Abstract
The inability to identify, isolate, and culture intestinal epithelial stem cells (IESCs) has been prohibitive to the study and therapeutic utilization of these cells. Using a Sox9EGFP mouse model, we demonstrate that Sox9EGFP fluorescence signatures can be used to differentiate between and enrich for progenitors (Sox9EGFPsubLo) and multipotent IESCs (Sox9EGFPlo). Sox9EGFPlo cells generate “organoids” in a recently defined culture system that mimics the native IESC niche. These organoids possess all four differentiated cell types of the small intestine epithelium, demonstrating the multipotent capacity of Sox9EGFPlo cells. Our results are consistent with the previously reported observation that single IESCs generate cryptlike units without a detectable mesenchymal cell component. A prospective search revealed that CD24 is expressed in the Sox9EGFPlo population and marks IESCs that form organoids in culture. CD24 represents the first cell surface marker that facilitates fluorescence-activated cell sorting enrichment of IESCs with widely available antibodies without requiring a specialized fluorescent reporter gene mouse model.
Keywords: SOX factors, intestinal epithelial culture
translational highlights Isolation and culture of intestinal epithelial stem cells (IESCs) remains a challenge. This paper uses differential expression levels of a Sox9EGFP reporter to FACS isolate IESCs, and as a tool to identify CD24 FACS gates that would allow enrichment for IESCs using widely available antibodies. Single “low-expressing” Sox9EGFP cells and CD24 cells gave rise to “organoids” containing all differentiated cell types of the epithelium. These data demonstrate that low-expressing Sox9EGFP are functional IESCs and that CD24 functions as a new cell surface marker permitting enrichment for IESCs from a nonfluorescent reporter gene mouse.
the intestinal epithelium is one of the most proliferative tissues in the mammalian organism. The entire monolayer of epithelium is replaced approximately every 7 days (6, 11), and this renewal process is driven by a pool of multipotent, self-renewing stem cells that are positioned between Paneth cells and also immediately above the Paneth cell compartment in the base of the crypt (4, 35). Typically, in the adult organism stem cells are thought to undergo an asymmetric division process that generates a larger pool of more rapidly dividing transit-amplifying populations (or progenitors) that are located higher up the crypt-villus axis toward the lumen (6, 11). Intrinsic and extrinsic signaling directs these progenitors to commit to one of four functionally distinct postmitotic cell types: the enterocyte (absorptive) (9), goblet cell (mucus producing) (7), enteroendocrine cell (hormone producing) (10), and Paneth cell (antimicrobial peptide producing) (8). With the exception of the Paneth cell population, the other three cell types migrate up the crypt-villus axis and are sloughed into the lumen at the villus tip. Paneth cells are born around cell position +5 (as numbered from the base of the crypt toward the lumen) and migrate down to the crypt base, where they secrete antimicrobial peptides into the crypt lumen (8). Normal gut homeostasis is critically reliant on the proper control of stem cell proliferation, division, and subsequent generation of the transit-amplifying progenitor pool.
Significant progress toward identifying, isolating, and manipulating the intestinal epithelial stem cell (IESC) has been hindered by the lack of specific IESC biomarkers and suitable culturing techniques. Recently, a number of putative IESC biomarkers have been identified, but relatively few of these biomarkers have been tested to confirm that cells expressing these biomarkers pass the test of multipotency and self-renewal, the two functional characteristics that define a stem cell. Three studies have employed Cre-recombinase-mediated genetic lineage-tracing technology, currently the most rigorous method to assess multipotency in vivo, to demonstrate that the biomarkers Lgr5, Bmi1, and CD133/prominin1 are expressed in cell populations that demonstrate multipotency and self-renewal capacity in the small intestine (4, 35, 39). The expression patterns of Lgr5, Bmi1, and CD133/prominin1 mark stem cell populations that are differentially positioned in distinct regions of the crypt. Lgr5 expression is restricted primarily to the crypt-based columnar cells (CBC) that are intercalated between the Paneth cells (4). Bmi1 also shows a highly restricted expression pattern that is primarily localized to the supra-Paneth cell region, the zone typically defined as cell position +4 (35). CD133/prominin1 exhibits a broad expression pattern encompassing the CBC positions through at least cell position +10 (39). Although all three of these stem cell populations demonstrate multipotent capacity in vivo, it is unclear whether they have functionally equivalent roles in the homeostasis of the intestinal epithelial monolayer.
We have recently shown that distinct levels of Sox9 expression mark putative IESCs based on enriched levels of Lgr5 mRNA in discrete cell populations (14). SOX9 is a transcription factor that not only marks stem/progenitor cells in various tissues but also has an established role in maintaining the multipotent and proliferative capacity of stem/progenitor populations (25, 26). Using fluorescence-activated cell sorting (FACS) of the dissociated small intestine epithelium from a Sox9EGFP reporter gene mouse model, we demonstrated that “low” levels of Sox9EGFP (termed Sox9EGFPlo) mark cells that are enriched for Lgr5 (14); moreover, we identified that “high” levels of Sox9EGFP (termed Sox9EGFPhi) mark postmitotic enteroendocrine cells (14). Expression of endogenous levels of Sox9 mRNA and SOX9 protein were consistent with the Sox9EGFPlo and Sox9EGFPhi levels, validating that the Sox9EGFP reporter gene faithfully recapitulates expression patterns of endogenous Sox9 (14). Although formal inducible Cre-mediated genetic lineage tracing studies using Sox9 as a biomarker have not yet been conducted in an adult experimental model, Cre-mediated lineage tracing during embryonic gut development suggests that Sox9 expression marks multipotent IESCs at embryonic day 17 (E17) (1). This embryonic lineage tracing study provides compelling evidence that Sox9 expression marks a population of IESCs in the embryo that may be conserved in the adult.
Given the CBC location and enriched Lgr5 expression in the Sox9EGFPlo population, we hypothesized that Sox9EGFPlo expression marks functional IESCs. To test this hypothesis we utilized a recently described novel in vitro assay that provides a culturing environment, which supports the generation of crypt/villus-like structures from a single Lgr5-expressing intestinal epithelial stem cell (37). In this study we also characterize genetic signatures of small intestine epithelial stem/progenitor populations exhibiting different Sox9 expression levels and explore the multipotent and self-renewal capacity of these different populations of Sox9-expressing crypt cells. In an attempt to identify a cell surface marker that can be used to enrich for multipotent IESCs without the requirement of a specialized fluorescent reporter gene animal model, we investigate the coexpression pattern of Sox9EGFPlo and the cluster-of-differentiation (CD) marker, CD24, a cell surface marker that has been reported to be a putative stem cell biomarker and is easily detected by use of widely available FACS antibodies (32, 45).
MATERIALS AND METHODS
Mice/genotyping.
The Sox9EGFP mouse line was originally generated as part of the GENSAT Brain Atlas Project (15) and contains genomic integration of a modified BAC (RP32–140D18) with ∼75.5 kb upstream and ∼151 kb downstream sequence to Sox9. Frozen Sox9EGFP mouse embryos were obtained from the Mutant Mouse Regional Resource Center (University of California-Davis) and reconstituted by transfer into foster mice. All mice are on the outbred CD-1 strain and were maintained as heterozygotes on the CD-1 genetic background. These mice breed normally and live to adulthood with no overt phenotypes due to the transgene. At ∼10 days postnatal, tail snips were viewed under an epifluorescent microscope fitted with filters for enhanced green fluorescent protein (EGFP) visualization. A high level of EGFP fluorescence compared with transgene-negative control mice was scored as positive for the Sox9EGFP transgene. All protocols for animal use were reviewed and approved by the University of North Carolina Institutional Animal Care and Use Committee.
