Abstract
Sarcomere length (SL) is an important determinant and indicator of cardiac mechanical function; however, techniques for measuring SL in living, intact tissue are limited. Here, we present a technique that uses two-photon microscopy to directly image striations of living cells in cardioplegic conditions, both in situ (Langendorff-perfused rat hearts and ventricular tissue slices, stained with the fluorescent marker di-4-ANEPPS) and in vitro (acutely isolated rat ventricular myocytes). Software was developed to extract SL from two-photon fluorescence image sets while accounting for measurement errors associated with motion artifact in raster-scanned images and uncertainty of the cell angle relative to the imaging plane. Monte-Carlo simulations were used to guide analysis of SL measurements by determining error bounds as a function of measurement path length. The mode of the distribution of SL measurements in resting Langendorff-perfused heart is 1.95 μm (n = 167 measurements from N = 11 hearts) after correction for tissue orientation, which was significantly greater than that in isolated cells (1.71 μm, n = 346, N = 9 isolations) or ventricular slice preparations (1.79 μm, n = 79, N = 3 hearts) under our experimental conditions. Furthermore, we find that edema in arrested Langendorff-perfused heart is associated with a mean SL increase; this occurs as a function of time ex vivo and correlates with tissue volume changes determined by magnetic resonance imaging. Our results highlight that the proposed method can be used to monitor SL in living cells and that different experimental models from the same species may display significantly different SL values under otherwise comparable conditions, which has implications for experiment design, as well as comparison and interpretation of data.
Keywords: two-photon imaging, Monte-Carlo simulation, magnetic resonance imaging, confocal microscopy, ventricular tissue slice
end-diastolic sarcomere length (SL) is a key determinant of cardiac mechanical function. It has implications beyond the adjustment of cellular contractility to local mechanical demand, e.g., by modulating mechanoelectric interactions and energetics, and it may be relevant for other aspects such as second messenger and drug actions on the heart.
There is a distinct lack of in vivo SL data, however, and current SL estimates are largely based on observations made in explanted and fixed tissue (10, 23, 36), or in isolated cells (24). Tissue fixation and histological processing are known to affect cardiac ultrastructural properties (7), whereas isolation of myocardial fragments for ex vivo investigations may alter the complex interaction of microarchitecture (17, 38), transmural pressure gradients (35), and, hence, change stress-strain distribution in the sample. This applies not only to excised tissue models and isolated cells [for which the tools to expose them to a controlled dynamically changing external mechanical environment are only beginning to emerge (12)] but also to isolated heart preparations. In addition to acute mechanical consequences of tissue extraction (e.g., removal of the pericardium), Langendorff-perfused hearts (whether arrested or beating) tend to show pronounced edema when exposed to perfusion with crystalloid solutions (28), and at present it is unclear how this affects SL during the course of a typical ex vivo experiment.
In this context, there are two elementary questions for research into cardiac mechanoelectrical behavior: 1) how can we relate experimental findings obtained ex vivo to conditions experienced in the intact system; and 2) how can we interrelate experimental observations made using different ex vivo model systems where cells are maintained in different mechanical environments.
Question 1 will benefit from the advent of suitable imaging techniques that can be used in the intact organism, such as the exciting prospect of catheter-tip confocal microscopy (18, 19). Question 2 calls for a systematic, side-by-side comparison of resting SL in different ex vivo preparations of the same species to integrate research findings and help to develop a framework of boundary parameters to inform high-resolution computer model development of cardiac anatomy and function (5, 22).
Several methods have been used to measure SL in intact ex vivo tissue. Laser (33) and X-ray diffraction techniques (20) have been applied to excised papillary muscle and trabeculae and, more recently, to Langendorff-perfused beating heart preparations (36). Diffraction-based techniques are suitable for measuring rapid changes in SL during contraction. However, for larger tissue samples, such as the whole heart, measured SL data provide a group average of (at least) several hundred cells and are not, therefore, well positioned to determine SL values as a function of local anatomic and/or stress-strain variations. Other established techniques are based on fast-Fourier transformation of light microscopic myocardial birefringence data (9) and work well for single cells, but are less suitable for most multicellular preparations.
More recent approaches, such as confocal microscopy (1, 2), second-harmonic generation microscopy (4), optical coherence tomography (32), or multiphoton microscopy (25), may hold the key to overcoming the above limitations. Here, we present a technique to support direct comparison of SL, measured in cardioplegically arrested cells using two-photon 1-(3-sulfonatopropyl)-4-{β-[2-(di-N-butylamino)-6-naphthyl]vinyl}pyridinium betaine (di-4-ANEPPS) fluorescence microscopy of rat isolated ventricular cardiomyocytes, ventricular tissue slices, and Langendorff-perfused whole hearts, and we provide an algorithm for semiautomated SL analysis in these model systems.
METHODS
The experimental design aims to minimize differences between experimental conditions for acutely isolated cells (referred to as in vitro) as well as for cells that remain in their normal tissue microenvironment (referred to as in situ) using tissue slices and Langendorff-perfused heart preparations. Identical imaging techniques and data analysis tools were applied to quantify SL in the different preparations at matching temperatures during cardioplegic arrest. Inherent limitations of these techniques are detailed in the discussion. All experiments had the required UK Home Office approval as designated by the Animals (Scientific Procedures) Act of 1986.
Cardiac Excision
Hearts, isolated from Sprague-Dawley rats (∼300 g) killed by cervical dislocation, were swiftly (within 150 s) mounted to a Langendorff system for coronary perfusion with normal Tyrode. After a 3- to 5-min wash, hearts entered either the protocol for cell isolation (N = 9), for cardiac slice preparation (N = 3), or for use in whole heart two-photon microscopy (N = 11) and magnetic resonance imaging (MRI, N = 4, part of N = 11 used for two-photon studies).
Cell Isolation
Rat ventricular myocyte isolation has been described in detail elsewhere (37). In short, hearts were initially Langendorff perfused with a modified Tyrode solution containing (in mM): 128.0 NaCl, 2.6 KCl, 2.0 CaCl2, 1.18 MgSO4, 1.18 KH2PO4, 10 HEPES, 20 taurine, 11 glucose, and 5,000 U/l heparin, subsequently with a cardioplegic solution (20.0 mM K+) to induce cardiac arrest. Upon induction of arrest, the perfusate was changed for 12 min to enzyme-containing solution (cardioplegic solution containing 0.24 mg/ml BlendZymes III, a blend of protease, dispase, and thermolysin; Roche). The predigested ventricular tissue was then harvested, minced, and gently agitated in 10 ml of the enzymatic solution. This solution was collected every 5 min (5 repeats), filtered, and centrifuged for 1 min at 18 g (Precision Durafuge 100). The supernatant was discarded, and the cell pellet was resuspended in cell culture medium (DMEM; Sigma). Cells were kept at room temperature until use.
