Abstract
The density of the organic matrix of bone substance is a critical parameter necessary to clinically evaluate and distinguish structural and metabolic pathological conditions such as osteomalacia in adults and rickets in growing children. Water and fat suppressed proton projection MRI (WASPI) was developed as a noninvasive means to obtain this information. In this study, a density calibration phantom was developed to convert WASPI intensity to true bone matrix density. The phantom contained a specifically designed poly(ethylene oxide)/poly(methyl methacrylate) blend, whose MRI properties (T1, T2 and resonance linewidth) were similar to those of solid bone matrix (collagen, tightly bound water, other immobile molecules), minimizing the need to correct for differences in T1 and/or T2 relaxation between the phantom and the subject. Cortical and trabecular porcine bone specimens were imaged using WASPI with the calibration phantom in the field of view as a stable intensity reference. Gravimetric and amino acid analyses were carried out on the same specimens after WASPI and the chemical results were found to be highly correlated (r2 = 0.98 and 0.95 respectively) to the WASPI intensity. By this procedure the WASPI intensity can be used to obtain the true bone matrix mass density in g cm−3.
Keywords: bone, MRI, WASPI, UTE ultrashort T2 imaging
INTRODUCTION
Osteomalacia is a condition in which adult patients have an impaired extent of mineralization of bone, and growing children have diminished calcification of growth cartilage (rickets). In addition, some patients have a reduced amount of total bone substance (osteoporosis). Patients with osteoporosis who also have a diminished calcium phosphate mineral content in bone due to osteomalacia may remain for many years misdiagnosed as only having osteoporosis, since radiographs and various serum and urine measurements frequently fail to distinguish the two conditions (1). Measuring the degree or extent of bone mineralization (mineral mass per unit volume of bone matrix) would be helpful for clinicians to confirm or exclude osteomalacia. Information on both 3D volumetric bone mineral density (BMD, mineral mass per unit volume of bone tissue) as well as bone matrix density (solid bone matrix mass per unit volume of bone tissue) is needed. Current X-ray based in vivo bone densitometry measurements, such as dual energy X-ray absorptiometry (DXA) and quantitative computed tomography (QCT), only provide either 2D BMD (DXA) or 3D BMD (QCT) respectively. Both of these measurements yield a lower than normal BMD score in osteomalacia or osteoporosis, and therefore fail to distinguish the conditions. Neither method provides information about bone matrix density (2). Recent studies have questioned the accuracy of DXA measurement, since the DXA BMD value can be contaminated by sizable soft tissue contributions and errors caused by its 2D projection mapping of what is always a 3D object (3). At present, in the clinic the degree of bone mineralization can only be obtained invasively by bone biopsy followed by histomorphometry of thin sections under optical microscopy. In the laboratory, chemical and gravimetric analyses of bulk bone specimens can also be used (4).
Solid state MRI (SMRI) has been developed in our lab to detect NMR signals from all but the most molecularly immobile constituents (5–7). SMRI may be considered a “zero TE” method, as neither spin nor gradient echoes are employed. Water and fat suppressed proton projection imaging (WASPI), a method that combines SMRI with suppression of fluid signals, has been developed recently to detect the solid bone matrix signal (8,9). Other researchers have also developed various MRI methods to study bone mineral and bone matrix. Bydder and co-workers (10) have developed ultra-short TE 31P and proton MRI to image the solid constituents of human bone. More recently, Wehrli and co-workers utilized a radial projection pulse sequence with a long-T2 soft tissue suppression to image the solid-like constituents of bone matrix, which they have assigned to cortical bone water (11). Idiyatullin and co-workers acquired time-domain signals during a swept RF excitation using time-shared manner to visualize hard tissues (12).
For any solid state MRI method to be useful in diagnosing osteomalacia, quantitative accuracy is fundamental, and must be validated. Calibration phantoms are essential for quantitative MRI measurements, since they provide stable, accurate and known references. The phantom media are typically aqueous solutions, aqueous gels, or oils, and are intended to characterize one or more MR image properties for conventional (fluid-state) MRI: spatial resolution, proton density, T1 or T2 measurement, RF homogeneity, diffusion anisotropy, signal-to-noise ratio (13–15) and new MRI contrast agents (16–18). There are phantoms specifically designed for fluid state density measurements (19–21), but no reports of phantoms designed for solid state density measurements to the best of our knowledge.
