxidant enzyme catalase (no. 23), which verifies previous in vivo and in vitro studies of alcohol exposure (9, 13). Although catalase activity in mitochondrial preparations has largely been assumed to be due to peroxisome contamination, recent reports indicate that catalase is present within rat liver mitochondria (26, 52). Relevant to this present study is that SAM significantly downregulates catalase protein. The SAM-mediated suppression of catalase suggests that the ethanol-dependent increase in this antioxidant enzyme may reflect an adaptive response to oxidative stress, which SAM blunts. Differential effects were observed in levels of the mitochondrial protein thiosulfate sulfurtransferase (TSST), a.k.a. rhodanese, with two separate protein “spots” identified as this protein (nos. 21 and 25). Of note, we observed that ethanol and SAM treatment caused a shift in the levels of the two detected TSST isoforms, e.g., TSST protein no. 21 was decreased whereas TSST protein no. 25 was significantly increased by SAM treatment (Figs. 3 and 4D and Table 3). Finally, we observed SAM-dependent decreases in two mitochondrial carboxylase enzymes: propionyl CoA carboxylase (no. 10) and pyruvate carboxylase (no. 15) (Fig. 4D).
OxPhos system proteins identified by BN-PAGE.
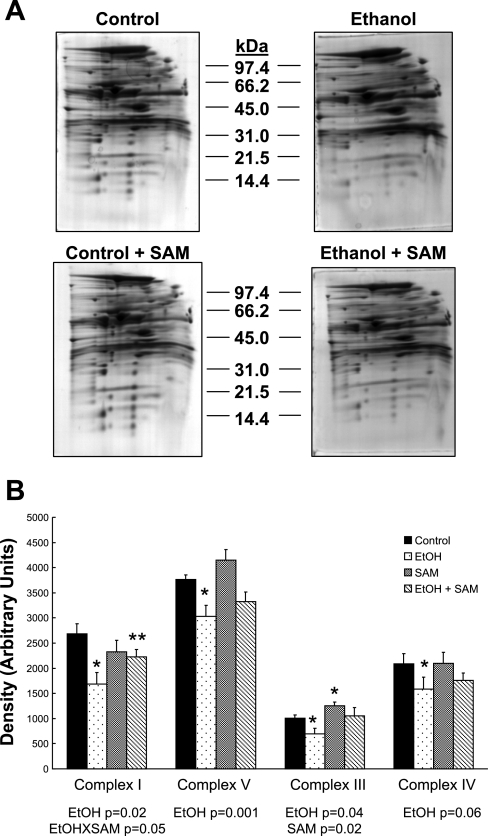
Analysis of OxPhos system proteins using 2D IEF/SDS-PAGE is hampered because many of the inner membrane proteins precipitate at the basic end of the IEF gel and therefore are poorly resolved (6). This is notable because only five OxPhos proteins were identified on 2D IEF/SDS-PAGE gels (see supplemental data, Table 1). To address this problem, BN-PAGE was used for analyses of the OxPhos system subproteome (57). Representative 2D BN-PAGE gels for each group are presented in Fig. 5A. On visual inspection it is apparent there is a decrease in OxPhos proteins in the ethanol gel (Fig. 5A, top right) compared with control (top left). Densitometry analysis of those proteins comprising intact complexes I, III, IV, and V showed a significant decrease in the content of OxPhos complexes in response to ethanol (Fig. 5B). Note that proteins “associated” with each complex, but not actual components of the complex, are also detected in these gels; however, these proteins were excluded from densitometry analysis. Important to this present study is the observation that the ethanol-dependent decrease in OxPhos complexes is partly attenuated by SAM cotreatment (Fig. 5A, compare bottom panels). There was a statistically significant increase in complex I protein in the ethanol + SAM group compared with ethanol (Fig. 5B). We also observed a statistically significant interaction for complex I and a significant effect of SAM for complex III (Fig. 5B). There were also numerical increases in complex IV and V content in the ethanol + SAM compared with ethanol alone that missed statistical significance.
Fig. 5.
Representative 2D blue native gel electrophoresis (BN-PAGE) gels of liver mitochondrial proteins. A: mitochondrial protein from livers of control, ethanol, control + SAM-, and ethanol + SAM-treated groups were subjected to BN-PAGE as described in materials and methods. B: comparison of the relative quantities of complexes I, V, III, and IV in liver mitochondria from the 4 treatment groups. Data represent means ± SE, n = 4 animals per each group; *P < 0.05 compared with control; **P < 0.05 compared with EtOH. Results from 2-factor ANOVA for each complex are reported in B.
To extend these investigations, we further examined these 2D BN-PAGE maps to identify changes in individual complex polypeptides. Of the 70+ proteins we routinely detect and can identify on 2D BN-PAGE gels (2), we were able to match 33 proteins in common across all gels from the four treatment groups. A master map of these proteins (Fig. 6A, circled proteins) and their IDs (Table 4) are provided. Moreover, the bar graphs in Fig. 6, B–E illustrate the abundances (i.e., densities) for each of the 33 identified proteins. Of these proteins, 19 were found to be significantly altered in abundance in response to ethanol, SAM, or both. A listing of the statistical analysis results and P values for these proteins is provided in Table 4. The protein ID numbers used in the following sections refer to proteins referenced in Fig. 6 and Table 4.
NADH dehydrogenase: complex I proteins.
