Abstract
Rett syndrome is an X-linked autism-spectrum disorder caused by mutations in MECP2, encoding methyl CpG-binding protein 2. Since the discovery of MECP2 mutations as the genetic cause of Rett syndrome, the understanding of MeCP2 function has evolved. Although MeCP2 was predicted to be a global transcriptional repressor of methylated promoters, large-scale combined epigenomic approaches of MeCP2 binding, methylation and gene expression have demonstrated that MeCP2 binds preferentially to intergenic and intronic regions, and sparsely methylated promoters of active genes. This review compares the evolution of thought within two ‘classic’ epigenetic mechanisms of parental imprinting and X chromosome inactivation to that of the MeCP2 field, and considers the future relevance of integrated epigenomic databases to understanding autism and Rett syndrome.
Keywords: autism, chromatin, epigenetic, imprinting, MeCP2, methylation, Rett syndrome, X chromosome inactivation
Autism is a common heterogeneous neurodevelopmental disorder with complex genetic etiology. Epigenetic mechanisms act at the interface of genetic and environmental risk factors in complex genetic disorders. Two well-characterized epigenetic mechanisms, parental imprinting and X chromosome inactivation, are involved in several autism-spectrum disorders, including Angelman, Prader–Willi, 15q duplication, Rett and Fragile-X syndromes. Rett syndrome (RTT) is an X-linked disorder representing one of the most frequent causes of severe intellectual disability in females. RTT infants develop normally until 6–18 months of age, but then develop progressive loss of neurodevelopmental milestones, similar to what is observed in regressive forms of autism. Clinical features include deceleration of head growth, loss of purposeful hand movements, ataxia, loss of verbalization skills, autistic features, seizures and respiratory dysfunction. However, following a stabilization phase of the disease, most RTT patients do not meet the criteria for autism.
Rett syndrome brain samples share some neuropathologic abnormalities at the cellular level with autistic patients. Foremost among these is a similar reduction in cell-body size and dendritic branching in specific neuronal populations in RTT and autism cortex [1,2]. In addition, dendritic spines appear to be immature in both RTT and autism brain [3,4]. However, in contrast to autism, reduced brain volume and diminished neuronal size are common features of RTT [5].
The genetic defect in RTT maps to Xq28 in the MECP2 gene [6]. Therefore, RTT involves epigenetic mechanisms in at least two levels. First, MECP2 encodes methyl CpG-binding protein 2, and is therefore a known recognition and regulatory protein of epigenetic marks in the genome. Second, MECP2 maps to the X chromosome and is subject to the epigenetic process of X chromosome inactivation, and RTT affects females who are heterozygous and mosaic for the expression of MECP2 mutations.
The prediction of the function and relevance of MeCP2 has evolved over the last decade from an enzymatic model of transcriptional repression of silent methylated genes [7,8], to an active dynamic player in activity-dependent neuronal responses and the transcriptional modulation of active genes [9-11]. The evolution of thought on MeCP2 as an epigenetically regulated epigenetic regulator has paralleled and integrated with the transition away from epigenetics simply as the study of ‘quirky’ exceptions such as imprinting and X chromosome inactivation. In the post-genomic era, the scope of epigenetic mechanisms has significantly broadened to include the entire genome, and our understanding of MeCP2 has kept pace with these developments.
Classic epigenetics in the post-genomic era
The term ‘epigenetics’ refers to heritable changes to DNA, chromatin or chromosomes that do not change the DNA sequence or chromosome structure, but can alter gene expression and phenotype. Examples of epigenetic modifications exist in multiple layers in the nucleus, from methylation of CpG dinucleotides [12], to histone modifications [13], chromatin remodeling [14] and higher order organization of chromatin loops [15] and chromosome territories [16]. While epigenetic modifications are frequently mitotically heritable and occasionally meiotically heritable, reversibility of modifications is also a key discriminating feature of epigenetic inheritance compared with genetic inheritance [17]. The two best ‘classic’ examples of epigenetic mechanisms, X chromosome inactivation and parental imprinting, both require the epigenomic reprogramming of epigenetic marks every generation in meiosis. In the next generation of dynamic environmental epigenetics and epigenomics, some surprising, but informative, lessons have been learned from these two textbook epigenetic mechanisms.
Parental imprinting refers to an inheritance pattern that breaks Mendel's rule of inheritance that an allele acts independently of the parental origin. For imprinted loci, genes are differentially expressed from the maternal and paternal chromosome; thus, a mutation or deletion can result in a different phenotypic outcome depending on the parent of origin. At the organismic level, parental imprinting was first discovered by the early mouse pronuclear transplantation experiments, in which parthenogenic zygotes (derived from only egg-derived or sperm-derived nuclei) failed to result in successful post-implantation development [18]. In humans, parental imprinting is a recognized mode of inheritance for several developmental disorders, including Prader–Willi (paternal 15q11–13 deficiency), Angelman (maternal 15q11–13 deficiency), Beckwith–Weideman (maternal 11p15.5 deficiency) and Silver–Russell (paternal 11p15.5) syndromes [19,20]. Imprinted genes tend to be clustered together in regions containing both maternally and paternally imprinted genes and shared non-genic regulatory sequences termed imprinting control regions (ICRs). Imprinted genes include protein-coding transcripts, as well as noncoding antisense RNA, microRNA and small nucleolar RNA [21]. Imprinted genes are clustered in regions distinguished by epigenetic differences in replication timing, chromatin accessibility and neighboring repeat sequences [22]. While differential promoter methylation was one of the earliest markers of imprinted genes such as SNRPN and IGF2/H19, it now appears that most imprinted transcripts do not show differential DNA methylation at their 5′ ends, but rather differentially methylated regions (DMRs) overlap with ICRs in influencing the imprinting of gene clusters instead of individual imprinted genes. Genome-wide screens of expression have been generally more useful in uncovering novel imprinted genes than those screening for DMRs [23]. Interestingly, a large fraction of transcripts in the human genome are monoallelically expressed in B-lymphoblastoid clonal cell lines without showing a parent-of-origin preference [24]. Thus, differential DNA methylation has evolved within the field of imprinting from a defining mark to only one of the many available epigenetic marks determining parental origin.