The purpose of this study is to develop a new type of proton calibration phantom, based on a solid polymeric material, to convert the WASPI image intensity of the solid bone matrix signal to true bone matrix density. Utilizing a polymer with MRI properties (T1, T2, and resonance line shape) similar to those of solid bone matrix minimizes the need to correct for differences in T1 and/or T2 relaxation between the phantom and the subject. Choosing a polymer system with properties that are stable over time (not subject to water loss, autolysis, microbial degradation, etc.), enables the analysis to compensate for small variations in scanner settings or performance, and also obviates the need to perform frequent (and complex and inconvenient) recalibrations with actual bone. By choosing an appropriate pair of polymers that form a homogeneous blend and then varying the blend proportions, it is possible to tune the magnetic resonance properties (to a limited extent) to specific T1 and T2 target values.
The WASPI signal arises from a portion of collagen protons, tightly bound water, and other motionally restricted molecules in bone matrix. Although the wide bandwidth acquisition of FID signals in WASPI will capture all proton signals present (even those with the shortest T2s), not all protons will have been excited because of practical limits on the bandwidth of the excitation pulse. Additionally, the very shortest T2 protons appear in WASPI images with low spatial resolution, and merely contribute to a diffuse background. Hereafter, the term “solid bone matrix” is used to refer to the constituents of solid bone tissue which are imaged by the WASPI pulse sequence. Our definition is thus based on MR signal observability, and is not intended to reflect absolute chemical composition. In particular, the shortest T2 material is not imaged at all, and protons of different chemical identity (molecularly mobile segments of proteins, motionally restricted water, etc.) are lumped together because their chemical shift differences are small compared to their spectral linewidths. Nevertheless, as we show in this study, the WASPI signal, when properly calibrated against an appropriate reference phantom, yields an accurate quantitative measure of bone matrix that correlates highly with traditional accepted in vitro chemical and biochemical measures of bone matrix.
METHODS
All animal specimens were obtained in full compliance with federal regulations and the guidelines of the respective animal care and use committees of the relevant institutions.
The right rear leg was harvested from a 4-year, 5-month-old female Yorkshire pig (Cummings School of Veterinary Medicine, Tufts University, North Grafton, MA, USA) about one hour after the animal was sacrificed and was transported on ice in less than 2 hours between the animal sacrifice site and the bone dissection lab.
Bone Specimens
Three specimens of bone sections 1.0 cm long × 1.0 cm wide × 0.45 cm thick were cut from the midshaft cortices of the pig femora (cortical bone specimens 1–3). Eight trabecular specimens of the same dimensions were obtained from the femoral neck and upper metaphyses of the same bone (trabecular bone specimens 1–8). Bone specimens were intact except that external soft tissues including the periosteum were completely dissected. The specimens were kept frozen before MRI scanning.
Bone marrow and intramedullary fat were extracted from the medullary cavity of the unused part of the same bone and put into a glass tube (the marrow specimen) to serve as a reference for water and fat suppression in WASPI experiments.
Bone Powder/Glass Powder Mixtures
In order to expand the range of bone matrix density values for MRI, gravimetric and chemical analysis, and thereby provide a more rigorous assessment of the methodology, five mixtures of porcine bone powder and powdered glass beads in varying ratios were prepared to give an incremental series of bone powder densities (bone/glass mixtures 1–5). The femora of a 2-month-old pig (Children’s Hospital, Boston, MA) were harvested less than one hour after the animal was sacrificed and kept frozen before further preparation. The midshaft cortices of the femora were stripped of soft tissue, and dried at room temperature. The dried bone specimens were ground with various amounts of borosilicate glass beads into powder using a SPEX 6700 Freezer/Mill (SPEX Industries Inc., Metuchen, NJ, USA) under liquid nitrogen. The mixtures were put into glass cylinders; a total of 5 mixtures were prepared and the volumes of the mixtures were found to be 0.374, 0.343, 0.363, 0.337, and 0.341 cm3 respectively.