Ventricular Tissue Slice Preparation
Live ventricular tissue slices were prepared using a modification of the method developed by Bussek et al. (6) In brief, hearts from three adult female rats were rapidly excised and stained with 5 μM di-4-ANEPPS (Invitrogen) for 5 min [delivered by coronary perfusion in normal Tyrode solution containing (in mM): 120 NaCl, 4.5 KCl, 22 NaHCO3, 1 MgCl2, 1.8 CaCl2, and 11 glucose; pH 7.4]. Ventricular tissue blocks were then dissected and mounted on the specimen holder of a high-precision vibratome (VT1000 S; Leica Microsystems), in ice-cold solution, supplemented with 10 mM 2,3-butanedione monoxime (BDM). Slices (350 μm) were cut in the plane of the epicardial surface (from the epicardial side) to optimize cell alignment (8) and stored in solution containing 10 mM BDM. Slices were imaged in normal Tyrode containing BDM or in cardioplegic solution (20 mM K+) to confirm lack of side effects caused by BDM.
Fluorescence Imaging
Isolated cells were loaded for 2 min with 5 μM di-4-ANEPPS in cardioplegic solution for subsequent imaging.
Whole hearts were dye loaded by coronary perfusion for 5 min with 5 μM di-4-ANEPPS in normal Tyrode. Hearts were then placed on a silicone cradle and gently stabilized by a nylon mesh placed on top of the tissue. A needle was inserted via the apex in the left ventricle to prevent perfusate accumulation (right ventricle drainage was ensured via the pulmonary artery, cut open just above the base of the heart). For imaging, hearts were cardioplegically arrested at room temperature, and coronary perfusion was maintained using constant pressure feed (90 mmHg).
Isolated cells, tissue slices, and Langendorff-perfused hearts were imaged with a TCS MP2 (Leica; Wetzlar) multiphoton microscope using 840 nm excitation and 400- to 700-nm collection wavelengths. Imaging depth in left ventricular tissue was up to 300 μm subepicardially, and images were collected at a range of depths and locations for up to 80 min after cardiac excision.
Isolated Cell SL Validation
To validate two-photon SL measurements, and to assess potential effects of di-4-ANEPPS on SL, a substudy was conducted in isolated cells using a commercially available brightfield SL measurement system (Myocyte Contractility System; IonOptix). Cells were either dye loaded (di-4-ANEPPS, loaded as described above) or exposed to dye-free vehicle solution. Cells were placed in a measurement chamber and aligned with the fast scan direction of the charge-coupled device, and SL was determined for each of the two groups by a fast-Fourier transform method in real time, as previously described (8).
Magnetic Resonance Imaging
In vivo cardiac MRI was performed as described before (31). Briefly, rats were anesthetized with 2.5% isoflurane in O2 and positioned supinely in a purpose-built cradle. Electrocardiogram (ECG) electrodes were inserted in the forepaws, and a wire loop was taped across the chest to measure respiration. The cradle was lowered in a 500-MHz vertical bore 11.7T MR system with a Bruker console running Paravision 2.1.1 and with a 60-mm birdcage coil. The ECG trigger level was adjusted so that data were acquired at end diastole.
A stack of contiguous 1-mm true short-axis ECG and respiration-gated gradient echo images (field of view 51.2 mm2, matrix size 256 × 256, echo time/repetition time 1.64/4.8 ms, 15° pulse) were acquired to cover the entire length of the left ventricle.
Subsequently, ex vivo MRI was performed on Langendorff-perfused hearts isolated from the same animals. Hearts were enclosed within a 20-mm NMR tube, perfused with cardioplegic high-K+ (20 mM) Tyrode solution at a constant pressure of 90 mmHg at 37°C (to compare with in vivo) using a purpose-built apparatus (21), and scanned using the above system with a 30-mm birdcage coil. A stack of contiguous 1-mm true short-axis fast-spin echo images (field of view 25.6 mm2, matrix size 256 × 256, echo time/repetition time 1/100 ms, 17.5° pulse) was acquired for up to 90 min from heart excision.
Image Analysis
MRI.
MRI data were analyzed using Image J, a public domain Java image processing program (http://rsbweb.nih.gov/ij/index.html/). Both the endo- and epicardial surfaces of the left ventricle were segmented in every slice, and the volumes enclosed by the respective three-dimensional contours were calculated (the septum was treated as part of the left ventricular wall). The volume enclosed by the endocardial surface was taken to represent cavity volume so that the difference between volume closed by the epicardial surface and cavity volume was taken to represent left ventricular myocardial volume (LVMV).
Two-photon microscopy.
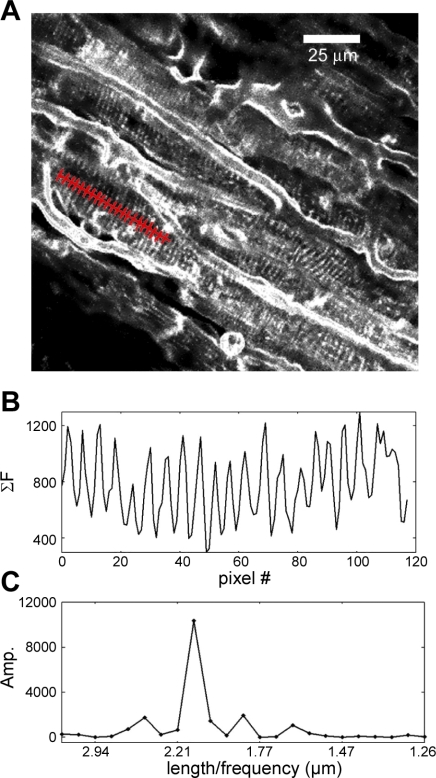
Two-photon microscopy images were collected over a wide range of magnifications and cell orientations. Data analysis methods were designed to support efficient extraction of SL information [see the Online Supplement for information on obtaining the software (Supplemental data for this article may be found on the American Journal of Physiology: Heart and Circulatory Physiology website.)]. In particular, custom software was developed that displays each image of the data series and parses scale and experimental time from metadata files generated by the Leica software. The user identifies cells in the image with clear striation patterns and draws (using a computer mouse or scan pad) a line within a cell's boundaries that follows the main axis of the cell (i.e., is roughly perpendicular to visible striation patterns). A fluorescence intensity profile for the path is obtained by summing pixel intensity values for three pixels, perpendicularly on either side of the path, along its entire length (Fig. 1). Pixel intensity is weighted by inverse of distance for a Moore neighborhood (radius = 1) to prevent aliasing. The software displays a chart of the intensity profile and its discrete Fourier transform (DFT). Based on the uniformity of the intensity profile, and the presence of a distinct frequency peak, the user selects whether to keep or discard the chosen path. The intensity profile is zero padded so that estimates of sarcomere period from the DFT are accurate to within 1%. Once the user has defined paths for all the cells in a data set with clearly visible sarcomere patterns, the path location information is saved and used in subsequent off-line error correction and analyses. Software tools for automated in-plane error correction of user-defined paths as well as filtering criteria for noisy and motion-distorted images are detailed in the Online Supplement Section 1.