For X chromosome inactivation, differential epigenetic marks can visually distinguish the two X chromosomes in mammalian females as the appearance of a Barr body. Random X chromosome inactivation occurs around the stage of implantation through the expression of a long noncoding RNA Xist which coats the inactive X chromosome, leading to the repressive epigenetic histone marks of macroH2A and H3K27 trimethylation [25]. While DNA hypermethylation has been a historical mark of the Barr body, hypermethylation is primarily limited to nongenic repetitive sequences of the X chromosome that make the internal core of the Barr body [26,27]. X chromosome inactivation is not absolute, as approximately 15% of X-linked genes escape X chromosome inactivation [28]. Microarray-based methylation analyses that exclude repetitive sequences have shown the surprising result that the active X chromosome alleles are hypermethylated in the gene bodies compared with inactive alleles [29]. The inactive alleles are hypermethylated in promoter regions, such as is observed in the common human X inactivation assay at the X-linked androgen receptor gene, suggesting that hypermethylation of the inactive X chromosome is limited to repeats and high CpG dense promoters [29,30].
The correlation of increased DNA methylation appearing in gene-dense and highly expressed regions has also held up in genome-wide microarray analyses, in which the majority of regions with the highest methylation were associated with genes, and the highest density of CpG methylation was in gene-dense R bands and subtelomeric regions [31]. Furthermore, genome-wide methylation of CpG sites within gene bodies correlated with increased, not decreased, expression [31], similar to the active X chromosome [29]. Paralleling the shift away from promoters as the primary site of epigenetic regulation, recent genome-wide analyses show that the human genome is predominantly methylated, with short unmethylated domains that are highly enriched for promoters, CpG islands and first exons [32]. In addition, despite the expectation of promoter hypermethylation in human cancers, most colon cancer and tissue-specific DNA methylation differences were observed in CpG island ‘shores’ up to 2-kb distant, rather than the CpG island promoters themselves [33]. Further studies have shown that enhancers are more variable in expression-regulating epigenetic marks than promoters [34], and the most dynamic of DNA methylation marks during embryonic stem cell differentiation were also outside of core promoter regions [35]. So the initially controversial epigenomic results from X-linked and imprinted genes are gaining broader acceptance by genome-wide studies.
Evolving role of MeCP2 in Rett syndrome
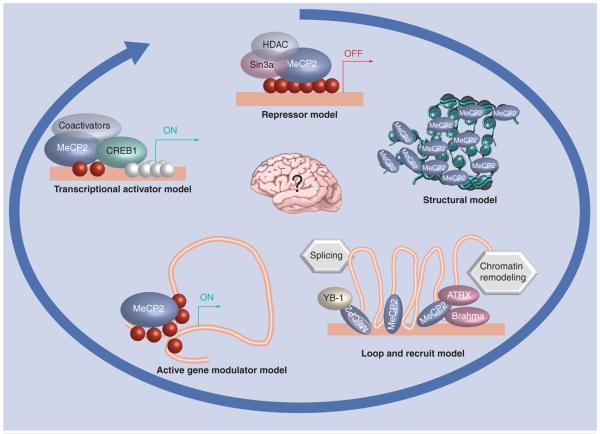
Since the discovery of MECP2 mutations as the genetic cause of RTT in 1999 [6], the understanding of MeCP2 function has evolved. Although MeCP2 was predicted to be a global transcriptional repressor of methylated genes, MeCP2 is only required during postnatal brain development, when expression is elevated [36]. The developmental regulation of MECP2 expression is highly complex, involving transcriptional increases as well as alternative splicing and alternative polyadenylation. MeCP2 selectively binds to methylated CpG residues and preferentially localizes to transcriptionally inactive heterochromatic regions of the nucleus. In the original ‘enzymatic’ model for MeCP2 function (Figure 1), MeCP2 interacts with the transcriptional repressor Sin3A and histone deacetylase (HDAC), providing a mechanism for the transcriptional repression of genes with methylated CpG sites [7,8]. MeCP2 has been shown to repress transcription of methylated promoters in vitro and SV40- or GAL4-containing promoters in vivo, and repression is partially reversed by an HDAC inhibitor [37,38]. Some studies have supported MeCP2 as a relevant repressor of methylated promoters of tumor suppressor genes [39-41].
Figure 1. Evolving models of MeCP2 have come full circle.
Models explaining the functional role of MeCP2 in the nucleus have evolved since the transcriptional repressor model of the 1990s. A structural model was later proposed based on studies showing MeCP2 associating with itself and DNA to form dense chromatin structures, consistent with its localization to nuclear heterochromatin. A ‘loop and recruit’ model incorporates a role for MeCP2 in chromatin loop structure, nuclear matrix binding, recruitment of RNA splicing and chromatin remodeling factors. An ‘active gene modulator’ model was supported by integrated epigenomic analyses showing MeCP2 binding to active gene promoters, but primarily to intronic and intergenic sites. And lastly, a transcriptional activator model was proposed based on the interaction of MeCP2 with the transcription factor CREB1. While the evolution of thought on MeCP2 has come full circle, the diversity of models is expected to reflect the diversity of roles for MeCP2 in vivo. The challenge for the field in the future is to determine which of the diverse MeCP2 roles are essential for the postnatal brain that lead to Rett syndrome when deficient. HDAC: Histone deacetylase.
However, mounting evidence has argued against the simple enzymatic model for the role of MeCP2 in repressing gene transcription in the brain, the tissue most relevant for RTT. MeCP2 does not form a stable complex with either Sin3a or HDAC when purified by size exclusion chromatography from rat brain [42]. Although MeCP2 was originally predicted to be a global silencer of methylated genes, methylated and imprinted genes remain silent in MECP2 mutant cell lines and Mecp2-deficient mice [43-45], in contrast to DNA methyltransferase (Dnmt1)-deficient mice [46]. Early gene-expression profiling experiments were mostly unsuccessful in identifying MeCP2 target genes regulated by methylated promoters in the brain and blood [47,48]. MeCP2 has been shown to be bound to several methylated promoters by chromatin immunoprecipitation with H19 [49], Bdnf [50,51], Sgk and Fkbp5 [52], Dlx5 and Dlx6 [53], Id1–3 [54], Crh [55], Fxyd1 [56,57] and Gtl2 [57,58], showing increased transcription (~twofold) in Mecp2-mutant tissue. Further studies, however, have demonstrated that Mecp2 deficiency does not reverse the paternal silencing of H19 [45] or maternal silencing of SNRPN/Snrpn [43,45], and Bdnf expression is actually lower in Mecp2-deficient brain [59], suggesting that the role of MeCP2 is not simple for even target genes with methylated promoters. Therefore, few neuronal target genes fit the simple enzymatic model for MeCP2.