Tissue dissociation/FACS.
To isolate intestinal crypt cells for FACS, small intestine epithelium was dissociated into single cells essentially as previously described (13) with the following modifications. For FACS experiments, mouse intestines were flushed with cold phosphate-buffered saline (PBS), cut open lengthwise in ∼10-cm-long pieces, and immersed in PBS/30 mM EDTA/1.5 mM DTT over ice for 20 min. The solution was disposed of and the tissue was shaken vigorously in fresh PBS/30 mM EDTA for ∼30 s before being incubated at 37°C for 10 min. Intact tissue was discarded and dissociated crypts and villi were pelleted at 2,500 rpm for 5 min. The cells were washed twice with cold PBS, resuspended in Hanks' buffered saline solution (HBSS) 0.3 U/ml dispase at 37°C, and shaken approximately every 2 min for 10 min. Then fetal bovine serum (FBS, 10%) and 100 μg DNaseI were added before the cells were passed through a 100-μm filter. Cells were pelleted at 2,500 rpm for 5 min and resuspended in 4 ml HBSS with 10% FBS, then passed through a 70-μm filter and combined with an additional 100 μg DNaseI. Equivalent numbers of cells from three animals were combined and passed through a 30-μm filter immediately prior to FACS.
Sox9EGFP and CD24-expressing cells were isolated by use of a MoFlo XDP FACS machine (Dako/Cytomation, Carpinteria, CA). Cells were collected in ice-cold Advanced DMEM/F12 supplemented with 10 μM Y27632 (Sigma, St. Louis, MO) and kept on ice throughout the sort. Forward-side scatter gating was used to exclude 99.8% of all dead cells and lymphocytes. Both forward scatter and side scatter height-width plots were used for doublet discrimination to maximize efficient single cell sorting. Sort gates for Sox9EGFP expression were defined by previous studies (14) with the inclusion of more precise parameters to include the Sox9EGFPsubLo population. Cells were sorted into the medium described above.
For CD24 or CD133 staining, 1 × 107 cells were isolated as described above and were stained with 5 μl rat anti-mouse CD24 preconjugated with Pacific blue (BioLegend, San Diego, CA) or anti-mouse CD133 preconjugated with allophycocyanin (APC) (BioLegend) in 2 ml Advanced DMEM/F12 (Invitrogen, Carlsbad, CA) for 1 h on ice. The cells were washed twice in Advanced DMEM/F12 prior to FACS. Gate decisions for CD24- or CD133-positive cells were made based on comparison with Pacific blue- or APC-conjugated isotype control stained cells.
Tissue culture.
Tissue culture methods were carried out essentially as previously described (37) with the following modifications. FACSed cells were transferred to 2.0-ml conical tubes and pelleted at 4,500 rpm for 5 min at 4°C before being resuspended at an approximate density of 2,000 cells/50 μl/well (24-well plate) in Matrigel (BD Biosciences, San Jose, CA) supplemented with 1 μM JAGGED-1 peptide (AnaSpec, San Jose, CA), 50 ng/ml EGF (R&D, Minneapolis, MN), 100 ng/ml NOGGIN (Peprotech, Rocky Hill, NJ), and 1 μg/ml R-SPONDIN1 (R&D). All mass-per-volume growth factor concentrations were calculated in respect to the 500-μl final volume of media per well. To facilitate ease of observation, 50-μl droplets of Matrigel/cell suspension were added per well to 24-well plates. After total polymerization, each formed droplet was overlaid with 500 μl Advanced DMEM/F12 containing N2 supplement (Invitrogen), B27 supplement minus vitamin A (Invitrogen), 10 mM HEPES (Invitrogen), and 10 μM Y27632 (anoikis inhibitor). Growth factors were added every other day at the same initial concentrations, with the exception of R-SPONDIN1, the dosage of which was reduced to 500 ng/ml for all time points following the initial plating. Medium was replaced every 4 days. Y27632 was withdrawn at culture day 4 with the first medium change and not included in any subsequent culture maintenance.
For passaging experiments, organoids were isolated from Matrigel with a pipette and transferred to Advanced DMEM/F12 with 10 μM Y27632 in 2.0-ml conical tubes. Single cells were obtained by mechanical trituration of isolated organoids followed by incubation in 0.3 U/ml dispase (BD Biosciences) for 20 min at 37°C combined with trituration every 5 min before medium was collected and cells were plated again in Matrigel, as described above.
cDNA preparation/real-time PCR analysis.
cDNA from ∼0.75–1.0 × 105 cells from each FACSed population (Sox9EGFPneg, Sox9EGFPsubLo, Sox9EGFPlo, and Sox9EGFPhi) was made by using RNAqueous Micro Kit (Ambion, Austin, TX) according to the manufacturer's protocols. Time between the death of the mouse to RNA extraction was kept to 3.5–4 h to ensure the highest quality of RNA. Real-time PCR was conducted for each sample in triplicate on ∼1/20,000 of the total amount of cDNA generated. Taqman probes [18S, HS99999901; Ascl2 Mm01268891_g1; Atoh1, Mm00476035_s1; CD133 Mm00477115_m1; CD24 Mm00782538_sH; ChgA Mm00514341_m1; cMyc Mm00487803_m1; CyclinD1 Mm03053889_s1; S100α4 (FSP), Mm00803371_m1; Hes1, Mm00468601_m1; Lactase Mm01285112_m1; Lgr5, Mm00438890_m1; Notch1, Mm00435245_m1; Olfm4 Mm01320260_m1; Smooth Muscle Actin (SMA), Mm01546133_m1; Sox9, Mm00448840_m1; Substance P, Mm00436880_m1] for each gene were obtained from Applied Biosystems (Pleasanton, CA) and used in reactions according to the manufacturer's protocol. 18S ribosomal RNA was amplified and used as the internal control gene for sample comparison. Delta cycle threshold values were calculated to obtain fold changes for sample comparison (27). All data points represent means ± SE from three separate experiments; statistical analysis was by one-way ANOVA and post hoc two-sample t-tests were then performed. A P value of <0.05 is considered statistically significant.
Immunostaining/microscopy.
For tissue preparation, small intestines were dissected from adult Sox9EGFP mice (>8 wk of age) and luminal contents were flushed out with PBS followed immediately by a single flush with freshly made 4% paraformaldehyde (PFA). The intestine was opened along the duodenal-ileal axis, and fixed for an additional 14–18 h at 4°C. The tissues were then prepared for cryosectioning by immersion in 30% sucrose solution for at least 24 h at 4°C. Tissues were then embedded in optimal cutting temperature (OCT) medium and frozen on dry ice. The OCT blocks were stored at −80°C until cryosectioning. Thin sections (8–10 μm) were cut on a cryostat and placed on positively charged microscope slides for staining and microscopy. This tissue preparation technique is critical for preserving the EGFP fluorescence.