Fig. 1.
Sarcomere length (SL) measurement software operation. A: image sets from 1-(3-sulfonatopropyl)-4-{β-[2-(di-N-butylamino)-6-naphthyl]vinyl}pyridinium betaine (di-4-ANEPPS)-labeled tissue are obtained using a two-photon microscope. A user-defined path, restricted by cell boundaries and entered nearly perpendicular to the striation pattern, is shown in red. B: intensity profile of the user-drawn path. Intensity values are summed for pixels perpendicular to the user path (3 pixels for each point) and plotted as shown (see methods). C: discrete Fourier transform (DFT) of the intensity profile in B.
Out of plane error estimation.
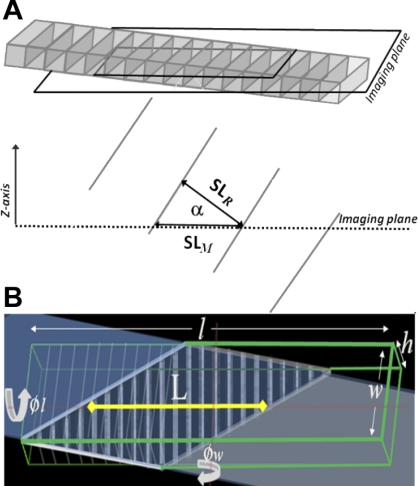
One of the key characteristics of two-photon fluorescence images of the intact heart is that the angle of the cell relative to the image plane is unknown, and any deviation off axis will cause an overestimate of SL. As shown in Fig. 2A, the real SL value (SLR) can be found from multiplying the measured SL value (SLM) by the cosine of the angle of the cell relative to the imaging plane (α). Although α is not known, its value is constrained by the cell's extent in the z-axis and length of the cell in the imaging plane (indicated by the analysis path). To visualize cell geometry and SLM in the image plane as a function of its angle, we constructed a simplified spatial model of a cell with a programatically definable striation pattern, dimensions [cell length (l), width (w), and height (h)], and orientation in three axes (given by ϕl, ϕw, and ϕh, which are the rotations around the length, width, and height axes, respectively, as show in Fig. 2B). The length and angle of the borders of the cell in the image plane (shown as a trapezoid in Fig. 2B) depend on the cell's rotation and position relative to the imaging plane. The cell's rotation relative to the imaging plane is constrained by the length of the cell in the image plane [defined as the longest line perpendicular to the striation patterns within cell boundaries (L) in Fig. 2B]. Rotation in ϕh results in an in-plane rotation and doesn't affect SLM. If the cell is rotated in ϕw only, the theoretical minimum SL can be directly determined from L and an estimated value for h. Because a cardiomyocyte's length is typically about an order of magnitude greater than its thickness, the measurement error is well constrained for large L. However, SLR is not as easily calculated if the cell is rotated in both ϕw and ϕl. Monte-Carlo simulations were used to estimate a bound on SLR from a set of SL measurements.
Fig. 2.
A: schematic illustration of how cell angle relative to the two-photon fluorescence image plane (α) affects apparent (measured) SL (SLM). The cell is rotated about the rotational axis through the cell center aligned with cell width. The cell's real SL (SLR) is a function of SLM and α. B: a diagrammatic representation of a cell (outlined by the green lines) with dimensions length (l) width (w), and height (h) and rotated by 15 and 25 degrees around the rotational axes through the cell center aligned with the cell width (ϕw) and length (ϕl), respectively, shown relative to the imaging plane (shaded section). The appearance of the cell in the image plane is given by the trapezoid (bold white lines).
Monte-Carlo estimates of SLM distribution.
We simulate the expected distribution of SLM, given a known distribution of SLR and random cell orientations relative to the image plane. The ratio of SLM to SLR and a maximum value for L are determined for the spatial cell model for all possible values ϕw and ϕl in one-degree increments and stored in a table. For each iteration of the simulation, a value of SLR is sampled from a normal distribution with known mean and SD and multiplied by a randomly sampled value for SLM/SLR from the table of stored values. The distribution of SLM as a function of minimum cell length in the image plane (Lmin) is found by filtering the results so that L > Lmin. Methodological details and simulation results are detailed in Online Supplement Section 2.
Statistical analysis of SL distributions.
Simulations indicate that, while mean SL varies with Lmin, SL mode (the SL value corresponding to the probability distributions peak) is relatively constant. The SL value for a distributions mode was found by fitting a continuous function to the distribution. The continuous function estimates of the underlying probability distributions for Monte-Carlo simulations were determined using kernel density estimation techniques [implemented with the Java BioComputing Library (11)], using a Gaussian kernel with bandwidth set to 1.06σn−1/5, where σ and n are the SD and the number of measurements, respectively (29, 34). Continuous function estimates for ex situ data were carried out with a fixed bandwidth of 0.06, corresponding to the maximum calculated bandwidth value of the whole heart and slice data sets to facilitate comparisons between these data sets. The mode of the density distributions was determined by finding the peak value with zero derivative of the continuous function.
RESULTS
Langendorff-Perfused Heart Measurements
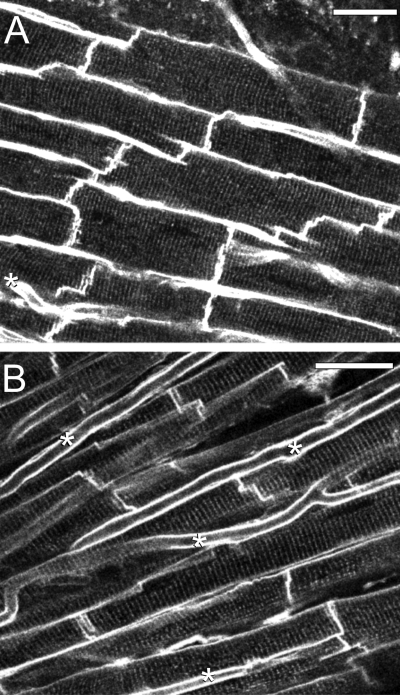
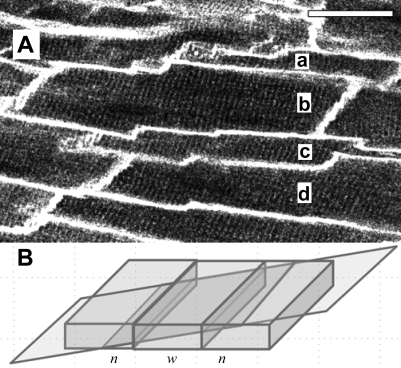
The majority (>75%) of individual images collected from di-4-ANEPPS-stained hearts contained distinct striation patterns and well-defined cell boundaries suitable for analysis. We found a range of morphological appearances that varied from densely packed cell assemblies (Fig. 3A) to more loosely arranged patterns (Fig. 3B). Densely packed cells were wide (15 μm or more), with relatively few and narrow “free” extracellular spaces between them, showing striation patterns that were in register between neighboring cells, and we assume that these may be representative of the cell arrangements within the layered structures of myocardium (17, 30). Images with more loosely associated cells were characterized by strand-like organization of predominantly narrower (<15 μm) myocytes, oriented at slightly different angles, with more prominent extracellular gaps between strands that also contained many capillaries. Striation patterns are in register within each strand, but not between neighboring cell strands, and we assume that these images represent an optical section with a transversal aspect relative to cardiac sheet structures. Many images showed cell organization patterns that were mixed or intermediate versions of the above extremes. Some images contained cells with nonrectangular boundaries, as well as repeating patterns of short-long-short or narrow-wide-narrow cell boundaries (Fig. 4). Repeating patterns of cells with apparently alternating dimensions are consistent with the optical sectioning plane having an oblique angle relative to a densely packed myocyte sheet, whereas nonrectangular boundaries are indicative of rotation relative to the cells' main axis (Fig. 2B).