Electrons from NADH enter the respiratory chain via complex I. Reducing equivalents from NADH oxidation are transferred to a flavin mononucleotide (FMN) and are passed through a series of iron-sulfur (Fe-S) centers, reducing ubiquinone to ubiquinol; an electron donor for complex III. Every electron pair transferred through complex I is coupled to the “pumping” of four H+s from the matrix to cytosolic side of the inner membrane. Figure 6B shows ethanol- and SAM-dependent alterations in those proteins classified in complex I. Of the nine complex I proteins identified, all were nuclear encoded with seven affected by ethanol, SAM, or both. These included three Fe-S center proteins (human genes: Ndufs1, 2, and 6), two flavin-containing proteins (human genes: Ndufv1 and 2), and four other proteins (human genes: Ndufa8, 10, and 12 and Ndufb4). In general, the ethanol-dependent decrease in the Fe-S center proteins (nos. 1, 3, and 9) was prevented by SAM coadministration, suggesting preservation of the electron-transferring capacity. Similar trends were observed for the flavin-containing proteins (nos. 2 and 5). Two proteins with less defined functions were also decreased by ethanol treatment, subcomplex 8 (no. 6) and 12 (no. 7) (human genes: Ndufa8 and 12) and preserved by SAM.
Cytochrome bc1 complex: complex III proteins.
This complex functions by transferring electrons from ubiquinol to cytochrome c, which is linked to the translocation of four H+s from the matrix to the intermembrane space. Figure 6D shows alcohol- and SAM-associated changes in complex III proteins. Although 7 subunits were detected with most showing a numerical decrease in response to ethanol, only subunits 7 (no. 22) and 8 (no. 23) were statistically altered by treatments. These findings are important because these subunits, along with cytochrome b, form the central core of the enzyme, with subunit 7 responsible for formation and/or stabilization of this key subcomplex (65). Therefore it is possible that the partial preservation of complex III in the ethanol + SAM group is related to maintenance of the central core subunits.
Cytochrome c oxidase: complex IV proteins.
The mammalian cytochrome c oxidase complex is the terminal enzyme of the respiratory chain and functions by transferring electrons from cytochrome c to molecular oxygen while pumping H+s across the inner membrane. Of the five complex IV proteins positively identified (Fig. 6E), all were decreased by ethanol consumption and include the mitochondrial encoded subunit II (no. 27) along with the nuclear encoded subunits of IV1 (no. 29), Va (no. 30), V1a (no. 31), and VIb (no. 32). Two unidentified proteins (Fig. 6E, nos. 28 and 33) speculated to be subunits III and VIII, respectively, were also decreased by chronic ethanol treatment. Importantly, SAM treatment prevented the loss of subunits II (no. 27) and IV1 (no. 29).
ATP synthase: complex V proteins.
The ATP synthase uses the energy of the electrochemical gradient stored across the inner membrane to synthesize ATP from ADP and Pi. Figure 6C shows the effect of ethanol and SAM treatment on complex V proteins. Of the five complex V proteins identified, all were nuclear encoded with two proteins showing a decrease in abundance in response to ethanol and one protein showing a SAM-mediated increase. Proteins decreased by chronic alcohol consumption were the d (no. 13) and g (no. 15) subunits of the F0 membrane portion of the enzyme. The gamma chain (no. 11) was elevated by SAM treatment.
Effect of ethanol and SAM on select gene transcripts.
To determine whether the effects of ethanol and SAM on proteins abundances correlated to alterations in gene transcripts, RT-PCR analysis was performed for a small subset of select genes whose proteins showed alterations in abundance from proteomic analyses. As shown in Table 5, the mRNA level of the chaperone Hspa5 was unaffected by ethanol, SAM, or both. The mRNA level of Dmgdh was increased by ethanol (control vs. ethanol, P = 0.015, t-test). Also, there was a significant effect of SAM on Hadh levels (control vs. control + SAM, P = 0.05, t-test). There was a statistically significant interaction of ethanol and SAM on mRNA levels of the ATP synthase subunit d (Atp5h), with SAM treatment attenuating an ethanol-dependent increase in Atp5h levels (ethanol vs. ethanol + SAM, P = 0.013, t-test). The mRNA level of the NADH dehydrogenase 1 α subcomplex 8 (Ndufa8) was unaffected by ethanol, SAM, or both.
Table 5.
Effect of ethanol and SAM on liver gene transcripts
| Treatment | Hspa5 | Dmgdh | Hadh | Atp5 h | Ndufa8 |
|---|---|---|---|---|---|
| Control | 0.88 ± 0.09 | 0.36 ± 0.05 | 0.62 ± 0.07 | 0.22 ± 0.05 | 0.26 ± 0.03 |
| Ethanol | 0.89 ± 0.03 | 0.57 ± 0.06 | 0.61 ± 0.03 | 0.35 ± 0.03 | 0.28 ± 0.03 |
| Control + SAM | 0.85 ± 0.15 | 0.42 ± 0.07 | 0.82 ± 0.10 | 0.22 ± 0.04 | 0.28 ± 0.05 |
| Ethanol + SAM | 0.90 ± 0.07 | 0.46 ± 0.04 | 0.69 ± 0.06 | 0.24 ± 0.03 | 0.26 ± 0.04 |
| ANOVA (P values)* | |||||
| Ethanol effect | 0.77 | 0.04 | 0.35 | 0.06 | 0.95 |
| SAM effect | 0.93 | 0.67 | 0.05 | 0.41 | 0.90 |
| Interaction (Ethanol × SAM) | 0.84 | 0.17 | 0.35 | 0.007 | 0.56 |
All transcript values are normalized to GAPDH transcripts and data are expressed as means ± SE for 6 pairs per group (21).