A structural model has also been proposed based on additional biochemical properties of MeCP2 [60]. The most convincing data showing a potential structural role for MeCP2 has come from in vitro chromatin compaction studies. MeCP2 incubated with either naked DNA or nucleosome-bound DNA forms tight elliptoid structures and complex intramolecular interactions between adjacent DNA strands [61]. The chromatin compaction properties of MeCP2 in this assay were much stronger than the known linker histones, and suggest that MeCP2 is capable of making tight bridges and connections between noncontiguous DNA molecules. Interestingly, the compaction activity was not dependent on methylation or the methyl-binding domain, suggesting that methylated CpGs may simply serve as a nucleation point for chromatin compaction of proximal chromatin [60]. The localization of MeCP2 to dense heterochromatin is consistent with this model.
However, subsequent studies have demonstrated that MeCP2 associates with chromatin remodeling factors Brahma [62] and ATRX [63], in additional to YB-1, a RNA splicing factor [64]. Since neither of the previously reviewed models for the function of MeCP2 is sufficient to explain the multiple and seemingly diverse roles recently ascribed to MeCP2, a third model dubbed ‘loop and recruit’ is proposed based on both recent and historical evidence. The best evidence for the involvement of MeCP2 in chromatin loop formation is a chromatin immunoprecipitation (ChIP)-walk and chromatin conformation capture (3C) study at the imprinted Dlx5/Dlx6 locus [53]. This study demonstrated that MeCP2 was required for silent chromatin looping, and increased expression of Dlx5 and Dlx6 was observed as a result of aberrant loop formation in Mecp2-deficient mouse brain. Increased DLX5 expression was also observed in a subsequent study of Mecp2-deficient mouse brain [65], but not in another [66]. This was somewhat surprising, considering that MeCP2 was isolated based on binding to methylated CpG sites [67], but actually has a higher affinity to oligonucleotides with methylated CpG sites adjacent to (AT)≥4 runs [68], a sequence found frequently at CpG island shores and chromatin loop bases. While these findings suggested important new roles for MeCP2, they are less consistent with a structural role for MeCP2 strictly in the dense inert heterochromatin, and more consistent with a role for MeCP2 in recruiting additional enzymes to more active regions of the nucleus, such as the nuclear matrix and heterochromatic boundaries.
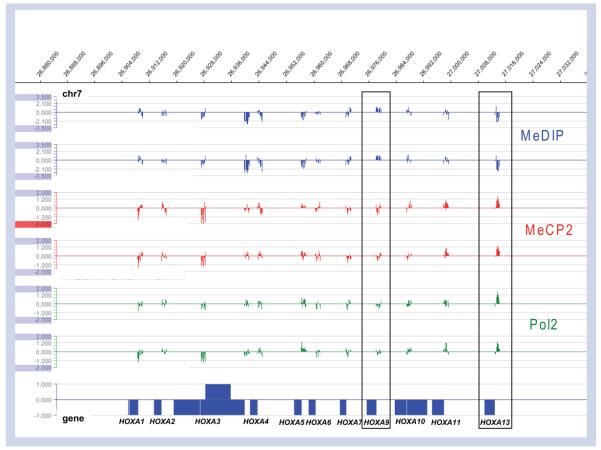
Even greater surprises came from targeted ChIP microarray (ChIP–chip) analysis of 26 Mb of selected imprinted and nonimprinted loci that revealed that 59% of MeCP2 sites were outside of gene boundaries, with only 5.9% in CpG islands [10]. Further analysis using an integrated genome wide promoter analysis of MeCP2 binding, CpG methylation and gene expression revealed that 66% of the strongest MeCP2-bound promoters are actively expressed, and only 6% of these show high levels of methylation. JUNB, an immediate early gene relevant to the pathogenesis of RTT, is one example of a highly active gene whose expression is modulated by distal and proximal MeCP2 binding to a partially methylated promoter. Further examples of the correlation between MeCP2 promoter binding and expression (revealed by Pol2 binding) and an inverse correlation with dense methylation is revealed at the HOXA gene cluster on human chromosome 7 (Figure 2). Tissue specificity of this gene cluster is epigenetically regulated and the active HOXA13 promoter has strong MeCP2 and Pol2 promoter association, but poor methylation, while the neuronally inactive HOXA9 promoter is highly methylated but de-enriched for MeCP2 and Pol2 (Figure 2, see boxes). These first epigenomic results for MeCP2 binding have challenged the previous paradigm that MeCP2 acts as a silencer of methylated gene promoters, and revealed a role for long-range modulation of active genes in neurons.
Figure 2. Combined epigenomic profiles at the HOXA gene locus exemplify the concordance of MeCP2 and gene activity with low methylation.
Data taken from genome-wide promoter array analyses from Yasui et al. [10] is mapped at the HOXA gene locus on human chromosome 7, for which tissue-specific expression differences correlate with epigenetic markers. HOXA9 is poorly expressed in neurons and has high promoter methylation (MeDIP, blue), low MeCP2 binding (red), and low Pol2 association (green). In contrast, the active HOXA13 promoter shows low methylation but high MeCP2 and Pol2 binding. Overall, there was a striking correlation between MeCP2 and Pol2 chromatin immuno precipitation signals throughout the genome, exemplified at this locus of variable gene activity.