Fig. 3.
Two-photon fluorescence images of di-4-ANEPPS-stained myocardium of a Langendorff-perfused rat heart displaying two different morphologies. A: cells in a densely packed tissue assembly. B: cells in a loosely packed tissue assembly. Capillaries (*) are more frequently visualized in loosely packed tissue. Scale bars are 30 μm in both panels.
Fig. 4.
A: cells a-d display an alternating narrow-wide-narrow pattern of apparent cell morphology (scale bar 30 μm). B: the pattern observed in A can be explained by the angle of the two-photon fluorescence image plane relative to tissue orientation in a modeled densely packed layer of cells, which yields a similar narrow-wide-narrow (n, w, n) pattern.
Monte-Carlo Estimates of Cell Angle Relative to the Imaging Plane
Since any deviation off-axis causes an overestimate of SL (Fig. 2A), we evaluated several approaches to estimating cell angle. Reconstructing three-dimensional structure from two-photon fluorescence image stacks proved to be problematic because of the duration needed to conduct multiple repeat scans in living tissue (motion between frames). Motion distortion, along with uncertainty of cellular dimensions, complicates efforts to assess cell angle based on visual cues such as striation angle relative to cell borders and periodic patterns in cell width or length (Fig. 4). We assume, therefore, that cell angle is random with respect to the imaging plane. Monte Carlo simulations are used to determine error estimates based on path length (Lmin) and sample size (see Online Supplement Section 2).
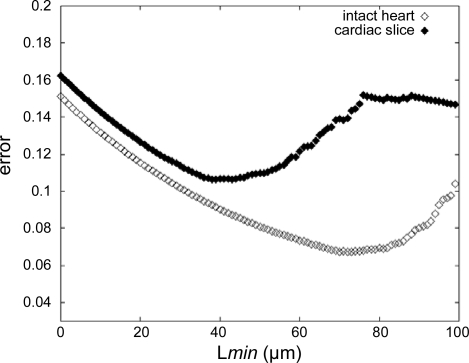
Figure 5 plots the magnitude of the bounds for the whole heart and cardiac slice measurements as a function of Lmin, where n is determined by the number of measurements with L > Lmin. The minimum of each curve indicates the optimal Lmin value for each data set. This optimal data set-specific Lmin is used to filter the whole heart and living slice data in subsequent analyses.
Fig. 5.
Error bounds estimate for the intact heart (◊) and cardiac slice (♦) preparations. The magnitude of the error is calculated based on the no. of actual measurements, available in a given data set, with longest line perpendicular to the striation patterns within cell boundaries (L) ≥ minimum cell length in the image plane (Lmin).
SL Measurements in the Intact Arrested Langendorff-Perfused Heart
Preliminary analysis of N = 11 experimental runs indicated that SL shows a 10% increase as a function of time (between 30 and 75 min after death) and that some data sets displayed variations in SL that may be caused by motion. We present data aggregated from the first 5 min of data collection for all 11 runs, followed by analysis of SL as a function of imaging time (t) for a subset of the experimental runs where data were collected for about 45 min (N = 4).
Initial SL Measurements
Mean SLM from the first 5 min of recording time (i.e., about 30 min after death) was 2.05 ± 0.18 μm (n = 300; N = 11 experiments). Means in individual experiments varied from 1.89 ± 0.06 to 2.36 ± 0.10 μm. SLM shows a dominant peak in SL frequency between 1.9 and 2.0 μm and a second lower peak between 2.3 and 2.4 μm. Although we do not rule out that a population of long SL cells exists in our preparation, further analysis (see Online Supplement Section 3) suggests that the second peak is caused by an interplay of tissue motion, the large difference in acquisition times for a single scan line and a whole frame, and the scanning direction of the two-photon microscope. If we filter the data set by excluding SLM measures obtained for cells where the analysis path falls outside of ± 30° of the fast scan axis of the microscope, we obtain a mean SL of 1.99 ± 0.12 μm. Additionally constraining the data to a path length of 72 μm or more, to minimize the out-of-plane SL measurement error, further lowers mean SL and variance. The underlying “true” mean for the whole heart preparation, based on an optimal Lmin of 72 μm (Fig. 5), is constrained between 1.90 and 1.97 μm. The results are summarized in Table 1.
Table 1.
Summary of sarcomere measurements as a function of exclusion criteria for the intact heart, cardiac slice, and single cell preparations in cardioplegic conditions
| Preparation | Exclusion Criteria | N | Mean ± SD, μm | Mode ± SD, μm | Lmin, μm | Bounds, μm | |
|---|---|---|---|---|---|---|---|
| Intact heart | None (all SLM <5 min) | 300 | 2.05 ± 0.18 | 1.95 ± 0.02 | |||
| +Angle ±30° from x-axis | 218 | 1.98 ± 0.12 | 1.95 ± 0.02 | ||||
| +Lmin | 162 | 1.99 ± 0.12 | 1.95 ± 0.02 | 72 | 1.90–1.97 | ||
| Cardiac slice | None (all SLM included) | 80 | 1.80 ± 0.13 | 1.78 ± 0.03 | |||
| +Lmin | 72 | 1.81 ± 0.13 | 1.79 ± 0.03 | 39 | 1.71–1.82 | ||
| Cells | None (all SL) | 313 | 1.71 ± 0.10 | 1.71 ± 0.02 |
The bandwidth for the kernel density estimations is set to 0.06 μm for all mode calculations to allow comparisons between mode values based on the highest bandwidth values calculated for whole heart and slice preparations. The minimum path length, Lmin, is calculated based on n values using Eq. S2 in the Online Supplement for preparations where the angle of the cell relative to the imaging plane is uncertain. SLM, measured sarcomere length (SL); N, no. of hearts.