Hspa5, heat shock 70 kDa protein 5 (a.k.a., GRP78); Dmgdh, dimethylglycine dehydrognase; Hadh, hydroxylacyl-CoA dehydrogenase (a.k.a., trifunctional protein); Atp5 h, ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d; Ndufa8, NADH dehydrogenase 1 α subcomplex 8.
Results from 2-factor ANOVA with ethanol effect (control vs. ethanol), SAM effect (absent vs. present), and interaction (Ethanol × SAM).
DISCUSSION
Mitochondrial dysfunction contributes, in part, to the initiation and progression of alcohol-induced liver injury as a failure to maintain hepatic ATP levels renders hepatocytes unable to support requisite adaptive repair mechanisms (5). Confirming previous studies (8), we observed that SAM treatment preserves mitochondrial respiratory capacity during chronic ethanol consumption. SAM administration alone increased respiratory rates in both control and ethanol groups over those observed in the non-SAM-treated groups (Table 2). Although the reason for this SAM-mediated effect is currently not known, it is likely that SAM supplementation enhances hepatocyte methylation-dependent reactions needed for proper mitochondrial maintenance (15). Indeed, the proteomic analyses performed in this study begin to shed light on these possible mechanisms and demonstrate a complex interaction of ethanol and SAM at the level of the mitochondrial proteome.
Studies have shown that chronic ethanol feeding impairs methionine metabolism resulting in decreased SAM/SAH ratio, which dysregulates methyltransferase reactions, polyamine synthesis, and transsulfuration (38). Consistent with this, we observed these chronic alcohol-mediated changes in SAM and SAH in liver. However, what is less well studied is the specific effect of chronic alcohol consumption on SAM and SAH in the mitochondrial compartment. Recent work showed decreased mitochondrial SAM in response to alcohol feeding with no change in SAH (22, 58). The mechanism for this decrease is not clear because mitochondrial SAM transport was unaltered by chronic alcohol (22). As in these previous reports, we observed no change in mitochondrial SAH in response to chronic alcohol. Nevertheless, we observed an alcohol-dependent increase in mitochondrial SAM that was further elevated by dietary SAM. Although elevated mitochondrial SAM was not unexpected in groups receiving extra dietary SAM, the increase in mitochondrial SAM by ethanol alone was unanticipated based on prior reports. Dissimilarities in results among studies may be related to multiple factors including differences in experimental design, species (e.g., rat vs. mouse), alcohol feeding duration, ethanol concentration and amount consumed, and preparation of mitochondria. However, similar to what has been shown in the literature for GSH, mitochondrial SAM depletion may not be an unwavering, constant characteristic of chronic alcohol exposure. Indeed, multiple papers report alcohol-dependent increases in liver mitochondrial GSH (8, 18, 36, 55). With this in mind, it is proposed that the observed increase in mitochondrial SAM may be an adaptive response within the hepatocyte to counteract alcohol-mediated stress and preserve the mitochondrial pool of SAM, which like GSH could be a critical factor for cell viability (37). Conservation of mitochondrial SAM may also contribute, in part, to depletion in total cellular SAM by alcohol. Notably, the impact of ethanol on methionine metabolism might also be quite dynamic. Thus time course studies are required to precisely document alcohol-mediated alterations in SAM and SAH during both early and late alcohol exposures. These studies could be patterned after those of Nagy and colleagues (51) illustrating the dynamic nature of cytokine responses during 1 day to 5 wk of ethanol exposure.
Although proteomics can be criticized for being intrinsically descriptive, we propose that proteomics is ideal for understanding SAM hepatoprotection at the level of the mitochondrion for several reasons. First, data presented herein support a central role of the mitochondrion in alcohol hepatotoxicity and SAM protection. Second, the mitochondrial proteome is well defined with the functions and amino acid sequences of the majority of mitochondrial proteins known. Third, specialized proteomic methods such as BN-PAGE allow for the identification of defects in assembly of multiprotein complexes in mitochondria and improve resolution of inner membrane proteins that cannot be analyzed by 2D IEF proteomics (6). Fourth, alterations in gene expression may not always result in the same change at the protein level. This might be especially true for mitochondrial proteins as chronic alcohol consumption negatively impacts protein turnover (20) and assembly of OxPhos complexes (61). For example, the chaperone protein GRP78 (gene: Hspa5) and NADH dehydrogenase 1 α subcomplex 8 (gene: Ndufa8) showed significant alcohol-dependent decreases in protein abundance (Tables 3 and 4) but no effect on gene expression (Table 5), whereas alcohol-dependent changes in gene expression were mirrored at the proteome level for DMGDH (Table 3). These disparities in results are important to note and strengthen the notion that gene expression results should not be used solely to predict protein abundance in the mitochondrial proteome or functional outcomes on metabolic pathways especially as ethanol and SAM are likely to alter posttranslational mechanisms affecting protein stability, turnover, modification, import, and complex assembly. Thus a comprehensive proteomics approach is a superior method to identify and characterize specific molecular targets and pathways through which metabolic stressors such as chronic alcohol consumption lead to hepatotoxicity and, more importantly, how therapeutics such as SAM provide benefit against mitochondrial dysfunction and liver disease from toxicant exposures.