Polymer Calibration Phantom
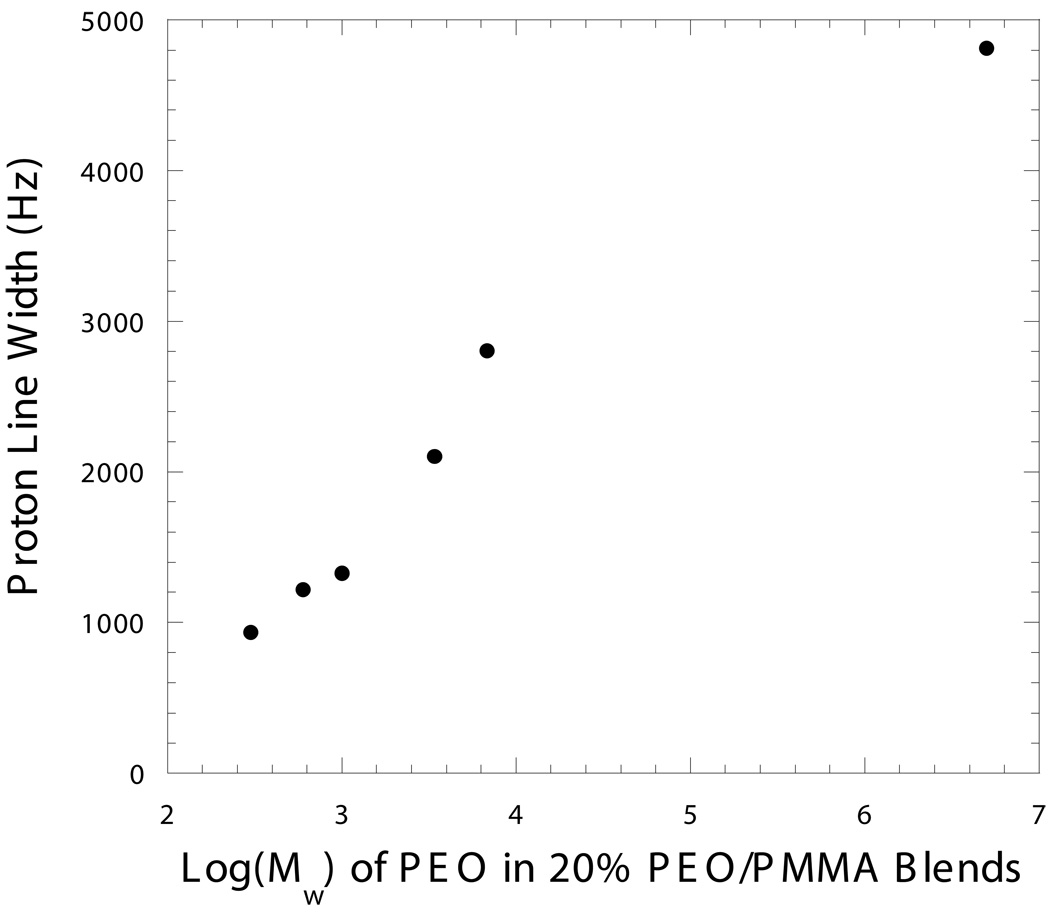
20% Poly(ethylene oxide)/80% poly(methyl methacrylate) (PEO/PMMA) blends were prepared as SMRI calibration materials for the intensity reference phantom. PEO terminated at one end by –OH and at the other end by –H was purchased from Sigma-Aldrich (St. Louis, MO, USA). The PEO had a weight-average molecular weight MW = 3,400 g/mol. The choice of this specific molecular weight of PEO was based on the observation presented in the Results section and Fig. 1. PMMA, MW = 35,000 g/mol, was purchased from Scientific Polymer Products (Ontario, NY, USA). The PMMA was greater than 79% syndiotactic. The blends were prepared by solvent casting from a chloroform solution. A mixture of polymers was dissolved in chloroform. The solvent was evaporated at room temperature. Subsequent annealing was performed in vacuo at 120 °C for 2 h to remove any traces of the solvent. The blends were vacuum-dried at room temperature for at least 2 days to further ensure the removal of all the solvent. Clear and transparent film samples were obtained.
Figure 1. 1H resonance linewidth of 20% PEO/PMMA blends vs. PEO weight-average molecular weight Mw.
Data were acquired with a 200 MHz Varian Mercury NMR spectrometer at 26 °C. The Mw of the PMMA component was 75,000. (Unpublished result from Haihui Cao’s thesis under the guidance of the late Prof. Alan A. Jones).