Organoids were prepared for immunostaining by aspirating culture media from Matrigel cultures and fixing entire contents of each well with 4% PFA for 14–18 h at 4°C. Following fixation, PFA was removed and replaced with 30% sucrose solution for at least 24 h at 4°C. Organoids were then removed from Matrigel with a micropipette, embedded in OCT medium, and frozen on dry ice. The OCT blocks were stored at −80°C until cryosectioning. Thin serial sections (8–10 μm) were cut on a cryostat and placed on positively charged microscope slides for staining and microscopy.
To remove OCT the sections were washed twice in PBS, followed by incubation in blocking medium (5% normal goat or donkey serum, in PBS-0.3% Triton-X100) for at least 30 min at room temperature (21–25°C). Primary antibodies were applied to the tissue sections in antibody staining solution (1% normal goat serum, in PBS-0.3% Triton X-100). Dilutions were as follows: αCD326/EpCAM (rat, 1:1,000, BioLegend, 118211), αLysozyme (rabbit, 1:1,000, Diagnostics Biosystems, Pleasanton, CA, RP 028), αMucin2 (rabbit; 1:100, Santa Cruz Biotechnology, Santa Cruz, CA, sc-15334), αSOX9 (rabbit, 1:1,000, Chemicon, Temecula, CA, AB5535), αSubstance P (rat; 1:500, Chemicon, MAB356), αsucrase isomaltase (goat; 1:100, Santa Cruz Biotechnology, sc-27603). All secondary antibodies (αRabbit-Cy3, Sigma, St. Louis, MO, C2306; αRabbit-Alexafluor 488, Molecular Probes, Eugene, OR, Z-25302; αRat-Alexafluor 555, Molecular Probes, Z-25305; αGoat-Cy3) were used at a 1:500 dilution in staining buffer. Nuclei were stained with Draq5 (1:10,000, Biostatus, San Diego, CA, BOS-889-001). Background staining was negligible as determined by nonspecific IgG staining.
Day-to-day development of organoids was tracked by light microscopy. Defects in culture plate plastic were utilized as “landmarks” to facilitate consistent observation of developing organoids. Epifluorescence images were captured on an Olympus IX70 fitted with an Olympus digital camera. Objective lenses used were ×20 and ×40 with numerical apertures of 0.55 and 1.40, respectively. All confocal images represent 1.0-μm optical sections unless otherwise noted. Objective lenses for the confocal images were ×40 to ×63 with a numerical aperture of 0.1.
RESULTS
A cell population enriched for IESC biomarkers can be distinguished from progenitor cells based on Sox9EGFP levels.
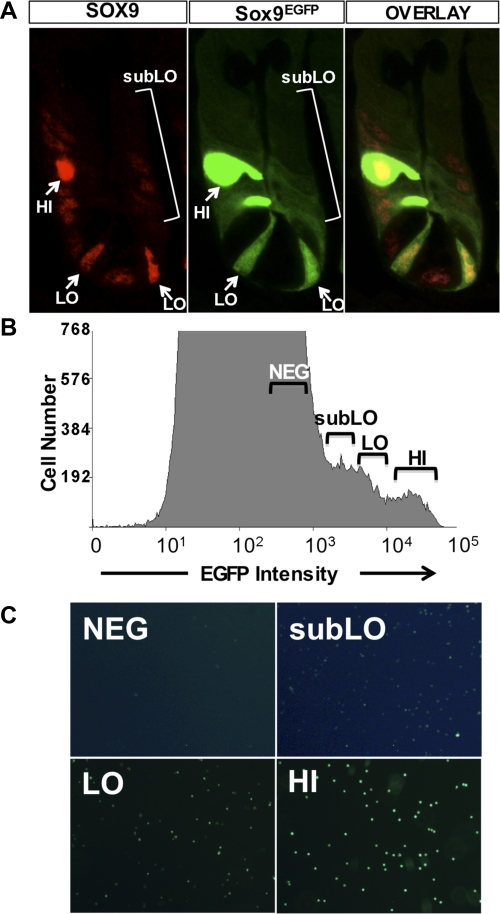
SOX9 is expressed at different levels in cells localized to the bottom region of the crypts (14, 43). Our previous study demonstrated that high levels of SOX9 marked mature enteroendocrine cells, and that lower levels of Sox9 marked a cell population that was enriched for the experimentally validated biomarker, Lgr5. Our initial report regarding Sox9 expression in the small intestine crypts did not describe a third population that can be visualized in all crypts of the small intestine by immunostaining for endogenous SOX9 and also by Sox9EGFP transgene expression (Fig. 1A). We have defined these cells as “sub-low” (subLo)-expressing cells based on low but detectable levels of SOX9 and Sox9EGFP and show that these cells localize to the transit-amplifying progenitor cell population (Fig. 1A and Supplemental Fig. S1; the online version of this article contains supplemental data). Using the mouse model in which Sox9 regulatory regions control expression levels of the EGFP reporter gene, we sought to define parameters that would allow specific separation and isolation of IESCs from progenitor populations. We reasoned that by further dividing low-expressing Sox9EGFP cells into two more precise categories, 1) Sox9EGFPlo levels and 2) the lowest detectable levels of Sox9EGFP, termed Sox9EGFPsubLo, we would potentially enrich and separate Sox9EGFPlo-expressing IESCs from Sox9EGFPsubLo transit-amplifying cells (Fig. 1B). FACS cells based on Sox9EGFPneg, Sox9EGFPsubLo, Sox9EGFPlo, and Sox9EGFPhi sort parameters demonstrated efficient separation of the four populations of Sox9EGFP-expressing cells (Fig. 1C).
Fig. 1.
Sox9EGFP is expressed at 3 different levels in the small intestinal epithelium. A: Sox9EGFP is expressed at variable levels, “HI,” “LO,” and “subLO,” in the crypts of the small intestine. HI levels are associated with postmitotic enteroendocrine cells, LO levels are associated with crypt-base columnar cells, “subLO” levels are associated with the transit-amplifying region of the crypt, and NEG levels are Sox9EGFP negative. Images represent ×1,260 original magnification. B: flow cytometric analysis indicates distinct Sox9EGFP expression levels. Gate parameters used to sort each population are indicated above each region of the histogram. C: postsort analysis indicates that single Sox9EGFP-expressing cells have been isolated based on enhanced green fluorescent protein (EGFP) status. Images represent ×200 original magnification. The image exposures for Sox9EGFPneg and Sox9EGFsubLo panels were doubled to produce images that would allow visualization of EGFP expression in these 2 populations.