In this study, we show novel effects of ethanol and SAM on chaperone proteins. GRP78, a member of the Hsp70 family, is involved in protein folding within the endoplasmic reticulum (ER) (46); however, it is also localized to the mitochondrial matrix and cytosol (12, 53). The alcohol-mediated decrease in GRP78 may explain the susceptibility of mitochondrial proteins to oxidative modification and inactivation. For example, oxidative modification to protein thiols is linked to protein inactivation following chronic alcohol (43, 62) including an alcohol-dependent cysteine modification in GRP78 that might be linked to inactivation of the protein (62). Of note, GRP78 overexpression inhibits ER-stress-induced SREBP-1c activation and decreases hepatic fat accumulation (28). On this basis, decreased GRP78 may contribute, in part, to the development of alcohol-dependent steatosis. Importantly, we observed that SAM prevents alcohol-dependent decreases in several chaperone proteins including GRP78, Hspd1, and PDI/T3BP while lessening alcohol-dependent steatosis. It is proposed that the SAM-mediated maintenance of these highly conserved chaperone proteins contributes to the partial preservation of mitochondrial function and reduction of steatosis in animals exposed to ethanol and SAM. Future studies are planned to investigate the SAM-dependent mechanism responsible for chaperone maintenance under conditions of alcohol-dependent metabolic and oxidative stress.
Recent proteomic data show that a betaine-supplemented ethanol diet preserves functionality of several key cytosolic enzymes of methionine metabolism including betaine homocysteine methyltransferase (BHMT) and methionine adenosyltransferase (30). Data presented herein also shows ethanol- and SAM-dependent alterations in the levels of mitochondrial proteins interconnected with methionine metabolism via choline and glycine metabolism; CDH, DMGDH, and SARDH. Choline is an essential nutrient that serves as a precursor in the synthesis of membrane and lipoprotein phospholipids and the neurotransmitter acetylcholine. It is also a key carbon donor for remethylation of homocysteine to methionine. Glycine plays a key role in collagen and glutathione synthesis. Briefly, choline is metabolized to betaine through two sequential reactions catalyzed by CDH and betaine ALDH. Betaine, in turn, functions as a methyl donor to remethylate homocysteine to methionine via the action of BHMT with production of dimethylglycine (DMG). DMGDH is a mitochondrial matrix enzyme that catalyzes the demethylation of DMG to sarcosine, which, in turn, is demethylated to glycine by SARDH. We observed an alcohol-dependent increase in DMGDH protein, which was normalized by SAM treatment (Fig. 4C, protein no. 17). The ability of SAM to prevent the ethanol-mediated increase in DMGDH is important; a recent study demonstrated that reduction of DMGDH by use of shRNA suppressed hypoxic cell death in cardiomyocytes (49). This finding suggests that DMGDH deserves additional attention as a possible risk factor for alcoholic liver disease. Moreover, we observed a SAM-dependent increase in CDH (Fig. 4C, protein no. 29). A SAM-dependent increase in CDH may have the desired consequence of increased betaine formation needed for remethylation of homocysteine to methionine, thereby blunting the toxicity of homocysteine to induce ER stress (25). Data from this study and others (30, 54) show that SAM or betaine treatments have the ability to attenuate many of the damaging effects of ethanol on the liver through modulation of key enzymes involved in methionine metabolism.
TSST, a highly abundant mitochondrial protein, is responsible for the detoxification of cyanide to thiocyanate with thiosulfate as the sulfur donor. However, the biological role of TSST has recently expanded to include sulfane sulfur transfer to other thiol groups, leading to persulfide group formation within protein and enzymes, thus representing a new posttranslational modification for protein thiols (45). In this present study, we observed that ethanol and SAM treatment caused a shift in the levels of the two detected TSST isoforms, e.g., TSST protein no. 21 was decreased whereas TSST protein no. 25 was significantly increased by SAM treatment (Figs. 3 and 4D). We propose that this shift in protein isoform levels might be due to an alteration in phosphorylation status such that the observed increase in the amount of the more basic, dephosphorylated isoform (protein no. 25) reflects conversion to active enzyme with a concomitant loss in the acidic, phosphorylated isoform (protein no. 21), which represents inactive TSST (34). Although the significance of ethanol and SAM to increase TSST expression and activity is currently not known, a recent study showed a biological interaction between the thiol-containing antioxidant lipoic acid (LA) and TSST, specifically that LA administration increased TSST activity in multiple tissues including liver with concomitant decrease in tissue markers of oxidative stress (11). Taken together, these results are intriguing and predict that future investigations focused on increased understanding of the impacts and interactions of sulfur-containing compounds such as SAM on sulfur metabolism in liver may lead to improved benefit of patients suffering from alcoholic liver disease.