Subsequent analysis of hypothalamus from both Mecp2-deficient and Mecp2 duplication mice by Chahrour et al. provided independent corroboration of the epigenomic MeCP2 analyses [11]. Gene-expression microarray profiling of these transgenic mice suggested that up to 85% of genes in the hypothalamus were upregulated by MeCP2, while the remainder were down-regulated. With few exceptions, the effect of Mecp2 deletion or Mecp2 duplication resulted in modest changes in target gene expression. Six potential target genes were selected for further study and were found to have varying levels of promoter methylation using bisulfite analysis and MeCP2 occupancy by ChIP. Interestingly, MeCP2 was shown to associate with the transcriptional activator CREB1 at the promoter of Sst, a gene upregulated in Mecp2 duplication mice, thereby suggesting a potential activation mechanism. These combined results suggest that MeCP2 has a more predominant role as a modulator of active genes, rather than a transcriptional repressor in vivo [10,11].
Autism: the complex genetic puzzle that can benefit from Rett syndrome epigenomics
Rett syndrome is the only one of the five pervasive developmental disorders (including autism) with a known genetic cause (Diagnostic and Statistical Manual of Mental Disorders, 4th Edition). In addition, MECP2 mutations or expression defects are observed in autism and other autism-spectrum neurodevelopmental disorders [69-74]. Therefore, investigations of the role of MECP2 in reading epigenomic marks in the developing brain have broad relevance to mental retardation and autism. Autism is characterized by severe impairments in social interaction and communication, combined with restrictive and repetitive interests and behaviors. The onset of symptoms generally occurs around 1–3 years of age, and is characterized by stereotyped mannerisms, abnormal preoccupations, lack of pragmatic language and imaginative play, impaired eye gaze and impaired joint attention. Males with autism outnumber females by around 4 to 1, and the frequency in the population has increased dramatically in recent decades to an estimated 1 in 150 children. Shifts in the interpretation of diagnostic criteria could not explain the increased prevalence, suggesting environment may affect the rate of autism [75]. However, twin studies have provided compelling evidence for a genetic origin of autism, as the concordance rate is 70–90% for monozygotic and 0–10% for dizygotic twins [76].
Despite the high heritability in monozygotic twins, autism has a heterogeneous etiology, with multiple genes and chromosomal regions suspected to be involved. Direct approaches to investigating the genetic etiology of autism have involved cytogenetic studies, linkage analyses, genome-wide association studies and, more recently, the analysis of copy number variations (CNVs). Cytogenetic methods have historically detected the fragile site caused by CGG repeat expansion at FMR1 responsible for Fragile X syndrome (FXS), as well as the approximately 12 Mb deletions of 15q11–13 that cause Prader–Willi, Angelman or 15q duplication syndromes. 15q11–13 rearrangements and FMR1 CGG expansions each account for approximately 1% of autism etiology by recent estimates [76]. Many linkage and association studies have also supported the heritance of autism with polymorphisms within 15q11–13. Homozygosity mapping of families with shared ancestry and autism revealed homozygous deletions of DIA1 upstream of NHE9, and a region upstream of PCDH10, all of which are involved in neural activity, suggesting that defective regulation of gene expression after neural activity may be shared between seemingly diverse autism mutations [77]. Complex genetic diseases are characterized by both polygenic inheritance (multiple genes contributing to the susceptibility) and locus heterogeneity (mutations in different genes causing the same phenotype). The investigation of autism genetics, like that of other neuropsychiatric disorders, has therefore moved from a prediction of ‘common disease, common variant’ to a ‘common disease, rare variant’ prediction that suggests that many alternative rare variants may disrupt overlapping genetic and epigenetic pathways [76,78].
Rapid developments in microarray technologies have spawned the discovery and greater appreciation of extensive CNV in the human genome, which are gains or losses of DNA segments ranging from several kb to several Mb [79-81]. While CNVs are polymorphic and often inherited in humans, a higher frequency of de novo rare CNVs are found in patients with autism and schizophrenia compared with unaffected controls [82-84]. CNVs mapping to 15q are among the most frequently observed in both disorders [82,84-86]. While progress in CNV detection and frequent occurrences in autism spectrum disorders is an exciting recent development, challenges to understanding causality of specific genes within specific CNVs remain [78]. Gains and losses of the same locus often leads to overlapping phenotypes, as is observed in 22q11.2, 7q11.23, 17p11.2 and 15q11–13 deletion and duplication syndromes with comorbid autism [78,87]. Complicating CNV genotype–phenotype studies even further are issues of variable penetrance or variable expressivity of the phenotype. For instance, while maternal duplication of 15q11–13 is associated with autism in 85% of cases [88], paternal 15q11–13 duplication has been observed in healthy unaffected individuals [89], as well as in cases with autism or language and social defects [90-93]. Within the patients with maternally derived idic(15), there is much clinical heterogeneity, even within patients with similar duplication breakpoints, including a subset of patients with Prader–Willi-like features [93]. Therefore, other genetic liabilities, nongenetic factors and epigenetics are considered to be important but uncharacterized determinants in determining the pathogenicity and expressivity of CNVs in autism-spectrum disorders [78].
In the last several years, several important molecular connections have been made between MeCP2 and autism-spectrum disorders, particularly those with an epigenetic basis. Reduced MeCP2 expression was observed in 79% of autism brain samples and correlated with aberrant methylation of the MECP2 promoter in male autism samples [94,95]. MeCP2 deficiency in mouse and human RTT and autism brain results in reduced expression of the Angelman gene UBE3A in some studies [45,96]. Aberrant methylation was observed around UBE3A in an autism brain sample [97]. MeCP2 binds to the maternally methylated imprinting control region in 15q11–13 [10,96,98]. Intriguingly, the nonimprinted GABAA receptor genes GABRB3, GABRA5 and GABRG3 in 15q11–13 show epigenetic dysregulation, resulting in monoallelic expression and reduced expression in a subset of RTT and autism [99], as well as 15q duplication [100] brain samples. An autism candidate and early response gene, EGR2, is a target of MeCP2 and is reduced in both RTT and autism brain samples [101]. In addition, Mecp2 mutant [102,103] and hypomorphic [104] mouse models show defects in social behavior, making them useful models for autism research.