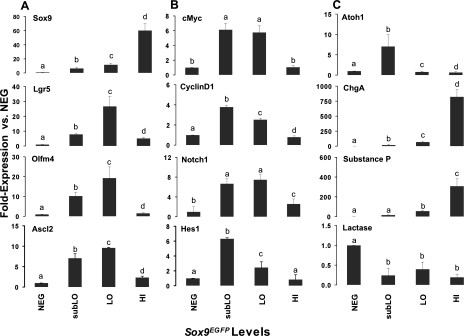
To determine the genetic profiles of each of these populations, we conducted real-time semiquantitative RT-PCR (sqRT-PCR) for “stemness” and lineage-specific genes. Sox9 mRNA levels were consistent with Sox9EGFP transgene levels, thus validating the Sox9EGFP-based FACS (Fig. 2A). Sox9EGFPlo cells show enriched expression for the validated IESC biomarker, Lgr5, as well as the two putative IESC biomarkers, Olfm4 and Ascl2, which have been shown to colocalize with the Lgr5 CBC population (42, 43) (Fig. 2A). Two hallmarks of stem/progenitor cell maintenance are active Wnt/β-catenin (23) and Notch-pathway signaling (31, 44). To determine whether these two pathways were active in Sox9EGFPlo cells, we assessed the expression levels of Wnt target genes, c-Myc and CyclinD1, and also Notch1 and its downstream target, Hes1 (Fig. 2B). Our results show increased expression of c-Myc/CyclinD1 and Notch1/Hes1 in the Sox9EGFPlo and Sox9EGFPsubLo populations, demonstrating intact signaling of these two pathways in both populations. Interestingly, Sox9EGFPsubLo cells were distinguishable from Sox9EGFPlo cells by marked upregulation of Atoh1 (Math1) (Fig. 2C). Atoh1 expression is an indicator of early stem cell differentiation into secretory lineages (46), consistent with a “progenitor cell” nature of the Sox9EGFPsubLo population. Analysis of the lineage-specific genes, Chromogranin A and Substance P, show that the Sox9EGFPhi sort parameters enrich for enteroendocrine cells. Increased Lactase expression in the Sox9EGFPneg population indicates this population is enriched for differentiated enterocytes (Fig. 2C). Together these results demonstrate that FACS based on Sox9EGFP expression levels allows for the efficient isolation and enrichment of distinct cell populations of the small intestine epithelium. These populations include, Sox9EGFPhi enteroendocrine cells, Sox9EGFPlo IESCs, Sox9EGFPsubLo progenitors/transit-amplifying cells, and Sox9EGFPneg differentiated enterocytes.
Fig. 2.
Gene expression analysis demonstrates isolation and enrichment of intestinal epithelial stem cells (IESCs), progenitors, and enteroendocrine cells by use of fluorescence-activated cell sorting (FACS). Semiquantitative RT-PCR conducted on FACSed NEG, subLO, LO, and HI cells demonstrates enrichment of IESC stem cell biomarkers in the LO populations (A), active Wnt/β-catenin and Notch1 signaling in subLO and LO populations (B), and enrichment of committed secretory progenitors in the subLO population and enteroendocrine cells in the HI population (C). Elevated lactase expression in the NEG population indicates this population is enriched for enterocytes. All data points represent means ± SE from 3 independent experiments; statistical analysis was by ANOVA and post hoc 2 sample t-tests were then performed. A P value <0.05 is considered statistically significant. Different letters above each bar represent data points that are statistically different from each other.
Single Sox9EGFPlo intestinal epithelial stem cells generate organoids and differentiate into Paneth cells, enteroendocrine cells, goblet cells, and enterocytes.
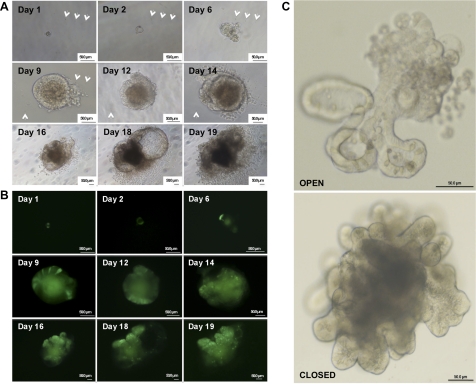
Sox9EGFPlo cells show enriched expression for established markers of multipotent IESCs, but we sought to establish whether these cells possess functional multipotent characteristics of IESCs. To assess whether Sox9EGFPlo cells have the capacity to give rise to crypt/villus-like structures containing all four postmitotic epithelial cell populations of the small intestine, we dissociated small intestine epithelium from Sox9EGFP mice, isolated Sox9EGFPlo cells by FACS, and plated cells into a three-dimensional (3D) culture system that has been recently described by Sato et al. (37). A single cell was identified and followed by brightfield and fluorescence microscopy from 24 h postplating to 19 days postplating to assess growth rate and morphological changes (Fig. 3 and Supplemental Fig. S5). At 24 h postplating, the single cells undergo cell division generating microcolonies, which are composed of cells that maintain Sox9EGFP expression (Fig. 3B). By 48 h postplating the microcolony develops a pseudolumen that is fully developed by 6 days postplating. The pseudolumen contains dead or apoptotic cells, which were indicated by punctate, fractured nuclear staining (data not shown), and high levels of autofluorescence. Clear segregation of cell types expressing distinct levels of Sox9EGFP was observed by day 9 and was coincident with the initial formation of crypt buds, which were fully developed by days 12–16 (Fig. 3C). Two gross morphologies of “organoids” (39) were observed, one defined by an “open” configuration where the dead and apoptotic cells were expelled into the Matrigel, and a second defined by a “closed” confirmation where the dead and apoptotic cells were expelled into a central luminal cavity (Fig. 3C). Organoids were allowed to grow in culture until they either became disorganized or grew large enough to come into contact with the bottom of the plate, which usually occurred between 16 and 20 days in culture. At this point, the organoids were dissociated by mechanical trituration and replated into the 3D culture system. The replated cells gave rise to multiple organoids for multiple passages (at least 6 to date), indicating the self-renewal capacity of the IESCs within the original organoid. Single Sox9EGFPlo cells were able to give rise to organoids with a well-defined pseudolumen (days 2–6) at an average incidence of 5% (5 organoids/100 FACSed Sox9EGFPlo cells plated); however, fully formed organoids with well-defined crypt units that were present at days 12–16 were more rare and were present at an average incidence of 1%. It is important to note that upon culturing Sox9EGFPneg, Sox9EGFPsubLo, and Sox9EGFPhi cells mature organoids never formed, further validating that Sox9EGFPlo sort parameters were effectively isolating the multipotent, self-renewing IESC population.
Fig. 3.
Single Sox9EGFPlo cells form complex organoids in vitro. A: single cells develop into large aggregates with crypt and villus-like structures over 7–10 days. The central pseudolumens of the single-cell-derived organoids expel apoptotic cells resulting from the sloughing of terminally differentiated epithelial cells. Arrows mark defects in culture plastic used as “landmarks” to track individual cells through early developmental phases into formed organoids. At day 9 the organoid grew large enough to obscure the tracking defects; therefore, additional tracking marks were identified (white arrows). B: epifluorescent images of organoids depicted in A. Note that the green pseudolumen is nonspecific autofluorescence and not EGFP fluorescence. Differential Sox9EGFP expression patterning remains evident throughout expansion in vitro. C: organoids form both open and closed morphologies, based on developmental positioning of the apoptotic pseudolumen. Apoptotic cells slough openly into Matrigel in the “open” morphology and are seen as a dense, dark region in the “closed” morphology. Ages of organoids are 12 days postsort (top) and 15 days postsort (bottom). Organoids are magnified ×300 (top) and ×200 (bottom).