In addition to ethanol- and SAM-dependent alterations in matrix proteins, several novel changes were observed in OxPhos proteins. For example, complex I, subcomplex 12 (a.k.a. B17.2) is thought to play a key chaperone-like role in complex I assembly (31, 60). Therefore, the ethanol-mediated decrease in these nuclear encoded subcomplexes may explain, in part, the overall decrease in total complex I protein in response to chronic ethanol exposure. Previously, we have shown that chronic alcohol also decreases cytochrome c oxidase activity and levels of the three mitochondrial encoded subunits (I, II, and III) that comprise the catalytic core (61). Emerging evidence shows similar alterations in some of the remaining ten nuclear encoded subunits (61). These subunits are thought to function in assembly, modulation of activity, and possible protection against oxidative damage in mitochondria (10). In the present study, we observed that SAM preserved levels of both mitochondrial and nuclear-encoded subunits of complex IV, thereby providing a molecular mechanism to explain the ability of SAM to preserve mitochondrial respiratory function.
For the first time, chronic ethanol was found to decrease levels of two nuclear-encoded ATP synthase subunits: d and g. The hydrophilic d subunit is essential for activity through direct interactions with the other subunits oligomycin-sensitivity conferring protein, b, and F6, of the peripheral stalk or “stator” of the complex (19). Decreased content of subunit d is predicted to contribute to an alcohol-mediated decrease in complex V activity based on functional studies by Cunningham and colleagues (42). Moreover, subunits g and e are small membrane-spanning hydrophobic F0 proteins typically referred to as the “dimer-specific” subunits functioning to stabilize supramolecular organization of ATP synthase into oligomeric structures within the inner membrane (19). Emerging data suggest that subunits g and e serve a structural function helping to model the curvatures needed to form cristae within the inner membrane (47). We propose that an alcohol-dependent decrease in subunit g may contribute, in part, to the loss of cristae and formation of megamitochondria, hallmark features of alcohol hepatotoxicity (4).
Accruing evidence supports the concept of “supramolecular” organization of complexes I, III, and IV within mammalian mitochondria, forming discrete functional units (63). Indeed, recent studies show the architecture of two separate “supercomplexes” in bovine mitochondria with stoichiometries of I1III2 and I1III2IV1 (56). Taking this into account, it is likely that the presence of complex IV, subunit IV outside its expected location on the 2D BN-PAGE map (Fig. 6A, complex IV lane, protein no. 29) with it crossing though the lanes of complex I, V, and III likely represents its association with complexes I and III within a supercomplex. This is supported by the observation that complex IV, subunit IV was 1 of 20 proteins identified as being part of an integrated supercomplex (56). Although preliminary, this observation highlights the importance of investigating these recently characterized higher order supercomplex structures of the respiratory chain and the potential role their disruption might contribute to pathologies with known mitochondrial defects such as alcoholic liver disease. Future studies are planned to examine whether supercomplex formation is disrupted in alcoholic mitochondria.
Previous to this present study, a limited number of liver proteins had been identified whose abundances were altered in response to chronic alcohol consumption. Using complementary proteomics approaches, we have expanded this growing database of proteins. This is the first comprehensive analysis of the liver mitochondrial proteome studied under conditions of ethanol exposure in combination with a promising hepatoprotective agent, SAM. Identification of these altered proteins provides critical insight into the mechanisms responsible for alcoholic mitochondrial dysfunction and reveals multiple new molecular targets that can be exploited for therapeutic investigations. Specifically, we show for the first time that ethanol and SAM have profound interactions on chaperone proteins, as well as methionine, choline, and glycine metabolism enzymes. We report novel observations of ethanol and SAM on pathways of persulfide and sulfane sulfur metabolism and have identified novel ethanol-mediated changes in several OxPhos system proteins, e.g., ATP synthase subunit g. These latter findings provide much needed insight into the precise molecular nature through which ethanol, SAM alone, or both modulate mitochondrial function and structure. Future studies aimed at isolating and characterizing the supramolecular complexes of the OxPhos system are needed to advance the concept that disrupted and/or misassembled supercomplexes contribute to disease pathogenesis in the alcoholic liver. Similarly, increased understanding of how SAM and other methionine metabolites regulate mitochondrial function in both normal and diseased states is required to advance our understanding of how SAM and related drugs can be used to safely and successfully treat individuals suffering with chronic liver diseases. In conclusion, this study demonstrates that a systems biology approach utilizing complementary proteomics technologies is a valuable tool that can be used to enhance ongoing mitochondrial studies aimed at elucidating the molecular events and pathways altered in complex, multifactor diseases such as alcoholic liver disease.
GRANTS
This work was supported in part by NIH grants AA15172 and DK73775 to S. M. Bailey. Operational funds came in part from the UAB Comprehensive Cancer Center Core Grant (P30 CA13148), the Purdue-UAB Botanicals Center for Age-Related Disease (P50 AT00477), the UAB Center for Nutrient-Gene Interaction in Cancer Prevention (U54 CA100949), the UAB Skin Disease Research Center (P30 AR050948), and the UAB Polycystic Kidney Disease Center (P30 DK74038).
DISCLOSURES
The authors have no conflicts of interest and no disclosures.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Orchid Chemicals and Pharmaceuticals, Tamil Nadu, India for the kind gift of SAM used in these studies. The authors also thank Dr. Kusum K. Kharbanda for advice regarding SAM and SAH HPLC studies, Dr. Grier Page for advice on statistical analyses, and Dr. Steve Barnes and Landon Wilson of the UAB Mass Spectrometry Shared Facility for mass spectrometry analyses. Mass spectrometers in the Shared Facility came from funds provided by National Center for Research Resources Grants S10 RR11329 and S10 RR13795 plus UAB Health Services Foundation General Endowment Fund.