The polymer film was ground into powder using the freezer/mill under liquid nitrogen. A fine uniform powder was obtained by sieving through a 200-mesh screen. Three cylindrical pellets of the polymer blend powder diluted with various amounts of silicon dioxide (quartz sand, 40–100 mesh) from Fisher Scientific (Waltham, MA, USA), were made under high pressure (2,000–6,000 lb) in a Carver (Wabash, IN, USA) model 3887 25-ton hydraulic press. Formed in this manner, the pellets have well defined geometric shapes, enabling the dimensions of each pellet to be accurately measured and the volume calculated. The polymer densities of the pellets were 0.56, 0.80 and 1.17 g cm−3 respectively. The pellets were used as a calibration phantom imaged alongside bone specimens in the WASPI experiments.
MRI Measurement
In this study, bone specimens as well as bone/glass mixtures were always imaged with the calibration phantom. SMRI and WASPI measurements were carried out using pulse sequences described in (9) with a Bruker 4.7 T 33 cm scanner equipped with a 400 mT/m gradient system (Bruker-Biospin, Billerica, MA, USA) and a home made RF probe with a 15 mm ID coil at room temperature. The 1H Larmor frequency was 200.13 MHz. SMRI (total proton content) data were acquired under the following protocol if not otherwise specified, including a first set of FID acquisitions under the regular gradient magnitude, and a second set of FID acquisitions under a lower gradient magnitude with fewer directions to recover the data points lost in the receiver dead time (9). The first data set was acquired under fixed 160 mT/m gradient magnitudes in 2934 directions at a sampling rate of 5 µs per complex point. The short hard pulse used to excite the signal was 8 µs in duration, the receiver dead time was 10 µs, the repetition time TR was 0.15 s and the FIDs were averaged over 16 acquisitions. The second data set were acquired with the same parameters in the first set of acquisitions except that the number of projection directions was 20, and the gradient magnitude was 80 mT/m with a sampling rate of 10 µs per complex point. The measurement time was approximately 2 hours including both the first and the second set of acquisitions. Eighty complex points of the FID in each projection direction were used in the reconstruction, effectively creating a 17 mm FOV in a 64 × 64 × 64 cubic lattice with a projection resolution of 0.57 mm.
WASPI data were acquired with pairs of 90° suppression pulses of 2.0 – 2.5 ms in duration, with frequencies set at the water and fat chemical shifts. Other acquisition parameters of the WASPI experiments were the same as the SMRI experiments.
MRI Data Processing
In this study, SMRI and WASPI images were first corrected for B1 inhomogeneity. A tube of aqueous MnSO4 (~10 mM, T2 ~1 ms) was imaged to calibrate the image intensity over the measurement volume. Since a small excitation pulse (8 µs, 12°–13° flip angle) was used in the experiments, the signal intensities of the images were assumed to be directly proportional to B1. For a flip angle as large as 15°, the error in B1 is only –1.1%. The intensity of each WASPI image was divided by that of the MnSO4 image pixel by pixel.
Definitions of terms and calculations used in data processing are given in the following. In these analyses we represent volumes V derived from images by counting the number of image pixels assigned to a particular substance, and we represent masses M by summing the intensities of pixels assigned to a particular substance. Image segmentation and assignment is performed on the basis of pixel intensity.
VBt (volume of bone tissue): The total number of pixels of bone tissue in the non-suppressed SMRI images. VBt was determined by selecting all pixels whose signal to noise ratio was above 20 and represents the volume of bone tissue.
VCi (volume of calibration pellet i): The total number of pixels of polymer pellet i (i = 1, 2, 3) in the calibration phantom in each WASPI image. VCi was determined in the above manner in only the WASPI images, since the pellets (which contain no fluid) are not visualized as accurately in the SMRI images (whose dynamic range tends to be dominated by fluid signals).
MBmW (mass of bone matrix expressed in terms of WASPI image intensity): The sum of WASPI intensities over all the pixels within the bone tissue volume VBt.
MCWi (mass of calibration pellet i expressed in terms of WASPI image intensity): The sum of WASPI intensities over all the pixels within the ith polymer pellet volume VCi of the calibration phantom.
Obtaining the bone matrix density requires three steps:
Step 1: The WASPI derived bone matrix density DBmW and the ith polymer pellet density DCWi were calculated in units of MRI intensity per pixel:
| [1] |
and
| [2] |
Step 2: The ith polymer pellet’s physical density DCPi in units of polymer g cm−3 was computed by dividing its physical mass (g) by its physical volume (cm3). A calibration curve of DCWi vs. DCPi was created by linear regression of these two sets of data for each WASPI image. DBmW (matrix density in image intensity units) was converted to DBmP , which is the matrix density in polymer mass density units, by using the formula found in the linear regression for that image.