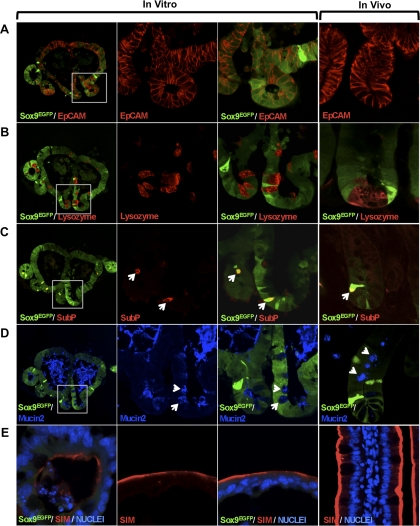
To determine whether Sox9EGFPlo cells in the organoid were able to give rise to the four postmitotic differentiated cell types of the small intestine epithelium, we sectioned a single organoid and used immunostaining to identify the cell lineages represented. The data indicate that cells in the organoid are epithelial in nature as demonstrated by positive immunostaining with epithelial cell adhesion molecule (EpCAM; Fig. 4A). The three secretory lineages were represented and identified by immunostaining for Lysozyme (Paneth cell; Fig. 4B); Substance P (enteroendocrine; Fig. 4C); and Mucin2 (goblet; Fig. 4D). An interesting observation was that high levels of Mucin2 were present in the pseudolumen, which is physiologically consistent with correct cellular polarity observed in normal gut function. Enterocytes were identified by immunostaining for the brush-border enzyme sucrase isomaltase (Fig. 4E). These data indicate that single Sox9EGFPlo IESCs have the capacity to function as multipotent IESCs in culture.
Fig. 4.
Sox9EGFPlo cells are capable of differentiation into all postmitotic cell types associated with the intestinal epithelium. A: epithelial cell adhesion molecule (EpCAM) staining indicates organoids cells are epithelial. B: lysozyme marks Paneth cells near the base of cryptlike structures. C: enteroendocrine cells are labeled with Substance P and also restricted to the base of cryptlike structures in the organoids. D: Mucin2 demonstrates the goblet cell phenotype in cryptlike structures. Secreted Mucin2 in the pseudolumen suggests that goblet cells are correctly polarized. E: sucrase isolmaltase (SIM) labels the apical ends of enterocytes lining the pseudolumen in the organoids. White boxes in the far left images (×100 original magnification) represent areas magnified in the center 2 columns (×800 original magnification). White arrows point to representative cells. Associated expression patterns in vitro correlate with those observed in vivo (far right). The organoid represented was cultured for 12 days.
One pressing question is whether contaminating mesenchymal cells, which may be attached to a Sox9EGFPlo IESCs and therefore undetectable by FACS doublet-discrimination parameters, are providing instructive signals and mitogens to the organoids. To address this question, we visually assessed postsort cells using fluorescence microscopy and never identified a cell aggregate containing a Sox9EGFP-negative cell. A more objective approach to answer this question was to section an entire organoid and immunostain for biomarkers of fibroblasts [fibroblast-specific protein (FSP)-S100α4] and myofibroblasts [smooth muscle actin (SMA)], the major cell types that have been proposed to support the stem cell niche and crypt formation in vivo (21). Although tissue sections of intact small intestine demonstrated robust immunostaining staining for fibroblast/myofibroblast markers, no staining was ever observed in 12- to 16-day-old organoids, indicating that cells marked by SMA or S100α4 do not appear to be supporting in vitro growth of organoids (data not shown).
CD24 can be used to isolate and enrich for IESCs that form organoids in culture.
To date, no cell surface marker that can be used to FACS-isolate normal IESCs has been identified. Although investigators have been searching for a single biomarker that exclusively marks stem cell populations, no single gene has been demonstrated to possess this characteristic. Prevailing evidence suggests that pure stem cell populations may be isolated from a larger pool of heterogeneous cells by using a combination of cell surface biomarkers that produce a genetic signature for the stem cell. For example, CD markers are a class of cell surface proteins that have been used to successfully FACS isolate multipotent stem cells from other epithelial tissues, such as the brain, pancreas, and mammary glands (3, 29, 32, 45). One common feature of all these epithelial stem cell populations is that CD24 is expressed on the cell surface and can be used in combination with FACS to enrich for stem cells from the heterogeneous cell isolates. CD24 is a short signal-transducing cell surface protein that was first characterized in B lymphocytes (17–19) and has been since shown to be a ligand for P-selectin, an adhesion molecule found on activated platelets and endothelial cells (34). The role of CD24 in the small intestine epithelium is currently not understood.
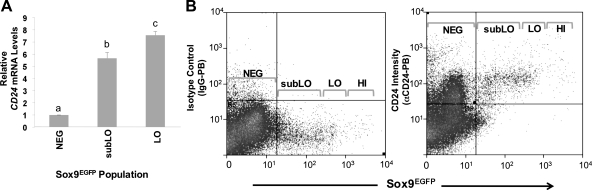
To explore the possibility of using CD24 as potential enrichment factor for IESCs, we conducted gene expression analysis for CD24 by real-time sqRT-PCR on Sox9EGFPlo and Sox9EGFPsubLo FACSed cells and compared their CD24 expression with CD24 expression in Sox9EGFPneg cells (Fig. 5A). The data show a five- to sevenfold increase in CD24 mRNA in Sox9EGFPsubLo and Sox9EGFPlo cells compared with Sox9EGFPneg cells (Fig. 5A). Likewise, flow cytometric analysis demonstrates CD24 protein is expressed in >99% of Sox9EGFP-positive cells (Fig. 5B). These data support the concept that CD24 is a potential candidate biomarker for isolating and enriching IESCs from nontransgenic tissue sources.
Fig. 5.
Small intestine epithelial stem/progenitor cells express CD24 mRNA and protein. A: semiquantitative RT-PCR (sqRT-PCR) demonstrates that CD24 is expressed 5.5- to 7.2-fold higher in the subLO and LO cells compared with Sox9EGFP-negative cells. All data points represent means ± SE from 3 independent experiments; statistical analysis was by ANOVA, and post hoc 2-sample t-tests were then performed. A P value <0.05 is considered statistically significant. Different letters above each bar represent data points that are statistically different from each other. B: flow cytometric analysis indicates that nearly all Sox9EGFP-positive cells express CD24 protein. Left: Pacific blue-conjugated isotype antibody control. αCD24-Pacific blue conjugated antibody. Right: gray brackets in the histograms represent the FACS gating parameters.
CD24 is also present on the surface of some non-IESC types; therefore, FACS gates must be clearly defined to specifically identify where the IESC population is located on a CD24/forward scatter FACS histogram. Since organoids could only be generated from IESCs isolated by using Sox9EGFPlo parameters, we reasoned that Sox9EGFPlo status could be used as a color-gating tool to identify where IESCs were located on a FACS histogram with only CD24 and forward scatter as variables. Color gating is an algorithmic feature of flow cytometry software that allows a defined parameter (such as EGFP expression status) to be mapped back onto cells on a FACS histogram that does not contain an EGFP variable. In other words, Sox9EGFPlo cells can be recognized on a CD24 vs. forward scatter histogram to reveal where the Sox9EGFPlo IESC population falls within CD24-expressing cells. The significance of using this strategy is that defined sort parameters (or “sort gates”) can be specifically drawn to exclude the majority of CD24-expressing cells that do not contain IESCs. The IESC-specific CD24 gates can then be used as a standard to isolate and enrich for IESCs from any mouse intestinal epithelial tissue source, not just from specialized fluorescent reporter gene mice.