Isolated Cell SL Measurements
Isolated cells were imaged using the same two-photon microscope and data analysis techniques used for the whole heart. SL measurements on isolated cells confirm that SL in isolated cells is shorter than in the intact isolated organ in cardioplegic conditions. Isolated cells have a mean SL of 1.71 ± 0.1 μm (n = 313; N = 9 isolations), which is significantly shorter than the SL found in the intact heart preparation (P < 0.001). In addition, in contrast to the whole heart data, the mode of the distribution is the same as the mean, and SLM does not change appreciably as a function of experimental time. Measurement bounds and results for a set Lmin are not given, since the cell angle relative to the image plane is restrained by cell attachment to the perfusion dish, abolishing significant out-of-plane errors.
In addition, SL in di-4-ANEPPS-loaded cells was compared with control cells exposed to dye-free vehicle (high-K Tyrode solution), both to determine whether di-4-ANEPPS affects SL and to validate our two-photon-based measurement protocol. Dye-loaded and unloaded cells showed no significant difference in SL (1.72 ± 0.1 μm, n = 40 vs. 1.71 ± 0.1 μm, n = 40, respectively). Furthermore, SL measured using the two-photon microscopy methods detailed here and those obtained using a bright field commercial system are not different.
Cardiac Slice SL Measurements
Images obtained from living cardiac slice preparations were not as clear as those obtained from the whole heart or isolated cell preparation and often showed localized distortions and damage caused by the slicing procedure. Accordingly, clear striations often were measurable only over relatively short path lengths compared with the whole heart and isolated cell preparations. The optimal Lmin for the cardiac slice data set was found to be 39 μm (Fig. 5), resulting in greater uncertainty in the measurement compared with the whole heart data set. The cardiac slice preparation displays SL that are relatively close to that of isolated cells, with a mean SL of 1.81 ± 0.13 μm (n = 72 measurements; N = 3 hearts involving 19 separate slices) in cardioplegic conditions. The cardiac slice data do not show such a marked dependence on path alignment with the fast scan direction of the detector, as is seen in the whole heart, suggesting that the cardiac slice is more effectively immobilized. However, variance in SL is higher than that seen in the whole heart. The increased variance may be the result of tissue damage or distortion by the slicing procedure. The mode for the distribution of SL from cardiac slice preparations is not significantly different from the mean (1.79 μm), suggesting that the majority of cells imaged were either “in-plane” or at a more restricted range of angles relative to the imaging plane. Results from the isolated cell and cardiac slice preparation are summarized in Table 1.
SL as a Function of Experimental Time
In isolated heart, SL was found to increase as a function of experiment duration. Mean SL increases rapidly between 30 (first observation) and 60 min after death. It then settles at a plateau about 10% above the first measurements. Figure 6A plots the mode from the aggregate results from four experiments, where SL is binned in time at intervals that constrain the magnitude of the bounds within 0.06 μm for Lmin = 85 μm (Eq. 3 in the Online Supplement). The mode increased from 1.96 μm [time (t) = 30 min, bounded between 1.92 and 1.98 μm] to 2.16 μm (t = 70 min, bounded between 2.12 and 2.18 μm). The mean SL increased from 2.01 ± 0.11 μm (t = 30 min) to 2.16 ± 0.07 μm (t = 70 min). No such increase in SL was seen in tissue slices or isolated cells.
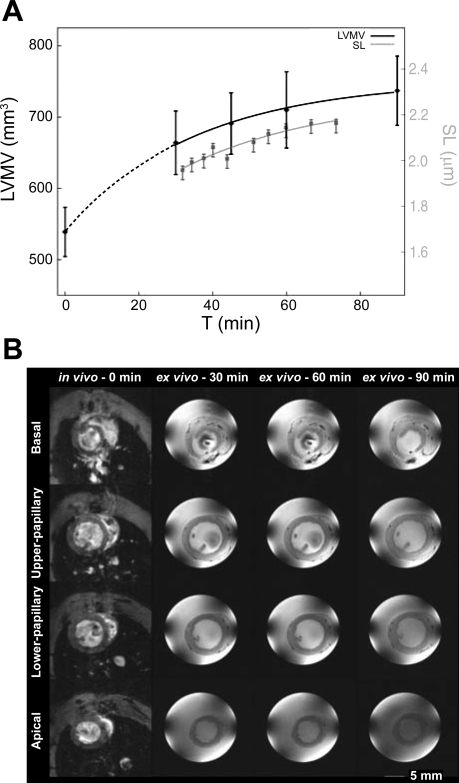
Fig. 6.
SL and tissue volume changes as a function of experimental time. A: the first SL measurement is taken at 30 min post-heart extraction (gray curve, see y-axis on right). Tissue volume shows a similar change with time (black line, see y-axis on left); the 0 time point measurement has been taken from the same heart before excision [in vivo magnetic resonance image (MRI) scan]. LVMV, left ventricular myocardial volume. B: MRI of a heart in vivo (first column) and ex vivo as a Langendorff-perfused cardioplegically arrested preparation [time (t) = 30, 60, and 90 min]. Images from different axial planes from basal (1st row) to upper papillary (2nd row) and lower papillary (3rd row) levels and near apical plane (4th row).
To test whether the gradual increase in SL in arrested Langendorff-perfused heart correlates with macroscopic changes in tissue volume associated with edema, we performed MRI studies to measure LVMV, both in vivo in the living rat at end diastole and ex situ in the arrested Langendorff-perfused rat heart preparation (Fig. 6B). LVMV increased by 24 ± 7% from in vivo to the first perfused heart measurement made 30 min after death (from 539 ± 35 to 664 ± 44 mm3; P < 0.05; Fig. 6A). A more gradual increase in LVMV by a further 11 ± 7% of the initial in vivo volume occurred between 30 and 90 min of perfusion (from 664 ± 44 to 737 ± 49 mm3; P < 0.05; Fig. 6A). For t > 30 min, the time course and amplitude of changes in LVMV in Langendorff-perfused hearts parallel the SL increase seen in the ex vivo whole heart. Experimental limitations (time required for heart extraction and preparation) prevent us from obtaining SL measurements or ex vivo MRI recordings at earlier ex vivo time points.
Local Differences in SL
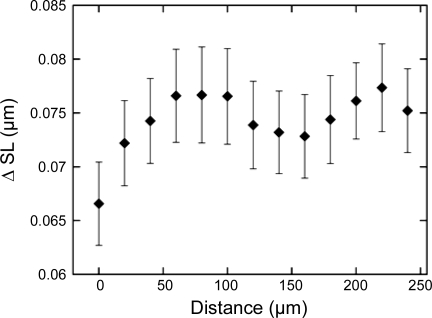
The distance between measurement sites within a single image has a small but measurable effect on SL. We compared the absolute difference in SL (ΔSL) measures as a function of distance between SL measurement sites within each image where multiple measurements were taken (Fig. 7). Distance between measurement sites is measured as the closest distance between user-defined analysis paths. Mean ΔSL for cells within 20 μm of each other are less than for cells farther apart (0.066 ± 0.062 μm, n = 6,609 vs. 0.075 ± 0.064 μm, n = 50,943, respectively, P < 0.001), but all of them are well bellow 0.01 µm, or 5% pf SL.