Step 3: Gravimetric and amino acid analyses were carried out on the specimens after MRI scanning. Correlations between DBmP and the actual matrix mass densities determined by gravimetric analysis (DBmG), and DBmP and the actual matrix protein densities determined by amino acid analysis (DBmA), were examined by linear regression using KaleidaGraph (Synergy Software, Reading, PA, USA), and conversion factors were obtained. The linear regression relations can be used to convert DBmP of additional bone specimens measured by WASPI under the same measurement conditions to matrix mass density DBmG or matrix protein density DBmA without performing gravimetric/amino acid analysis.
Gravimetric Analysis
Bone Specimens
After thawing, surface water on each bone specimen was removed by blotting lightly with filter paper and the wet weight was obtained. The volume of each bone specimen was measured with an AccuPyc 1330 pycnometer (Micromeritics Instrument Corporation, Norcross, Georgia, USA). The bone specimen was then ground into a powder using the freezer/mill under liquid nitrogen. The bone powder was completely transferred to FEP (Teflon fluorinated ethylene propylene) centrifuge tubes by rinsing with dry ethanol, and the transfer solvent was evaporated under a nitrogen gas flow. The fat in the bone powder was extracted using 2:1 (v/v) chloroform/methanol with shaking overnight in the cold. The extracted powder was spun down at 10 k RPM for 30 min. The supernatant was transferred to tared glass vessels and the solvent evaporated under flowing nitrogen to obtain the extracted lipid weight. The fat content was calculated as (fat weight)/(wet weight). The extracted bone powder was completely transferred to tared crucibles by rinsing with dry ethanol, and the transfer solvent was evaporated under a nitrogen gas flow. The bone powder was vacuum-dried at 50–100 mTorr at 70 °C for 3 days to constant weight (to obtain the mass of dry bone), ashed at 600 °C for 12–16 hours, and weighed again (to obtain the mass of mineral). The free water content was calculated as (wet weight − mass of dry bone − fat)/(wet weight). The weight loss after ashing (mass of dry bone − mass of ash) is the total organic content of the bone plus tightly bound water. The bone matrix density DBmG was calculated by dividing the total organic content of the bone by the specimen volume [(mass of dry bone − mass of mineral)/volume].
Bone /Glass Mixtures
One half of each cortical bone/glass mixture was weighed for gravimetric analysis. The fat extraction wasn’t performed on the mixtures because the fat content of the cortical bone was found to be low, as expected for young (2-month-old) pigs. The analysis was otherwise the same as described above and the corresponding density DBmG was determined.
Amino Acid Analysis
Bone Specimens
About 2–5 mg (analyzed weight) of the dry powder (after fat extraction and vacuum-drying for 3 days at 70 °C) of each bone specimen was placed in a vacuum hydrolysis tube and suspended in 6 N HCl. The tube was then flushed with nitrogen, evacuated and sealed with a torch. Hydrolysis was performed at 110 °C for 18–24 hours. Following hydrolysis, the sample was allowed to cool and the HCl was evaporated under vacuum. The residue was washed with distilled water and evaporated once again to dryness. The sample was then resuspended in dilution buffer and analyzed by a Beckman (Ramsey, Minnesota, USA) Model 7300 amino acid analyzer. The amino acids were separated by ion-exchange chromatography, followed by post column derivitization using ninhydrin for detection. Ninhydrin-reactive components were mixed with DMSO-ninhydrin reagent and allowed to react at the temperature of boiling water. Signals at 440 and 570 nm wavelengths were integrated and the concentration of each ninhydrin-reactive component was recorded. The total amount of the protein in the analyzed weight and the amount of the protein per weight unit of dry bone was obtained. Therefore the total amount of the protein in the whole bone specimen was calculated by multiplying the amount of the protein per unit weight of dry bone by the mass of the dry bone. The bone matrix protein density DBmA was obtained by dividing the total amount of the protein in the whole bone specimen by the bone tissue volume.
Bone/Glass Mixtures
The other half of each bone/glass mixture was weighed. The mixtures were not dried to avoid the possibility of the drying process altering bone mass fraction and causing error in the bone mass calculation. The mixture was hydrolyzed in 6 N HCl for 18–24 hours at 110 °C. Following hydrolysis, the solution was transferred quantitatively to a volumetric flask and distilled water was pooled into the volumetric flask up to the mark. The concentrations of the solutions were determined. One milliliter of each solution was filtered to remove any trace of the glass powder. The same procedures described above were performed. The bone matrix density DBmA was obtained by dividing the total amount of the protein in the mixture by the volume.