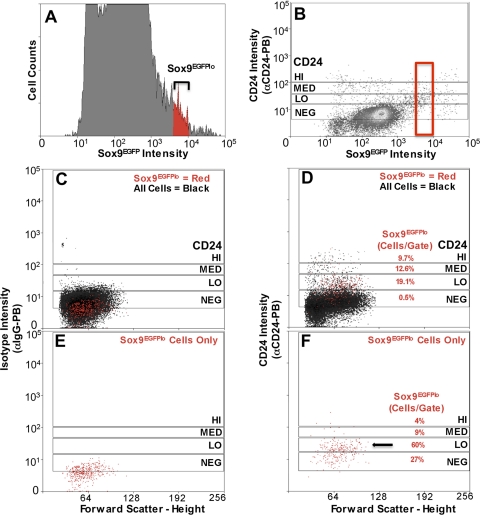
Sox9EGFPlo intensities (Fig. 6A, red events) that produced enrichment of IESCs and generation of organoids were used to define sort gates on a bivariate histogram of CD24-stained intestinal epithelial cells from a Sox9EGFP mouse (Fig. 6, C–F, red events). A bivariate EGFP/CD24 histogram indicated there were four discernable populations, which were used to define sort gates (CD24-high, medium, low, and negative) (Fig. 6B). Color-gating only the Sox9EGFPlo cells onto the CD24/FSC histogram shows that a majority (60%) of all Sox9EGFPlo cells fall into the CD24lo gate (Fig. 6, D and F; red events) representing 19% of all cells within the CD24lo sort parameter (Figs. 6F and 7A). CD24med and CD24hi gates contain significantly less Sox9EGFPlo cells, 9 and 4%, respectively. A significant number of Sox9EGFPlo cells (27%) fell in the CD24neg sort parameter; however, this represented only 0.5% of all cells within that gate, indicating a deenrichment of IESCs in the CD24neg population (Figs. 6, D and F, and 7A).
Fig. 6.
Flow cytometric analysis shows that a majority Sox9EGFPlo cells fall within CD24lo expression pattern. A: Sox9EGFP expression of dissociated small intestine epithelial cells on a univariate EGFP histogram. Black brackets indicate gate parameters. Color gating (red) allows visualization of Sox9EGFPlo population in histograms A–F. B: Sox9EGFP/CD24 bivariate histogram used to define the 4 CD24 gate parameters [negative (NEG), low (LO), medium (MED), and high (HI)]. C and E: IgG control antibody indicates there is no significant nonspecific staining. D and F: αCD24-PB (Pacific blue) antibody labels 73% of Sox9EGFPlo cells. D: percentages represent the number of Sox9EGFPlo-expressing cells in each gate. Mean percentage ± SE for each population are as follows: NEG, 1.67 ± 0.25%; LO, 25.63 ± 0.95%; MED, 12.23 ± 1.48%; HI, 9.33 ± 0.91%. E and F: just Sox9EGFPlo cells color backgated onto the CD24/FSC histogram to highlight their distribution on the histogram. F: percentages represent the relative number of all Sox9EGFPlo cells in each gate. Mean percentage ± SE for each population are as follows: NEG, 50.7 ± 5.37%; LO, 42.47 ± 4.34%; MED, 10.23 ± 1.16%; HI, 0.50 ± 0.00%.
Fig. 7.
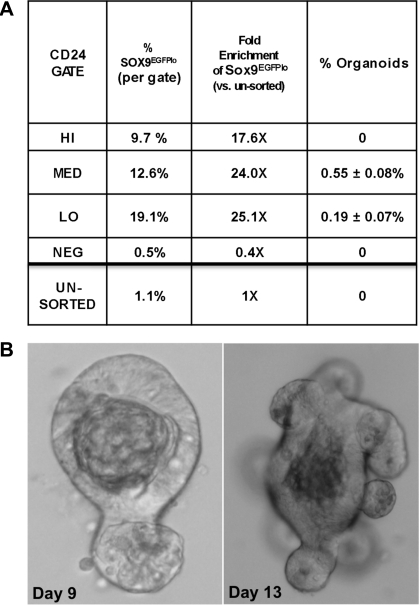
CD24 can be used as a biomarker to isolate and enrich for single cells capable of producing sustainable, differentiated organoids. A: table representing the enrichment of putative IESCs by using CD24 expression levels as sort criterion. Percent Sox9EGFPlo cells/gate = number of Sox9EGFPlo cells/total number of cells per gate. Fold enrichment, percent Sox9EGFPlo cells per gate/percent Sox9EGFPlo cells in all sorted cells. CD24lo sort parameters demonstrate the highest potential for enrichment of IESCs. There is a ×25.1 enrichment for Sox9EGFPlo cells in the CD24lo sort parameters compared with ungated cells. Organoid generating IESCs can be isolated only with the CD24lo (5 organoids in every 1,000 cells) and CD24med (1 organoid in every 1,000 cells) sort parameters. B: representative organoids derived from single CD24lo-expressing cells (left, day 9; right, day 13).
As proof-of-concept, the same color-gating strategy was used to determine whether CD133, a validated IESC marker by lineage tracing, would also be a candidate cell surface marker for the isolation of Sox9EGFPlo IESCs. CD133 has a broad expression pattern that encompasses the CBC cells up the crypt axis to the transit-amplifying cell zone (39). sqRT-PCR and flow cytometric analyses indicate that CD133 mRNA and protein are expressed in the Sox9EGFPsubLo and Sox9EGFPlo populations (Supplemental Fig. S2). Color gating demonstrates that the Sox9EGFPlo population (94.8%) expresses CD133 on the surface; however, the CD133-APC fluorescence shift is too low to enable significant separation of the Sox9EGFPlo IESCs from CD133-negative cells (Supplemental Fig. S3). If the sort gates indicated on the CD133/FSC histogram were used to isolate Sox9EGFPlo IESCs, only 10.7% of the cells would be Sox9EGFPlo cells representing approximately half the enrichment factor compared with using CD24 as a FACS marker.
To identify which CD24 gate contained cells with functional IESC characteristics, we introduced the four cell populations (CD24 negative, high, medium, and low) into organoid culture conditions. Organoids were generated only from CD24lo and CD24med populations at an incidence of 1:200 (CD24lo) and 1:1,000 (CD24med) (Fig. 7, A and B). There was no overt difference in the morphology or growth between organoids derived from cells isolated from the CD24lo and CD24med gates. No organoids formed from CD24neg or CD24hi populations, indicating that the cells in these two populations do not have stem cell capacity in culture conditions. Organoids derived from CD24lo and CD24med cells demonstrated multipotency (Supplemental Fig. S4) and have been able to be expanded in culture for at least four passages to date.
DISCUSSION
In this study, we report that Sox9EGFPlo expression levels can be used to isolate IESCs that are functionally defined by their ability to form organoids in vitro, self-renew, and contain all four postmitotic cell types of the small intestine epithelium. Additionally, we show that distinct levels of CD24 expression enrich for IESCs and can be used to efficiently FACS and isolate IESCs from nontransgenic intestinal tissue. CD24 is, to our knowledge, the first reported cell surface marker that can be used to FACS isolate and enrich for IESCs with functional characteristics.