REFERENCES
- 1.Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci 101: 51–64, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Andringa K, King A, Bailey S. Blue native-gel electrophoresis proteomics. Methods Mol Biol 519: 241–258, 2009 [DOI] [PubMed] [Google Scholar]
- 3.Andringa KK, Bajt ML, Jaeschke H, Bailey SM. Mitochondrial protein thiol modifications in acetaminophen hepatotoxicity: effect on HMG-CoA synthase. Toxicol Lett 177: 188–197, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai M, Leo MA, Nakano M, Gordon ER, Lieber CS. Biochemical and morphological alterations of baboon hepatic mitochondria after chronic ethanol consumption. Hepatology 4: 165–174, 1984 [DOI] [PubMed] [Google Scholar]
- 5.Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic Biol Med 32: 11–16, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Bailey SM, Landar A, Darley-Usmar V. Mitochondrial proteomics in free radical research. Free Radic Biol Med 38: 175–188, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Bailey SM, Patel VB, Young TA, Asayama K, Cunningham CC. Chronic ethanol consumption alters the glutathione/glutathione peroxidase-1 system and protein oxidation status in rat liver. Alcohol Clin Exp Res 25: 726–733, 2001 [PubMed] [Google Scholar]
- 8.Bailey SM, Robinson G, Pinner A, Chamlee L, Ulasova E, Pompilius M, Page GP, Chhieng D, Jhala N, Landar A, Kharbanda KK, Ballinger S, Darley-Usmar V. S-adenosylmethionine prevents chronic alcohol-induced mitochondrial dysfunction in the rat liver. Am J Physiol Gastrointest Liver Physiol 291: G857–G867, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Bardag-Gorce F, French BA, Dedes J, Li J, French SW. Gene expression patterns of the liver in response to alcohol: in vivo and in vitro models compared. Exp Mol Pathol 80: 241–251, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Barrientos A, Gouget K, Horn D, Soto IC, Fontanesi F. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim Biophys Acta 1793: 97–107, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilska A, Dudek M, Iciek M, Kwiecien I, Sokolowska-Jezewicz M, Filipek B, Wlodek L. Biological actions of lipoic acid associated with sulfane sulfur metabolism. Pharmacol Rep 60: 225–232, 2008 [PubMed] [Google Scholar]
- 12.Brookes PS, Pinner A, Ramachandran A, Coward L, Barnes S, Kim H, Darley-Usmar VM. High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics 2: 969–977, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Cederbaum AI. Cytochrome P450 2E1-dependent oxidant stress and upregulation of anti-oxidant defense in liver cells. J Gastroenterol Hepatol 21, Suppl 3: S22–S25, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Charlton M. Branched-chain amino acid enriched supplements as therapy for liver disease. J Nutr 136: 295S–298S, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Chen R, Fearnley IM, Palmer DN, Walker JE. Lysine 43 is trimethylated in subunit C from bovine mitochondrial ATP synthase and in storage bodies associated with batten disease. J Biol Chem 279: 21883–21887, 2004 [DOI] [PubMed] [Google Scholar]
- 16.Coleman WB, Cunningham CC. Effects of chronic ethanol consumption on the synthesis of polypeptides encoded by the hepatic mitochondrial genome. Biochim Biophys Acta 1019: 142–150, 1990 [DOI] [PubMed] [Google Scholar]
- 17.Cornwell PE, Morgan SL, Vaughn WH. Modification of a high-performance liquid chromatographic method for assay of homocysteine in human plasma. J Chromatogr 617: 136–139, 1993 [DOI] [PubMed] [Google Scholar]
- 18.Deaciuc IV, Fortunato F, D'Souza NB, Hill DB, Schmidt J, Lee EY, McClain CJ. Modulation of caspase-3 activity and Fas ligand mRNA expression in rat liver cells in vivo by alcohol and lipopolysaccharide. Alcohol Clin Exp Res 23: 349–356, 1999 [PubMed] [Google Scholar]
- 19.Devenish RJ, Prescott M, Rodgers AJ. The structure and function of mitochondrial F1F0-ATP synthases. Int Rev Cell Mol Biol 267: 1–58, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Donohue TM, Jr, Cederbaum AI, French SW, Barve S, Gao B, Osna NA. Role of the proteasome in ethanol-induced liver pathology. Alcohol Clin Exp Res 31: 1446–1459, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Esfandiari F, Medici V, Wong DH, Jose S, Dolatshahi M, Quinlivan E, Dayal S, Lentz SR, Tsukamoto H, Zhang YH, French SW, Halsted CH. Epigenetic regulation of hepatic endoplasmic reticulum stress pathways in the ethanol-fed cystathionine beta synthase-deficient mouse. Hepatology In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez A, Colell A, Caballero F, Matias N, Garcia-Ruiz C, Fernandez-Checa JC. Mitochondrial S-adenosyl-l-methionine transport is insensitive to alcohol-mediated changes in membrane dynamics. Alcohol Clin Exp Res 33: 1169–1180, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Ruiz C, Morales A, Colell A, Ballesta A, Rodes J, Kaplowitz N, Fernandez-Checa JC. Feeding S-adenosyl-l-methionine attenuates both ethanol-induced depletion of mitochondrial glutathione and mitochondrial dysfunction in periportal and perivenous rat hepatocytes. Hepatology 21: 207–213, 1995 [DOI] [PubMed] [Google Scholar]