Fig. 7.
Difference between SL measurements (ΔSL) as a function of distance between measurement sites within an image. Distance is calculated as the minimum distance between user-defined measurement paths. Error bars are in units of SE.
DISCUSSION
We describe a method for visualizing sarcomeric striation patterns in living tissue and provide a software solution for extracting SL data from large image sets (see the Online Supplement). Two-photon microscopy of di-4-ANEPPS-loaded tissue proved to be a reliable method for visualizing striations with commonly available laboratory techniques. We were able to routinely capture hundreds of images per experiment, each containing several discrete cells with well-defined striation patterns (Fig. 3). The developed software provides semiautomated methods for efficient measurement of SL in these image data sets. Monte-Carlo simulations, using a simplified geometrical cell model, give estimates for bounds of SL measurement error and guidelines for the minimum number of measurements required for estimating the real mean SL from image data.
Determining SL Bounds in the Whole Heart Preparation
The principal difficulty in determining the underlying mean SL from optically sectioned images is that the orientation of a cells' main axis, relative to the image plane (α), is not known, and large α can result in an overestimate of SL. However, there are two lines of reasoning that suggest that the majority of measurements are taken at relatively small angles between the imaging plane and long cell axis. First, images are not taken at random; rather, the operator seeks out fields of view where striations are visible over a large portion of the image. Because the myocardium appears to be organized in layered sheets that are four to five cells thick (17), this condition is most often met when the image plane is close to parallel with such a myocyte sheet. Second, even if field of view selection is random, packed structures as shown in Fig. 3A will yield a proportionately higher number of measurements than images with loosely arranged cells. It is likely, therefore, that proportionately more SL measurements are taken at relatively shallow angles relative to the cells' primary axis. If so, the bounds estimates determined from Monte-Carlo simulations are overly pessimistic, and the SLR mean is closer to the measured mode of the SL distribution.
Isolated Heart SL Increases as a Function of Time Ex Vivo
Analysis of two-photon fluorescence images demonstrated that mean SL was not fixed in isolated arrested Langendorff-perfused heart but increased as a function of experimental time, most notably within the first half-hour. This has consequences for both single cell research (since most cell isolation procedures involve Langendorff perfusion for 45–60 min for enzymatic digestion) and for whole organ studies, which typically are conducted over a 1- to 2-h time course. It is important to note that methods exist to reduce edema in Langendorff-perfused preparations, ranging from blood perfusion techniques to the addition of osmotic agents in perfusion solutions (28). However, crystalloid solutions are the most commonly used perfusate in Langendorff-based experimental studies for arrhythmia research.
The Dynamics of SL Increase Mirrors Edema-Driven Tissue Volume Increases
MRI studies on Langendorff-perfused hearts indicate that the tissue volume increases with a similar time course and magnitude as SL increases. Although it is not possible to relate cell length (and SL) to MRI-derived tissue volume data directly, the results suggest that, on average, most myocytes are elongated at a rate and by an extent that is similar to the development of edema. This finding suggests that mechanical stress caused by the increase in myocardial volume resulting from edema might affect SL, but we do not rule out the possibility that the SL increase is caused by other, unrelated mechanisms. The use of live animal MRI enabled the measurement of heart volume in vivo, giving an effective t = 0 measurement that is missing from the SL data sets. Cardiac tissue volume increases dramatically (>20%) during the first half-hour ex vivo. Experimental limitations (because of the length of time it takes to extract and mount the heart preparation) prevent us from determining more accurately the time course for myocardial swelling. The myocardial volume may increase smoothly between t = 0 and 30 min (as is shown in Fig. 6A). Alternatively, the heart may experience a sudden increase in volume upon removal from the chest cavity (because of relaxation of anatomic constraints) and increase more gradually thereafter.
It is interesting to speculate that the large initial (t < 30 min) increase in tissue volume accounts for the differences in SL between the Langendorff-perfused heart and isolated cells. If so, SL in isolated cells may better reflect SL in vivo than the intact isolated heart preparation (which would also imply that the residual strain in the resting heart may be largely accounted for by the extracellular matrix, keeping myocytes at their force-neutral set point seen after isolation from extracellular mechanical constraints). This is an attractive idea for further research, since it would assume that cardiomyocytes are able to adjust to their mechanical environment in a way that would keep them unstrained at or near “slack diastolic dimensions” of the heart. Although “teleologically attractive” (strain inducing cell hypertrophy until the new set point is reached), this hypothesis must be subjected to stringent experimental assessment. Of note, a “backwards extrapolation” of SL change dynamics to t = 0 (in Fig. 6, for example) would indeed point toward a SL ballpark level that is close to isolated cell values.
Comparison with SL Found Using Other Methods
The mean SL for our isolated and cardioplegically arrested cells (1.71 ± 0.10 μm) is shorter than the bulk of mean end-diastolic SL values reported by other groups for contracting cells, which vary from 1.83 ± 0.12 μm (24) to 1.93 ± 0.10 μm (16) in the same species. Whereas some of the variation in results between laboratories may be caused by experimental differences, such as solution tonicity (24), an additional contribution is likely to arise from the fact that rat cardiomyocytes gain calcium during diastole [as opposed to other animals, such as rabbit or guinea pig (3)]. This diastolic calcium balance is affected by the mechanical environment (13), which may add to the differences observed between experimental preparations.
Furthermore, SL measurement in cardiac preparations is complicated by differences in sarcomere spacing within individual cells. SL differences within cells may be attributed to mechanical interference of the environment with cell structures or by dynamic effects. Sarai et al. (26) report large variations in SL within individual cells (1.8–2.0 μm) and observe changes in individual sarcomeres over time that they attribute to local calcium release [sparks; note that their generation is affected, in turn, by the mechanical environment (14)]. Although the spectral-based method used here does not give information on individual SL, the SD of SL in single cells found using our method is equivalent to that found using other techniques.
The SL measurements we obtained for myocardium in the perfused and arrested ventricle are similar to SL measurements from postmortem histological studies of the intact slack rat heart [1.96 ± 0.09 μm (10)]. X-ray diffraction techniques give an estimated value of 2.0–2.1 μm for SL at end-diastole (36); however, the heart is perfused and under pressure in this case, so the results are not directly comparable. The lack of long SL in histological studies may be caused by dehydration of the tissue during the staining process, but parallel histology and live two-photon or confocal studies are required to confirm this.