Although we have previously reported two distinct levels of Sox9 expression in the small intestine epithelium (14), in this study we describe a third level of Sox9 expression termed Sox9EGFPsubLo. Localization of Sox9EGFPsubLo cells within the transit-amplifying zone of the crypt, FACS enrichment for genetic markers of dividing progenitor cells, and the lack of multipotent and self-renewal capacity of Sox9EGFPsubLo cells suggest that sub-low levels of Sox9 mark transit-amplifying progenitors. Using Sox9EGFPsubLo expression status as a cell isolation parameter will facilitate future studies on the small intestine epithelial progenitor population.
An emerging concept in the literature is that Sox-transcription factors function in a dose-dependent manner to affect competence, potency, and proliferation of stem and progenitor cell populations (30, 38, 41). Results from this present study and our previous work indicate that high levels of Sox9 are associated with nondividing cells, and a decreasing gradient of Sox9 is associated with stem/progenitor populations characterized by increased proliferative capacities (14). The control of proliferation by Sox9 is in part exerted through modulating Wnt/β-catenin signaling (2, 5, 14). Sox9 has been reported to be a downstream target of Wnt signaling and appears to function by suppressing Wnt signaling in a negative feedback loop mechanism (2, 5). High levels of transiently expressed SOX9 in a nontransformed intestinal epithelial cell line halt proliferation indicating that SOX9 influences expression of proliferation genes (14); however, the exact mechanism by which SOX9 exerts control of proliferation is not understood and warrants further investigation into downstream effector genes. The ability to specifically sort the more rapidly dividing transit-amplifying progenitor cells from the slower dividing IESCs of the small intestine epithelium will facilitate these investigations.
The seminal study describing the culture conditions for supporting the development of IESCs in vitro reports that single Lgr5-expressing cells are capable of building crypt-villus structures without a mesenchymal niche (37). A fibroblastic cell component has traditionally been thought to be essential for crypt-villus development (20, 22, 28, 40); therefore, it was interesting that stereotypical crypt-villus units could form in the absence of a supporting cellular niche. The 3D culture conditions that facilitate crypt-villus development in culture do, however, provide many critical components similar to those that a mesenchymal cell might contribute to an IESC niche, including laminin enriched extracellular matrix (36), growth factors, and Wnt agonists (37). Under culture conditions that mimic an in vivo IESC niche, Sox9EGFPlo- and CD24-expressing IESCs do not appear to require instruction from a mesenchymal cell component to expand and build crypt-villus units in culture. Immunostaining for SMA or FSP failed to demonstrate positive staining in serial sections through Sox9EGFPlo IESC-derived organoids (data not shown). To rule out the possibility that identification of a small population of fibroblasts and/or myofibroblasts was beyond the threshold of detection of the immunostaining technology, we conducted sqRT-PCR on RNA isolated from both isolated crypts and postsorted Sox9EGFPlo cells. sqRT-PCR analysis also failed to detect SMA or FSP from either sample after 40 cycles of PCR, indicating that there is no detectable contamination of a fibroblastic cell in the FACS-sorted cells (data not shown). These data provide strong evidence that generation of crypt-villus units derived from Sox9EGFPlo cells do not require a fibroblastic cell component. This is consistent with the proposal that in this 3D culture system IESCs are able to generate a crypt-villus axis by differential intrinsic responsiveness of intestinal epithelial cells to Wnt signals rather than from extrinsic stimuli (37).
To date, FACS of single IESCs that express experimentally validated markers of IESCs has required a specialized transgenic reporter-gene mouse expressing a fluorescent protein (4, 14). A previously reported method for isolation of a putative IESC population from nontransgenic mice was via side population (SP) sorting (12). Although the SP faction was shown to be from the crypt base, and to be enriched for putative IESC markers (16), SP cells have not been demonstrated to be capable of either replication or differentiation. We report here that CD24 is a cell surface biomarker that can be used to FACS enrich for cells with functional IESC characteristics (multipotency and self-renewal), and possess the ability to form crypt/villus-like units in vitro from a nontransgenic mouse. Low CD24 expression has been used as a marker to FACS isolate stem cells from the pancreas, brain, and mammary tissue (3, 29, 32, 34, 45). The expression pattern of CD24 in the normal small intestine epithelium is unknown; however, there are reports that CD24 is associated with epithelial cells in colonic adenocarcinomas (24, 33). With CD24 used as a FACS biomarker for IESCs, organoids only developed from cells isolated from CD24lo and CD24med gates with an incidence of 0.5 and 0.1%, respectively. This represents a 2- to 10-fold reduction in organoid formation compared with using Sox9EGFPlo expression alone as the sort parameter.
Recent lineage-tracing studies in mice suggest that CD133 expression marks a subset of crypt cells that exhibit IESC characteristics (39). The use of CD133 as a cell surface marker for FACS enrichment of these IESCs was not tested in this study. Our data suggest that CD133 is not an efficient cell surface marker for enrichment of murine IESCs with antibody reagents currently available (Supplemental Fig. S3). Putative IESCs marked by low levels of Sox9EGFP did exhibit expression of CD133 mRNA and protein (Supplemental Fig. S2); however, flow cytometry indicated that CD133 expression levels appeared to be below a threshold that would allow separation and enrichment by FACS (Supplemental Fig. S3).
Although the most rigorously validated biomarkers Lgr5, Bmi1, CD133, and Sox9 appear to mark IESC populations, none of these markers has been shown exclusively to mark IESCs. For instance, like Sox9EGFP, Lgr5EGFP is expressed at high and low levels in the crypt base. It was demonstrated that low-expressing Lgr5EGFP cells rarely gave rise to organoids (37), suggesting that Lgr5 also marks a non-IESC population. Furthermore, only 6% of Lgr5EGFPhi cells demonstrated self-renewal and multipotent capacity (37). Although the low number of Lgr5EGFPhi cells demonstrating functional IESC characteristics could be the result of isolation and culture conditions, it is also possible that not all of these cells are IESCs. We propose that a combinatorial approach using multiple genetic markers will provide a more robust genetic signature for the IESC and allow for a more pure population of IESCs to be isolated by FACS. In this regard, we used CD24- and CD133 antibodies to costain crypt cell preparations with the goal of further identifying a more pure population of IESCs. After mapping the Sox9EGFPlo cells back onto the CD24 vs. CD133 histogram there was no further enrichment of the Sox9EGFPlo cells or any unique cell population compared with using CD24-antibody alone (data not shown). This result might be due to the low fluorescent signature of the CD133-stained cells. To rule out the possibility that CD133 can be used an efficient IESC enrichment marker in mice, a more robust CD133 antibody must be developed. As this study demonstrates, the Sox9EGFP mouse model combined with color-gating flow cytometry provides a powerful tool to screen other known and uncharacterized cell surface markers for their capacity to mark the IESC population. The ability to use CD24 as an IESC enrichment factor represents a new and transformative approach to IESC isolation, as well as to the study of IESC maintenance, expansion, and differentiation.