- 24.Guo R, Zhong L, Ren J. Overexpression of aldehyde dehydrogenase-2 (ALDH2) attenuates chronic alcohol exposure-induced apoptosis, change in Akt and Pim signaling in liver. Clin Exp Pharmacol Physiol 36: 463–468, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology 124: 1488–1499, 2003 [DOI] [PubMed] [Google Scholar]
- 26.Jiang XS, Dai J, Sheng QH, Zhang L, Xia QC, Wu JR, Zeng R. A comparative proteomic strategy for subcellular proteome research: ICAT approach coupled with bioinformatics prediction to ascertain rat liver mitochondrial proteins and indication of mitochondrial localization for catalase. Mol Cell Proteomics 4: 12–34, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Jou J, Choi SS, Diehl AM. Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin Liver Dis 28: 370–379, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferre P, Foufelle F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest 119: 1201–1215, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kharbanda KK, Rogers DD, 2nd, Mailliard ME, Siford GL, Barak AJ, Beckenhauer HC, Sorrell MF, Tuma DJ. A comparison of the effects of betaine and S-adenosylmethionine on ethanol-induced changes in methionine metabolism and steatosis in rat hepatocytes. J Nutr 135: 519–524, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Kharbanda KK, Vigneswara V, McVicker BL, Newlaczyl AU, Bailey K, Tuma D, Ray DE, Carter WG. Proteomics reveal a concerted upregulation of methionine metabolic pathway enzymes, and downregulation of carbonic anhydrase-III, in betaine supplemented ethanol-fed rats. Biochem Biophys Res Commun 381: 523–527, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol 27: 4228–4237, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber CS. S-adenosyl-l-methionine and alcoholic liver disease in animal models: implications for early intervention in human beings. Alcohol 27: 173–177, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Lieber CS, DeCarli LM. The feeding of alcohol in liquids: two decades of applications and update. Alcohol Clin Exp Res 6: 523–531, 1982 [DOI] [PubMed] [Google Scholar]
- 34.Maduh EU, Baskin SI. Protein kinase C modulation of rhodanese-catalyzed conversion of cyanide to thiocyanate. Res Commun Mol Pathol Pharmacol 86: 155–173, 1994 [PubMed] [Google Scholar]
- 35.Mantena SK, King AL, Andringa KK, Eccleston HB, Bailey SM. Mitochondrial dysfunction and oxidative stress in the pathogenesis of alcohol- and obesity-induced fatty liver diseases. Free Radic Biol Med 44: 1259–1272, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mari M, Cederbaum AI. CYP2E1 overexpression in HepG2 cells induces glutathione synthesis by transcriptional activation of gamma-glutamylcysteine synthetase. J Biol Chem 275: 15563–15571, 2000 [DOI] [PubMed] [Google Scholar]
- 37.Mari M, Morales A, Colell A, Garcia-Ruiz C, Fernandez-Checa JC. Mitochondrial glutathione, a key survival antioxidant. Antioxid Redox Signal 11: 2685–2700, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Lopez N, Varela-Rey M, Ariz U, Embade N, Vazquez-Chantada M, Fernandez-Ramos D, Gomez-Santos L, Lu SC, Mato JM, Martinez-Chantar M. S-adenosylmethionine and proliferation: new pathways, new targets. Biochem Soc Trans 36: 848–852, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Mato JM, Camara J, Fernandez de Paz J, Caballeria L, Coll S, Caballero A, Garcia-Buey L, Beltran J, Benita V, Caballeria J, Sola R, Moreno-Otero R, Barrao F, Martin-Duce A, Correa JA, Pares A, Barrao E, Garcia-Magaz I, Puerta JL, Moreno J, Boissard G, Ortiz P, Rodes J. S-adenosylmethionine in alcoholic liver cirrhosis: a randomized, placebo-controlled, double-blind, multicenter clinical trial. J Hepatol 30: 1081–1089, 1999 [DOI] [PubMed] [Google Scholar]
- 40.Mato JM, Lu SC. Role of S-adenosyl-l-methionine in liver health and injury. Hepatology 45: 1306–1312, 2007 [DOI] [PubMed] [Google Scholar]
- 41.Minino AM, Heron MP, Smith BL. Deaths: preliminary data for 2004. Natl Vital Stat Rep 54: 1–49, 2006 [PubMed] [Google Scholar]
- 42.Montgomery RI, Coleman WB, Eble KS, Cunningham CC. Ethanol-elicited alterations in the oligomycin sensitivity and structural stability of the mitochondrial F0. F1 ATPase. J Biol Chem 262: 13285–13289, 1987 [PubMed] [Google Scholar]
- 43.Moon KH, Hood BL, Kim BJ, Hardwick JP, Conrads TP, Veenstra TD, Song BJ. Inactivation of oxidized and S-nitrosylated mitochondrial proteins in alcoholic fatty liver of rats. Hepatology 44: 1218–1230, 2006 [DOI] [PubMed] [Google Scholar]
- 44.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115: 629–640, 2003 [DOI] [PubMed] [Google Scholar]