Study Limitations
There are two potential limitations associated with labeling cells with the voltage-sensitive dye di-4-ANEPPS. First, di-4-ANEPPS stains T tubules, which are largely coaligned with the z-lines of sarcomeres. However, T tubule-sarcomere alignment may be compromised in certain situations (permanently during cell disintegration; transiently during rapid pacing) so that SL values may not always represent true z-to-z distances. Given the absence of mechanical activity and near-steady-state behavior of samples, this limitation is unlikely to have affected the reported data. Second, the dye is known to cause phototoxic damage to cells via the release of reactive oxygen species (ROS) that may alter cell length. If present, this effect would be more pronounced in isolated cell preparations, since whole tissue is known to be more resistant to ROS damage and surrounding unilluminated tissue would act as structural support. However, we observe no significant changes in SL in brightly illuminated unstained vs. stained isolated cell preparations (see results), which suggests that ROS damage does not play a role in determining SL over the measurement periods used in this study.
Two-photon imaging of sarcomere striation patterns is only viable for arrested hearts because of the long time (>1.6 s) needed to scan a 512 × 512 image. To allow interrelation of findings within this study, matching experimental conditions (cardioplegic arrest) were applied to all three model systems used; this limits the extent to which SL data can be compared outside this study (i.e., to SL obtained from beating cells). Fast line scan methods can potentially be used to measure SL at millisecond resolution along one axis [see Aistrup et al. (1) for a related example]; however, cardiac contraction would dramatically change the measurement site location because of the space scales involved, unless the tissue was fixed to the objective, not the bath. The utility of our method is of limited use for researchers interested in obtaining images to characterize dynamic SL changes during cardiac contraction, but the image analysis algorithms and tools provided would be suitable for application to images obtained at different stages of contraction, if and when that becomes possible.
The relatively slow scan rate of the two-photon microscope posed additional constraints on the data. Despite being in an arrested state, images may contain spatial distortions because of motion. Because these distortions tend to manifest themselves as apparent local increases or decreases in SL, the analysis software discards all spectra with multiple or indistinct peaks as a way of removing artifactual data caused by motion. However, more gradual changes in sample position may theoretically change SLM while having relatively sharp frequency peaks. We minimize this potential source of error by excluding cells aligned far from the line scan axis (Online Supplement Section 3).
Filtering data sets by Lmin may bias measurement by preferentially including a greater percentage of larger cells. The Monte-Carlo simulations oversimplify cardiac histoanatomy by assuming a fixed cell dimension. It is clear that a range of cell lengths exists in the myocardium. However, because Lmin is always considerably less than the typical length of a cell (Lmin varies between 39 and 85 μm in this study compared with 140 μm assumed cell length; see Refs. 23 and 27) and filtering by Lmin does not appreciably change SL (SL increases by <1%; Table 1), we believe the effect this potential source of bias has on the data set is low.
Filtering by Lmin may also introduce bias, since cells with large angles relative to the image plane are discarded. Because apparent cell length is highest for cells aligned with the imaging plane and the imaging plane is always roughly perpendicular to the heart surface, the filtered data set includes proportionately more data points from tissue with this orientation. It is assumed that the SL measurements for cells in this plane are representative of the population; if not, SL measurements may be biased.
The present setup did not allow us to control the location of individual measurements or to easily change the orientation of the heart during the course of the experiment. We believe that this is the main cause of large variation in SL observed between animals and expect that we would observe a more even distribution of SL within one experiment if we imaged at a larger number of locations with optimized orientation to the fast scan axis of the microscope.
In conclusion, a key benefit of the methods presented here is that the same imaging and analysis tools can be used to directly compare SL in different popular experimental models. We observe large SL differences between isolated cells and perfused whole heart, the two most frequently used experimental systems in basic cardiac research, collected from the same species under similar conditions. Because SL plays a key role in myocyte function and because mechanical conditions further affect many other aspects of cardiac cell behavior (15), we expect that findings from these models may display systematic functional differences. Quantification of such model-dependent differences is a critical first step toward integrating experimental data between the cell and organ levels and for projecting from basic research findings to pathophysiological relevance.
GRANTS
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council, the British Heart Foundation (BHF), the European Union NormaCOR project, and the E. P. Abraham Cephalosporin Fund. P. Camelliti held a Junior Research Fellowship at Christ Church, Oxford; G. Picton was supported by an educational grant by the Frances & Augustus Newman Foundation; P. Kohl is a BHF Senior Research Fellow.
DISCLOSURES
No conflicts of interest are declared by the authors.
Supplementary Material
ACKNOWLEDGMNTS
We thank Fleur Mason for expert technical support; Philip Cobden from Professor Richard Vaughan-Jones' laboratory for provision of isolated cells; and Drs. Michiel Helmes, Martin Fink, and Martin Bishop for valuable discussions and comments on experimental design and data analysis.
REFERENCES
- 1.Aistrup GL, Shiferaw Y, Kapur S, Kadish A, Wasserstrom JA. Mechanism underlying the formation and dynamics of subcellular calcium alternans in the intact rat heart. Circ Res 104: 639–649, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Bassien-Capsa V, Fouron JC, Comte B, Chorvatova A. Structural, functional and metabolic remodeling of rat left ventricular myocytes in normal and in sodium-supplemented pregnancy. Cardiovasc Res 69: 423–431, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Bers DM. Ca influx and sarcoplasmic reticulum Ca release in cardiac muscle activation during postrest recovery. Am J Physiol Heart Circ Physiol 248: H366–H381, 1985 [DOI] [PubMed] [Google Scholar]
- 4.Boulesteix T, Beaurepaire E, Sauviat MP, Schanne-Klein MC. Second-harmonic microscopy of unstained living cardiac myocytes: measurements of sarcomere length with 20-nm accuracy. Opt Lett 29: 2031–2033, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Burton RAB, Plank G, Schneider JE, Grau V, Ahammer H, Keeling SJ, Lee J, Smith NP, Gavaghan D, Trayanova P, Kohl N. 3-Dimensional models of individual cardiac histo-anatomy: tools and challenges. Ann NY Acad Sci 1380: 301–319, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bussek A, Wettwer E, Christ T, Lohmann H, Camelliti P, Ravens U. Tissue slices from adult mammalian hearts as a model for pharmacological drug testing. Cell Physiol Biochem 24: 527–536, 2009 [DOI] [PubMed] [Google Scholar]