GRANTS
This work was funded by the National Institutes of Health, 1-K01-DK080181-01, the American Gastroenterological Association Research Scholar Award, the North Carolina Biotechnology Center Grant, and the UNC-Chapel Hill the Center for Gastrointestinal Biology and Disease, 5P30DK034987 (S. T. Magness).
DISCLOSURES
No conflicts of interest are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the University of North Carolina (UNC) Center for Gastrointestinal Biology and Disease Imaging and Histology COREs (5P30DK034987), the UNC Neuroscience Confocal Imaging Facility (P30 NS045892-04), the UNC Chapel Hill Mutant Mouse Regional Resource Center, and the UNC Flow Cytometry CORE. We thank Dr. Toshiro Sato for a detailed protocol containing additional reagents not included in the Nature article (37). In addition, we thank Michael Cronce for technical support and Drs. P. Kay Lund, Susan Henning, Christopher Dekaney, Andrea Portbury, and Laurianne van Landeghem for useful discussions.
REFERENCES
- 1.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, de Crombrugghe B. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci USA 102: 14665–14670, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Devel 18: 1072–1087, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100: 3983–3988, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, Bibeau F, Scherer G, Joubert D, Hollande F, Blache P, Jay P. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol 178: 635–648, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjerknes M, Cheng H. The stem-cell zone of the small intestinal epithelium. III. Evidence from columnar, enteroendocrine, and mucous cells in the adult mouse. Am J Anat 160: 77–91, 1981 [DOI] [PubMed] [Google Scholar]
- 7.Cheng H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. II. Mucous cells. Am J Anat 141: 481–501, 1974 [DOI] [PubMed] [Google Scholar]
- 8.Cheng H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. IV. Paneth cells. Am J Anat 141: 521–535, 1974 [DOI] [PubMed] [Google Scholar]
- 9.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am J Anat 141: 461–479, 1974 [DOI] [PubMed] [Google Scholar]
- 10.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. III. Entero-endocrine cells. Am J Anat 141: 503–519, 1974 [DOI] [PubMed] [Google Scholar]
- 11.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat 141: 537–561, 1974 [DOI] [PubMed] [Google Scholar]
- 12.Dekaney CM, Fong JJ, Rigby RJ, Lund PK, Henning SJ, Helmrath MA. Expansion of intestinal stem cells associated with long-term adaptation following ileocecal resection in mice. Am J Physiol Gastrointest Liver Physiol 293: G1013–G1022, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Dekaney CM, Rodriguez JM, Graul MC, Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology 129: 1567–1580, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Gulati AS, Ochsner SA, Henning SJ. Molecular properties of side population-sorted cells from mouse small intestine. Am J Physiol Gastrointest Liver Physiol 294: G286–G294, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Hough MR, Rosten PM, Sexton TL, Kay R, Humphries RK. Mapping of CD24 and homologous sequences to multiple chromosomal loci. Genomics 22: 154–161, 1994 [DOI] [PubMed] [Google Scholar]
- 18.Kay R, Rosten PM, Humphries RK. CD24, a signal transducer modulating B cell activation responses, is a very short peptide with a glycosyl phosphatidylinositol membrane anchor. J Immunol 147: 1412–1416, 1991 [PubMed] [Google Scholar]
- 19.Kay R, Takei F, Humphries RK. Expression cloning of a cDNA encoding M1/69-J11d heat-stable antigens. J Immunol 145: 1952–1959, 1990 [PubMed] [Google Scholar]
- 20.Kedinger M, Duluc I, Fritsch C, Lorentz O, Plateroti M, Freund JN. Intestinal epithelial-mesenchymal cell interactions. Ann NY Acad Sci 859: 1–17, 1998 [DOI] [PubMed] [Google Scholar]
- 21.Kedinger M, Lefebvre O, Duluc I, Freund JN, Simon-Assmann P. Cellular and molecular partners involved in gut morphogenesis and differentiation. Phil Trans R Soc Lond 353: 847–856, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kedinger M, Simon-Assmann P, Bouziges F, Haffen K. Epithelial-mesenchymal interactions in intestinal epithelial differentiation. Scand J Gastroenterol 151: 62–69, 1988 [DOI] [PubMed] [Google Scholar]
- 23.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 19: 379–383, 1998 [DOI] [PubMed] [Google Scholar]
- 24.Lim SC, Oh SH. The role of CD24 in various human epithelial neoplasias. Pathol Res Pract 201: 479–486, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci USA 100: 9360–9365, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori-Akiyama Y, van den Born M, van Es JH, Hamilton SR, Adams HP, Zhang J, Clevers H, de Crombrugghe B. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 133: 539–546, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am J Physiol Cell Physiol 277: C183–C201, 1999 [DOI] [PubMed] [Google Scholar]
- 29.Pruszak J, Ludwig W, Blak A, Alavian K, Isacson O. CD15, CD24 and CD29 define a surface biomarker code for neural lineage differentiation of stem cells. Stem Cells 27: 2928–2940, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development (Cambridge, UK) 134: 2521–2531, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep 9: 377–383, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rietze RL, Valcanis H, Brooker GF, Thomas T, Voss AK, Bartlett PF. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature 412: 736–739, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Sagiv E, Memeo L, Karin A, Kazanov D, Jacob-Hirsch J, Mansukhani M, Rechavi G, Hibshoosh H, Arber N. CD24 is a new oncogene, early at the multistep process of colorectal cancer carcinogenesis. Gastroenterology 131: 630–639, 2006 [DOI] [PubMed] [Google Scholar]
- 34.Sammar M, Aigner S, Altevogt P. Heat-stable antigen (mouse CD24) in the brain: dual but distinct interaction with P-selectin and L1. Biochim Biophys Acta 1337: 287–294, 1997 [DOI] [PubMed] [Google Scholar]
- 35.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasaki T, Giltay R, Talts U, Timpl R, Talts JF. Expression and distribution of laminin alpha1 and alpha2 chains in embryonic and adult mouse tissues: an immunochemical approach. Exp Cell Res 275: 185–199, 2002 [DOI] [PubMed] [Google Scholar]
- 37.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265, 2009 [DOI] [PubMed] [Google Scholar]
- 38.Seymour PA, Freude KK, Dubois CL, Shih HP, Patel NA, Sander M. A dosage-dependent requirement for Sox9 in pancreatic endocrine cell formation. Devel Biol 323: 19–30, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snippert HJ, van Es JH, van den Born M, Begthel H, Stange DE, Barker N, Clevers H. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology 136: 2187–2194, 2009 [DOI] [PubMed] [Google Scholar]
- 40.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature 414: 98–104, 2001 [DOI] [PubMed] [Google Scholar]
- 41.Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Devel 20: 1187–1202, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137: 15–17, 2009 [DOI] [PubMed] [Google Scholar]
- 43.Van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M, Clevers H. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912, 2009 [DOI] [PubMed] [Google Scholar]
- 44.Van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435: 959–963, 2005 [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Hu J, Zhao D, Wang G, Tan L, Du L, Yang J, Hou L, Zhang H, Yu Y, Zhang H, Deng H, Ding M. NestinnegCD24low/- population from fetal Nestin-EGFP transgenic mice enriches the pancreatic endocrine progenitor cells. Pancreas 31: 385–391, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294: 2155–2158, 2001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.