Epigenetic mechanisms such a DNA methyl ation can be altered by environmental changes [17,105], and are an important interface between genetic and environmental risk factors in complex disorders such as autism. In addition to being environmentally sensitive, epigenetic mechanisms are also often stochastic, and thus may potentially explain variable penetrance observed with some CNVs. The identification of 68 strong MeCP2 binding sites within the 13 Mb region of 15q11–13 [10] are expected to be important in sorting out the epigenetic factors influencing the most common CNV found in autism. Furthermore, epigenetic mechanisms are predicted to be involved in neuronal homeostasis in which neurons are dynamically responsive to environmental signals, but perturbations in epigenetic pathways result in neurons with attenuated response to change [87]. Therefore, there is a critical need for understanding the roles of MeCP2 in epigenetically regulated pathways involved in normal neuronal homeostasis, as well as the multiple ways genetic, environmental and stochastic events alter epigenetic pathways and neurobehavioral phenotype.
Future perspective
Rapid progress in the field of epigenomics is due to a timely combination of novel testable hypotheses and technological innovation. In the course of just a few years, techniques such as ChIP-chip, ChIP sequencing and genome-wide DNA methylation analyses have revealed surprising insights into the human epigenome. Also, techniques for probing the chromatin structure of the genome in vivo, such as chromosome conformation capture (3C) and chromosome conformation capture on microarray (4C) are also expected to improve epigenomic understanding of the human brain and cultured neurons [106,107]. Ultimately, the ability to overlay multiple epigenomic landscapes with genetic information of structural variation and gene expression is expected to drive the field of epigenomics and enable future discoveries relevant to epigenetics, MeCP2 and autism-spectrum neurodevelopmental disorders.
The ongoing advancements in next-generation sequencing technologies are expected to have equal if not greater impact on uncovering the human epigenome as on understanding individual genomes. While relatively new epigenetic techniques such as ChIP-chip, DNA methylation ChIP (MeDIP)-chip and reduced representation bisulfite sequencing represent vast improvement over older methods, they have inherent limitations in that they are unable to assay repetitive DNA, which comprises over half of the human genome. Current high-throughput Solexa sequencing can produce tens of millions of sequences per run. However, the read length of these sequences is typically 35–70 base pairs making them often difficult to align to genomic sequences [108]. Competing technologies such as the Roche 454 platform are able to produce read lengths in the 250–500 base pair range, making them potentially more suitable for assaying the epigenome, but with an output of only approximately 400,000 reads per run [108]. Therefore, sequencing technologies still under development provide the best potential for advancing epigenomic research, as they would combine high throughput with long read lengths [109]. Given this potential, it is hard to predict what the whole epigenome with regards to MeCP2 binding, DNA methylation and chromatin organization will ultimately resemble.
Executive summary.
Rett syndrome is a genetic autism-spectrum disorder affecting epigenetic pathways
■ Mutations in the X-linked gene MECP2 cause Rett syndrome in females.
■ Due to the epigenetic mechanism of X chromosome inactivation, Rett syndrome affects females who are heterozygous and mosaic for the expression of MECP2 mutations.
■ MECP2 encodes methyl CpG-binding protein 2, a known epigenetic factor.
Epigenomic studies have enlightened the ‘classic’ epigenetic mechanisms observed in imprinting & X chromosome inactivation
■ While once considered a silencing mark of imprinted and X-inactivated gene promoters, DNA methylation is observed globally and extensively throughout the mammalian genome, and CpG islands are predominantly unmethylated.
■ Only a small subset of imprinted genes show allele-specific differential methylation at their promoters.
■ Genes on the active X chromosome are actually hypermethylated compared with the inactive allele, while hypomethylation is exclusively observed at the promoters of genes on the active X chromosome.
■ Not all X-linked genes are subject to X chromosome inactivation.
■ DNA methylation is observed in actively expressed regions genome-wide.
While originally predicted to be a global silencer of methylated genes, the role of MeCP2 in neurons has evolved
■ MECP2 expression is highest in brain, is regulated by complex developmental signals and is subject to alternate splicing, alternate polyadenylation and post-translational modifications.
■ MeCP2 is predicted to have multiple roles, including transcriptional repression, structural compaction of chromatin, chromatin looping, chromatin remodeling, splicing and transcriptional activation.
■ Combined epigenomic approaches of MeCP2 binding, methylation and gene expression have demonstrated that MeCP2 binds preferentially to intergenic and intronic regions and sparsely methylated promoters of active genes.
MeCP2 & methylation in the complex genetic & epigenetic etiologies of autism
■ While autism is strongly heritable, most cases of autism are expected to be due to a combination of genetic, environmental and epigenetic factors.
■ Genetic disorders on the autism spectrum affecting epigenetic pathways include Rett, Angelman, Prader-Willi and 15q duplication syndromes.
■ There is mounting evidence for epigenetic alterations leading to reduced expression of MeCP2 and other epigenetically regulated genes in idiopathic autism.
What the field currently lacks are unbiased high-resolution maps of DNA methylation in human and mouse brain to compare to MeCP2 and histone modification ChIP analyses. While both MeDIP and restriction enzyme-based approaches have been quite useful for early glimpses of the methylation landscape of the human genome, the gold standard for DNA methylation remains direct bisulfite sequencing. Such an approach of direct sequencing bisulfite-treated DNA has been successful in Arabidopsis [110,111], but the mammalian genome presents a larger challenge because of its much larger size and global methylation of the vast majority of the genome [12]. Reduced representative bisulfite sequencing has been successfully applied for high-throughput DNA methyl ation profile in mouse cell lines and tissues, but the approach of using MspI fragments biased the coverage to promoter regions [35]. As the epigenetic interest arises in sequences distal to promoter regions, new approaches that allow direct bisulfite sequencing on a genomic scale covering inter and intra-genic sequences, as well as repetitive sequences, will be instrumental in the future progress of understanding the role of DNA methylation and MeCP2 binding in long-range interactions of neuronal gene expression.
While sequencing the mammalian epigenome appears challenging enough with the ‘fifth base’ of 5-methylcytosine, a recent report of a ‘sixth base’ of 5-hydroxymethylcytosine further complicates the interpretation of genome-wide methyl ation analyses [112,113]. Kriaucionis and Heintz identified 5-hydroxmethylcytosine in 0.6% of nucleotides in Purkinje neurons at a level 40% as abundant as 5-methylcytosine and specific enrichment in brain tissues over non-neuronal tissues and cell lines [112]. Bisulfite treatment is predicted to deaminate cytosine when it is not modified, so bisulfite sequencing is not likely to distinguish 5-methylcytosine from 5-hydroxymethylcytosine. But MeCP2 has a much lower affinity to 5-hydroxymethylcytosine than 5-methylcytosine in vitro [114]. While the biologic function of 5-hydroxymethylcyto-sine has yet to be realized, it will be important in future epigenomic studies to be able to distinguish between these two cytosine modifications that are both highly enriched in brain compared with other tissues.