- 45.Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol 2: 185–194, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett 581: 3641–3651, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J 21: 221–230, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purohit V, Abdelmalek MF, Barve S, Benevenga NJ, Halsted CH, Kaplowitz N, Kharbanda KK, Liu QY, Lu SC, McClain CJ, Swanson C, Zakhari S. Role of S-adenosylmethionine, folate, and betaine in the treatment of alcoholic liver disease: summary of a symposium. Am J Clin Nutr 86: 14–24, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Pyo JO, Nah J, Kim HJ, Chang JW, Song YW, Yang DK, Jo DG, Kim HR, Chae HJ, Chae SW, Hwang SY, Kim SJ, Kim HJ, Cho C, Oh CG, Park WJ, Jung YK. Protection of cardiomyocytes from ischemic/hypoxic cell death via Drbp1 and pMe2GlyDH in cardio-specific ARC transgenic mice. J Biol Chem 283: 30707–30714, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rinaldo P. Fatty acid transport and mitochondrial oxidation disorders. Semin Liver Dis 21: 489–500, 2001 [DOI] [PubMed] [Google Scholar]
- 51.Roychowdhury S, McMullen MR, Pritchard MT, Hise AG, van Rooijen N, Medof ME, Stavitsky AB, Nagy LE. An early complement-dependent and TLR-4-independent phase in the pathogenesis of ethanol-induced liver injury in mice. Hepatology 49: 1326–1334, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salvi M, Battaglia V, Brunati AM, La Rocca N, Tibaldi E, Pietrangeli P, Marcocci L, Mondovi B, Rossi CA, Toninello A. Catalase takes part in rat liver mitochondria oxidative stress defense. J Biol Chem 282: 24407–24415, 2007 [DOI] [PubMed] [Google Scholar]
- 53.Sammut IA, Harrison JC. Cardiac mitochondrial complex activity is enhanced by heat shock proteins. Clin Exp Pharmacol Physiol 30: 110–115, 2003 [DOI] [PubMed] [Google Scholar]
- 54.Santamaria E, Avila MA, Latasa MU, Rubio A, Martin-Duce A, Lu SC, Mato JM, Corrales FJ. Functional proteomics of nonalcoholic steatohepatitis: mitochondrial proteins as targets of S-adenosylmethionine. Proc Natl Acad Sci USA 100: 3065–3070, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sastre J, Minana JB, Alguacil P, Malet G, Gomez-Cambronero L, Marin JA, Vina JR, Vina J. Chronic ethanol feeding causes oxidative stress in rat liver mitochondria. Prevention by S-adenosylmethionine. Free Radic Res 30: 325–327, 1999 [Google Scholar]
- 56.Schafer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J. Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem 281: 15370–15375, 2006 [DOI] [PubMed] [Google Scholar]
- 57.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199: 223–231, 1991 [DOI] [PubMed] [Google Scholar]
- 58.Song Z, Zhou Z, Song M, Uriarte S, Chen T, Deaciuc I, McClain CJ. Alcohol-induced S-adenosylhomocysteine accumulation in the liver sensitizes to TNF hepatotoxicity: possible involvement of mitochondrial S-adenosylmethionine transport. Biochem Pharmacol 74: 521–531, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sykora P, Kharbanda KK, Crumm SE, Cahill A. S-adenosyl-l-methionine co-administration prevents the ethanol-elicited dissociation of hepatic mitochondrial ribosomes in male rats. Alcohol Clin Exp Res 33: 1–9, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Triepels R, van den Heuvel L, Loeffen J, Smeets R, Trijbels F, Smeitink J. The nuclear-encoded human NADH:ubiquinone oxidoreductase NDUFA8 subunit: cDNA cloning, chromosomal localization, tissue distribution, and mutation detection in complex-I-deficient patients. Hum Genet 103: 557–563, 1998 [DOI] [PubMed] [Google Scholar]
- 61.Venkatraman A, Landar A, Davis AJ, Chamlee L, Sanderson T, Kim H, Page G, Pompilius M, Ballinger S, Darley-Usmar V, Bailey SM. Modification of the mitochondrial proteome in response to the stress of ethanol-dependent hepatotoxicity. J Biol Chem 279: 22092–22101, 2004 [DOI] [PubMed] [Google Scholar]
- 62.Venkatraman A, Landar A, Davis AJ, Ulasova E, Page G, Murphy MP, Darley-Usmar V, Bailey SM. Oxidative modification of hepatic mitochondria protein thiols: effect of chronic alcohol consumption. Am J Physiol Gastrointest Liver Physiol 286: G521–G527, 2004 [DOI] [PubMed] [Google Scholar]
- 63.Vonck J, Schafer E. Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim Biophys Acta 1793: 117–124, 2009 [DOI] [PubMed] [Google Scholar]
- 64.Welch KD, Wen B, Goodlett DR, Yi EC, Lee H, Reilly TP, Nelson SD, Pohl LR. Proteomic identification of potential susceptibility factors in drug-induced liver disease. Chem Res Toxicol 18: 924–933, 2005 [DOI] [PubMed] [Google Scholar]
- 65.Zara V, Conte L, Trumpower BL. Evidence that the assembly of the yeast cytochrome bc1 complex involves the formation of a large core structure in the inner mitochondrial membrane. FEBS J 276: 1900–1914, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Li X, Mueller M, Wang Y, Zong C, Deng N, Vondriska TM, Liem DA, Yang JI, Korge P, Honda H, Weiss JN, Apweiler R, Ping P. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics 8: 1564–1575, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.