- 7.Choy JS, Mathieu-Costello O, Kassab GS. The effect of fixation and histological preparation on coronary artery dimensions. Ann Biomed Eng 33: 1027–1033, 2005 [DOI] [PubMed] [Google Scholar]
- 8.deBoer TP, Camelliti P, Ravens U, Kohl P. Myocardial tissue slices: organotypic pseudo-two dimensional models for cardiac research & development. Fut Med 5: 425–430, 2009 [DOI] [PubMed] [Google Scholar]
- 9.Gannier F, Bernengo JC, Jacquemond V, Garnier D. Measurements of sarcomere dynamics simultaneously with auxotonic force in isolated cardiac cells. IEEE Trans Biomed Eng 40: 1226–1232, 1993 [DOI] [PubMed] [Google Scholar]
- 10.Grimm AF, Lin HL, Grimm BR. Left ventricular free wall intraventricular pressure-sarcomere length distributions. Am J Physiol Heart Circ Physiol 239: H101–H107, 1980 [DOI] [PubMed] [Google Scholar]
- 11.Gront D, Kolinski A. Utility library for structural bioinformatics. Bioinformatics 24: 584–585, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Iribe G, Helmes M, Kohl P. Force-length relations in isolated intact cardiomyocytes subjected to dynamic changes in mechanical load. Am J Physiol Heart Circ Physiol 292: H1487–H1497, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Iribe G, Kohl P. Axial stretch enhances sarcoplasmic reticulum Ca2+ leak and cellular Ca2+ reuptake in Guinea pig ventricular myocytes: experiments and models. Prog Biophys Mol Biol 97: 298–311, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Iribe G, Ward CW, Camelliti P, Bollensdorff C, Mason F, Burton RAB, Garny A, Morphew M, Hoenger A, Lederer WJ, Kohl P. Axial stretch of rat single ventricular cardiomyocytes causes an acute and transient increase in Ca2+ spark rate. Circ Res 104: 787–795, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohl P, Bollensdorff C, Garny A. Mechano-sensitive ion channels in the heart: experimental and theoretical models. Exp Physiol 91: 307–321, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Krueger J, Forletti WD, Wittenberg BA. Uniform sarcomere shortening behavior in isolated cardiac muscle cells. J Gen Physiol 76: 587–607, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeGrice IJ, Smaill BH, Chai LZ, Edgar SG, Gavin JB, Hunter JP. Laminar structure of the heart: ventricular myocyte arrangement and connective tissue architecture in the dog. Am J Physiol Heart Circ Physiol 269: H571–H582, 1995 [DOI] [PubMed] [Google Scholar]
- 18.Liu JTC, Mandella MJ, Ra H, Wong LK, Solgaard O, Kino GS, Piyawattanametha W, Contag CH, Wang TD. Miniature near-infrared dual-axes confocal microscope utilizing a two-dimensional microelectromechanical systems scanner. Optics Lett 32: 256–258, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llewellyn ME, Barretto RPJ, Delp SL, Schnitzer MJ. Minimally invasive high-speed imaging of sarcomere contractile dynamics in mice and humans. Nature 454: 784–788, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsubara I. Xray diffraction studies of the heart. Annu Rev Biophys Bioeng 9: 81–105, 1980 [DOI] [PubMed] [Google Scholar]
- 21.Murray AJ, Lygate CA, Cole MA, Carr CA, Radda GK, Neubauer S, Clarke K. Insulin resistance, abnormal energy metabolism and increased ischemic damage in the chronically infarcted rat heart. Cardiovasc Res 71: 149–157, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Plank G, Burton RAB, Hales P, Bishop M, Mansoori T, Bernabeu M, Garny A, Prassl AJ, Bollensdorf C, Mason F, Rodriguez B, Grau V, Schneider J, Gavaghan D, Kohl P. Generation of histo-anatomically representative models of the individual heart: tools and application. Phil Trans Royal Soc A 367: 2257–2292, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez EK, Hunter WC, Royce MJ, Leppo MK, Douglas AS, Weisman HF. A method to reconstruct myocardial sarcomere lengths and orientations at transmural sites in beating canine hearts. Am J Physiol Heart Circ Physiol 263: H293–H306, 1992 [DOI] [PubMed] [Google Scholar]
- 24.Roos KP, Brady AJ, Tan ST. Direct measurement of sarcomere length in isolated cardiac cells. Am J Physiol Heart Circ Physiol 242: H68–H78, 1982 [DOI] [PubMed] [Google Scholar]
- 25.Rubart M. Two-photon microscopy of cells and tissue. Circ Res 95: 1154–1166, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Sarai N, Kihara Y, Izumi T, Mitsuiye T, Matsuoka S, Noma A. Nonuniformity of sarcomere shortenings in the isolated rat ventricular myocyte. Jpn J Physiol 52: 371–381, 2002 [DOI] [PubMed] [Google Scholar]
- 27.Satoh H, Delbridge LM, Blatter LA, Bers DM. Surface:volume relationship in cardiac myocytes studied with confocal microscopy and membrane capacitance measurements: species-dependence and developmental effects. Biophys J 70: 1494–1504, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serna DL, Powell LL, Kahwaji C, Wallace WC, West J, Cogert G, Smulowitz P, Steward E, Purdy RE, Milliken JC. Cardiac function after eight hour storage by using polyethylene glycol hemoglobin versus crystalloid perfusion. ASAIO J 46: 547–552, 2000. [DOI] [PubMed] [Google Scholar]
- 29.Silverman BW. Density Estimation for Statistics, and Data Analysis London, UK: Chapman and Hall, 1986 [Google Scholar]
- 30.Smaill BH, LeGrice IJ, Hooks DA, Pullan AJ, Caldwell BJ, Hunter PJ. Cardiac structure and electrical activation: models and measurement. Clin Exp Pharmacol Physiol 31: 913–919, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Stuckey DJ, Carr CA, Tyler DJ, Aasum E, Clarke K. Novel MRI method to detect altered left ventricular ejection and filling patterns in rodent models of disease. Magn Reson Med 60: 582–587, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Sun CW, Wang YM, Lu LS, Lu CW, Hsu IJ, Tsai MT, Yang CC, Kiang YW, Wu CC. Myocardial tissue characterization based on a polarization-sensitive optical coherence tomography system with an ultrashort pulsed laser. J Biomed Opt 11: 054016, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Tombe PP, ter Keurs HE. Sarcomere dynamics in cat cardiac trabeculae. Circ Res 68: 588–596, 1991. [DOI] [PubMed] [Google Scholar]
- 34.Turlach B.1993 Bandwidth Selection in Kernel Density Estimation: A Review. Discussion Paper 9317, Institut de Statistique Louvain la Neuve, Belgium: UCL, 1993 [Google Scholar]
- 35.Tyberg JV. Mechanical modulation of cardiac function: role of the pericardium. In: Cardiac Mechano-Electric Feedback and Arrhythmias: From Pipette to Patient, edited by Kohl P, Sachs F, Franz MR. New York, NY: Elsevier, 2005, p. 208–217 [Google Scholar]
- 36.Yagi N, Shimizu J, Mohri S, Araki J, Nakamura K, Okuyama H, Toyota H, Morimoto T, Morizane Y, Kurusu M, Miura T, Hashimoto K, Tsujioka K, Suga H, Kajiya F. X-ray diffraction from a left ventricular wall of rat heart. Biophys J 86: 2286–2294, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto T, Swietach P, Rossini A, Loh SH, Vaughan-Jones RD, Spitzer KW. Functional diversity of electrogenic Na+-HCO3− cotransport in ventricular myocytes from rat, rabbit and guinea pig. J Physiol Lond 562: 455–475, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young AA, Legrice IJ, Young MA, Smaill BH. Extended confocal microscopy of myocardial laminae, and collagen network. J Microsc 192: 139–150, 1998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.