Therefore, the evolution of thought of the role of MeCP2 in the brain and its relevance to autism-spectrum disorders is not complete. While epigenomic approaches have certainly opened up the field of view for epigenetic research, there are still inherent biases in the methodologies that are likely to lead to incomplete answers. However, the explosion of new genomic and epigenomic information suggests that interactions of MeCP2 with the entire epigenome will soon be identifiable for human and mouse brain tissue. Armed with a more global perspective of MeCP2 and its interactions with methylated DNA, improved understanding and therapies for RTT, and perhaps a subset of autism cases, are certain to follow.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Armstrong DD, Dunn K, Antalffy B. Decreased dendritic branching in frontal, motor and limbic cortex in Rett syndrome compared with trisomy 21. J. Neuropathol. Exp. Neurol. 1998;57(11):1013–1017. doi: 10.1097/00005072-199811000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Raymond GV, Bauman ML, Kemper TL. Hippocampus in autism: a Golgi analysis. Acta Neuropathol. 1996;91(1):117–119. doi: 10.1007/s004010050401. [DOI] [PubMed] [Google Scholar]
- 3.Belichenko PV, Oldfors A, Hagberg B, et al. 3-D confocal microscopy of cortical pyramidal dendrites and afferents. Neuroreport. 1994;5(12):1509–1513. [PubMed] [Google Scholar]
- 4.Pickett J, London E. The neuropathology of autism: a review. J. Neuropathol. Exp. Neurol. 2005;64(11):925–935. doi: 10.1097/01.jnen.0000186921.42592.6c. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong DD. Neuropathology of Rett syndrome. J. Child Neurol. 2005;20(9):747–753. doi: 10.1177/08830738050200090901. [DOI] [PubMed] [Google Scholar]
- 6.Amir RE, Van den Veyver IB, Wan M, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl- CpG-binding protein 2. Nat. Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 7.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 8.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl- CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 9■.Zhou Z, Hong EJ, Cohen S, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52(2):255–269. doi: 10.1016/j.neuron.2006.09.037. Demonstrated that MeCP2 is phosphorylated in response to neuronal activity, resulting in a change to Bdnf expression and neuronal maturation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10■■.Yasui DH, Peddada S, Bieda MC, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl Acad. Sci. USA. 2007;104(49):19416–19421. doi: 10.1073/pnas.0707442104. This chromatin immunoprecipitation (ChIP)-chip and epigenomic analysis of MeCP2 binding sites in tiled genomic regions and promoters genome-wide challenged the expectation that MeCP2 would be primarily bound to inactive promoters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11■■.Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. This study showed that MeCP2 associates with the transcriptional activator CREB1 and, for the majority of identified target genes, activates rather than represses expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein E, Hake SB. The nucleosome: a little variation goes a long way. Biochem. Cell Biol. 2006;84(4):505–517. doi: 10.1139/o06-085. [DOI] [PubMed] [Google Scholar]
- 14.Zaidi SK, Young DW, Choi JY, et al. The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 2005;6(2):128–133. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galande S, Purbey PK, Notani D, et al. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 2007;17(5):408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Cremer T, Cremer M, Dietzel S, et al. Chromosome territories – a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18(3):307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8(4):253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37(1):179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 19.Eggermann T. Silver–Russell and Beckwith–Wiedemann syndromes: opposite (epi)mutations in 11p15 result in opposite clinical pictures. Horm. Res. 2009;71(Suppl. 2):30–35. doi: 10.1159/000192433. [DOI] [PubMed] [Google Scholar]
- 20.Lalande M, Calciano MA. Molecular epigenetics of Angelman syndrome. Cell. Mol. Life Sci. 2007;64(7–8):947–960. doi: 10.1007/s00018-007-6460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters J, Robson JE. Imprinted noncoding RNAs. Mamm. Genome. 2008;19(7–8):493–502. doi: 10.1007/s00335-008-9139-4. [DOI] [PubMed] [Google Scholar]
- 22.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 2007;19(3):281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Maeda N, Hayashizaki Y. Genome-wide survey of imprinted genes. Cytogenet. Genome Res. 2006;113(1–4):144–152. doi: 10.1159/000090826. [DOI] [PubMed] [Google Scholar]
- 24■.Gimelbrant A, Hutchinson JN, Thompson BR, et al. Widespread monoallelic expression on human autosomes. Science. 2007;318(5853):1136–1140. doi: 10.1126/science.1148910. Epigenomic study showing that monoallelic expression is not limited to imprinted genes. [DOI] [PubMed] [Google Scholar]
- 25.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 26.Chaumeil J, Le Baccon P, Wutz A, et al. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20(16):2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemson CM, Hall LL, Byron M, et al. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl Acad. Sci. USA. 2006;103(20):7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 29■.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315(5815):1141–1143. doi: 10.1126/science.1136352. Epigenomic study that countered the dogma that genes on the active X chromosome are hypomethylated compared with the inactive allele. [DOI] [PubMed] [Google Scholar]
- 30■.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39(4):457–466. doi: 10.1038/ng1990. A DNA methylation ChIP (MeDIP) analysis of promoters genome-wide showing that dense CpG islands are mostly unmethylated, even when inactive. [DOI] [PubMed] [Google Scholar]
- 31■.Rauch TA, Wu X, Zhong X, et al. A human B cell methylome at 100-base pair resolution. Proc. Natl Acad. Sci. USA. 2009;106(3):671–678. doi: 10.1073/pnas.0812399106. A gene-biased methylated DNA enrichment method on genome tiling arrays shows the correlation of dense methylation in active gene-dense R bands and a correlation of gene body methylation with gene activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32■.Rollins RA, Haghighi F, Edwards JR, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16(2):157–163. doi: 10.1101/gr.4362006. A combined restriction enzyme-based approach that covered repetitive and nonrepetitive sequences, and demonstrated that the human genome is globally and extensively methylated, with short hypomethylated domains enriched in gene promoters and CpG islands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33■.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. Tissue-specific methylation differences are primarily found at the ‘shores’ of CpG islands, not within CpG islands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35■■.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. A successful high-throughput bisulfite sequencing analysis of mammals using reduced representation of primarily CpG islands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaSalle JM. Paradoxical role of methyl-CpG- binding protein 2 in Rett syndrome. Curr. Top. Dev. Biol. 2004;59(61–86) doi: 10.1016/S0070-2153(04)59003-8. [DOI] [PubMed] [Google Scholar]
- 37.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88(4):471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 38.Yu F, Thiesen J, Stratling WH. Histone deacetylase-independent transcriptional repression by methyl- CpG-binding protein 2. Nucleic Acids Res. 2000;28(10):2201–2206. doi: 10.1093/nar/28.10.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller C, Readhead C, Diederichs S, et al. Methylation of the cyclin A1 promoter correlates with gene silencing in somatic cell lines, while tissue-specific expression of cyclin A1 is methylation independent. Mol. Cell Biol. 2000;20(9):3316–3329. doi: 10.1128/mcb.20.9.3316-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konduri SD, Srivenugopal KS, Yanamandra N, et al. Promoter methylation and silencing of the tissue factor pathway inhibitor-2 (TFPI-2), a gene encoding an inhibitor of matrix metalloproteinases in human glioma cells. Oncogene. 2003;22(29):4509–4516. doi: 10.1038/sj.onc.1206695. [DOI] [PubMed] [Google Scholar]
- 41.Ramachandran K, Gopisetty G, Gordian E, et al. Methylation-mediated repression of GADD45α in prostate cancer and its role as a potential therapeutic target. Cancer Res. 2009;69(4):1527–1535. doi: 10.1158/0008-5472.CAN-08-3609. [DOI] [PubMed] [Google Scholar]
- 42.Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodelling complex. J. Biol. Chem. 2004;279(45):46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- 43.Balmer D, Arredondo J, Samaco RC, et al. MECP2 mutations in Rett syndrome adversely affect lymphocyte growth, but do not affect imprinted gene expression in blood or brain. Hum. Genet. 2002;110(6):545–552. doi: 10.1007/s00439-002-0724-4. [DOI] [PubMed] [Google Scholar]
- 44.Gartler SM, Varadarajan KR, Luo P, et al. Normal histone modifications on the inactive X chromosome in ICF and Rett syndrome cells: implications for methyl-CpG binding proteins. BMC Biol. 2004;2(1):21. doi: 10.1186/1741-7007-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45■.Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum. Mol. Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. Analysis of mouse and human brain samples that showed an overlapping pathway of Rett syndrome, 15q11–13 imprinted disorders and idiopathic autism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 47.Tudor M, Akbarian S, Chen RZ, et al. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc. Natl Acad. Sci. USA. 2002;99(24):15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Traynor J, Agarwal P, Lazzeroni L, et al. Gene expression patterns vary in clonal cell cultures from Rett syndrome females with eight different MECP2 mutations. BMC Med. Genet. 2002;3(1):12. doi: 10.1186/1471-2350-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuks F, Hurd PJ, Wolf D, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278(6):4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 50.Chen WG, Chang Q, Lin Y, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 51.Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 52.Nuber UA, Kriaucionis S, Roloff TC, et al. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum. Mol. Genet. 2005;14(15):2247–2256. doi: 10.1093/hmg/ddi229. [DOI] [PubMed] [Google Scholar]
- 53■.Horike S, Cai S, Miyano M, et al. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37(1):31–40. doi: 10.1038/ng1491. Demonstrated a new role for MeCP2 in the formation of silent chromatin loops in an imprinted locus relevant to GABA synthesis. [DOI] [PubMed] [Google Scholar]
- 54.Peddada S, Yasui DH, LaSalle JM. Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum. Mol. Genet. 2006;15(12):2003–2014. doi: 10.1093/hmg/ddl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGill BE, Bundle SF, Yaylaoglu MB, et al. Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc. Natl Acad. Sci. USA. 2006;103(48):18267–18272. doi: 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng V, Matagne V, Banine F, et al. FXYD1 is an MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum. Mol. Genet. 2007;16(6):640–650. doi: 10.1093/hmg/ddm007. [DOI] [PubMed] [Google Scholar]
- 57.Jordan C, Li HH, Kwan HC, et al. Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets. BMC Med. Genet. 2007;8:36. doi: 10.1186/1471-2350-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urdinguio RG, Lopez-Serra L, Lopez-Nieva P, et al. Mecp2-null mice provide new neuronal targets for Rett syndrome. PLoS ONE. 2008;3(11):e3669. doi: 10.1371/journal.pone.0003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang Q, Khare G, Dani V, et al. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49(3):341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 60.Bowen NJ, Palmer MB, Wade PA. DNA damage repair and transcription chromosomal regulation by MeCP2: structural and enzymatic considerations. Cell Mol. Life Sci. 2004;61(17):2163–2167. doi: 10.1007/s00018-004-4177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Georgel PT, Horowitz-Scherer RA, Adkins N, et al. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003;278(34):32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 62.Harikrishnan KN, Chow MZ, Baker EK, et al. Brahma links the SWI/SNF chromatin- remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005;37(3):254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 63.Nan X, Hou J, Maclean A, et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl Acad. Sci. USA. 2007;104(8):2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young JI, Hong EP, Castle JC, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl Acad. Sci. USA. 2005;102(49):17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyano M, Horike S, Cai S, et al. DLX5 expression is monoallelic and Dlx5 is up-regulated in the Mecp2-null frontal cortex. J. Cell Mol. Med. 2008;12(4):1188–1191. doi: 10.1111/j.1582-4934.2008.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schule B, Li HH, Fisch-Kohl C, et al. DLX5 and DLX6 expression is biallelic and not modulated by MeCP2 deficiency. Am. J. Hum. Genet. 2007;81(3):492–506. doi: 10.1086/520063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 68.Klose RJ, Sarraf SA, Schmiedeberg L, et al. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19(5):667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 69.Lam C, Yeung W, Ko C, et al. Spectrum of mutations in the MECP2 gene in patients with infantile autism and Rett syndrome. J. Med. Genet. 2000;37(12):E41. doi: 10.1136/jmg.37.12.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dotti MT, Orrico A, De Stefano N, et al. A Rett syndrome MECP2 mutation that causes mental retardation in men. Neurology. 2002;58(2):226–230. doi: 10.1212/wnl.58.2.226. [DOI] [PubMed] [Google Scholar]
- 71.Carney RM, Wolpert CM, Ravan SA, et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr. Neurol. 2003;28(3):205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- 72.Kleefstra T, Yntema HG, Nillesen WM, et al. MECP2 analysis in mentally retarded patients: implications for routine DNA diagnostics. Eur. J. Hum. Genet. 2004;12(1):24–28. doi: 10.1038/sj.ejhg.5201080. [DOI] [PubMed] [Google Scholar]
- 73.Hammer S, Dorrani N, Dragich J, et al. The phenotypic consequences of MECP2 mutations extend beyond Rett syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 2002;8(2):94–98. doi: 10.1002/mrdd.10023. [DOI] [PubMed] [Google Scholar]
- 74.Van Esch H, Bauters M, Ignatius J, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am. J. Hum. Genet. 2005;77(3):442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hertz-Picciotto I, Delwiche L. The rise in autism and the role of age at diagnosis. Epidemiology. 2009;20(1):84–90. doi: 10.1097/EDE.0b013e3181902d15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135(3):391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morrow EM, Yoo SY, Flavell SW, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321(5886):218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cook EH, Jr., Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455(7215):919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 79.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 80.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 81.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 82.Stone JL, O'Donovan MC, Gurling H, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller DT, Shen Y, Weiss LA, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J. Med. Genet. 2008;46(4):242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Christian SL, Fantes JA, Mewborn SK, et al. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13) Hum. Mol. Genet. 1999;8(6):1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- 87.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455(7215):912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hogart A, Wu D, Lasalle JM, et al. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol. Dis. 2008 doi: 10.1016/j.nbd.2008.08.011. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cook EH, Jr., Lindgren V, Leventhal BL, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am. J. Hum. Genet. 1997;60(4):928–934. [PMC free article] [PubMed] [Google Scholar]
- 90.Mohandas TK, Park JP, Spellman RA, et al. Paternally derived de novo interstitial duplication of proximal 15q in a patient with developmental delay. Am. J. Med. Genet. 1999;82(4):294–300. [PubMed] [Google Scholar]
- 91.Mao R, Jalal SM, Snow K, et al. Characteristics of two cases with dup(15)(q11.2-q12): one of maternal and one of paternal origin. Genet. Med. 2000;2(2):131–135. doi: 10.1097/00125817-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 92.Veltman MW, Thompson RJ, Craig EE, et al. A paternally inherited duplication in the Prader-Willi/Angelman syndrome critical region: a case and family study. J. Autism. Dev. Disord. 2005;35(1):117–127. doi: 10.1007/s10803-004-1039-1. [DOI] [PubMed] [Google Scholar]
- 93.Depienne C, Moreno-De-Luca D, Heron D, et al. Screening for genomic rearrangements and methylation abnormalities of the 15q11-q13 region in autism spectrum disorders. Biol. Psychiatry. 2009;66(4):349–359. doi: 10.1016/j.biopsych.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 94.Nagarajan RP, Hogart AR, Gwye Y, et al. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1(4):172–182. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Samaco RC, Nagarajan RP, Braunschweig D, et al. Multiple pathways regulate MeCP2 expression in normal brain development and exhibit defects in autism-spectrum disorders. Hum. Mol. Genet. 2004;13(6):629–639. doi: 10.1093/hmg/ddh063. [DOI] [PubMed] [Google Scholar]
- 96.Makedonski K, Abuhatzira L, Kaufman Y, et al. MeCP2 deficiency in Rett syndrome causes epigenetic aberrations at the PWS/AS imprinting center that affects UBE3A expression. Hum. Mol. Genet. 2005;14(8):1049–1058. doi: 10.1093/hmg/ddi097. [DOI] [PubMed] [Google Scholar]
- 97.Jiang YH, Sahoo T, Michaelis RC, et al. A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A. Am. J. Med. Genet. 2004;131(1):1–10. doi: 10.1002/ajmg.a.30297. [DOI] [PubMed] [Google Scholar]
- 98.Thatcher K, Peddada S, Yasui D, et al. Homologous pairing of 15q11–13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Hum. Mol. Genet. 2005;14:785–797. doi: 10.1093/hmg/ddi073. [DOI] [PubMed] [Google Scholar]
- 99.Hogart A, Nagarajan RP, Patzel KA, et al. 15 q11–13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum. Mol. Genet. 2007;16(6):691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hogart A, Leung KN, Wang NJ, et al. Chromosome 15q11–13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number. J. Med. Genet. 2009;46(2):86–93. doi: 10.1136/jmg.2008.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Swanberg SE, Nagarajan RP, Peddada S, et al. Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism. Hum. Mol. Genet. 2009;18(3):525–534. doi: 10.1093/hmg/ddn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moretti P, Bouwknecht JA, Teague R, et al. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum. Mol. Genet. 2005;14(2):205–220. doi: 10.1093/hmg/ddi016. [DOI] [PubMed] [Google Scholar]
- 103.Moretti P, Levenson JM, Battaglia F, et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J. Neurosci. 2006;26(1):319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Samaco RC, Fryer JD, Ren J, et al. A partial loss of function allele of methyl-CpG-binding protein 2 predicts a human neurodevelopmental syndrome. Hum. Mol. Genet. 2008;17(12):1718–1727. doi: 10.1093/hmg/ddn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod. Toxicol. 2007;23(3):297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 106.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods. 2007;4(11):895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 107.Simonis M, Klous P, Splinter E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38(11):1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 108.von Bubnoff A. Next-generation sequencing: the race is on. Cell. 2008;132(5):721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 109.Ansorge WJ. Next-generation DNA sequencing techniques. N. Biotechnol. 2009;25(4):195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 110.Lister R, O'Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112■■.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. A sixth base in the mammalian genome of 5-hydroxymethylcytidine is identified and found to be enriched in brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Valinluck V, Tsai HH, Rogstad DK, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]