RESULTS
Polymer Phantom
Single-pulse proton NMR spectra of typical 20% PEO/PMMA blends at room temperature showed a broad featureless resonance. The full widths at half height of the resonances of 20% PEO/PMMA blends measured at 26 °C containing various PEO molecular weights are shown in Fig. 1. It was observed that the linewidth became broader as the PEO molecular weight increased.
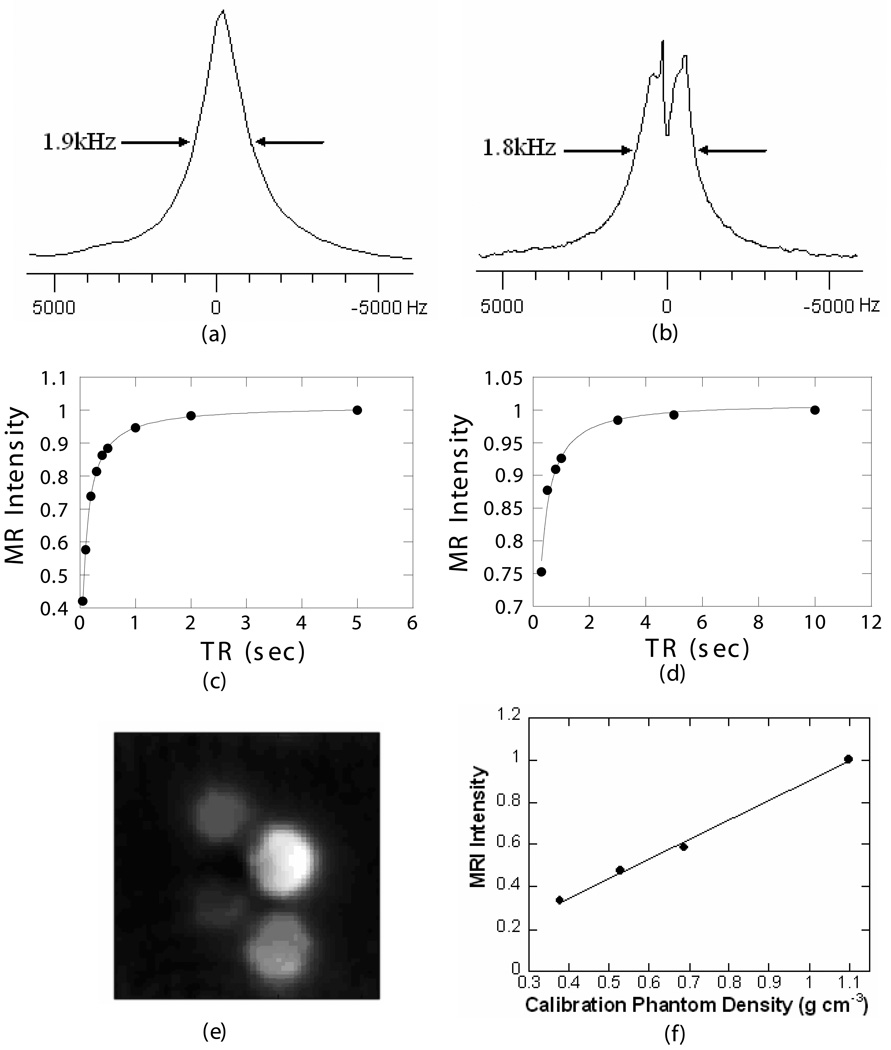
For a 20% PEO/PMMA blend containing PEO with Mw = 3,400 g/mol, the full width at half height was around 1.9 kHz at room temperature (Fig. 2a), similar to that of the 1.8 kHz observed in the WASPI signal of bone specimens without projection gradients (Fig. 2b). The T2* of the same blend was on the order of 100 µs, comparable to that of solid bone matrix. The polymer spectrum was obtained by a single pulse experiment since neither water nor fat need to be suppressed, while the solid bone matrix has to be obtained by a water and fat suppressed experiment. The low power long duration water and fat suppression pulses might cause some loss in the solid bone matrix signal, which can be manifest as a dip at the center of the resonance (8).
Figure 2. Comparison of polymer blend and porcine bone proton relaxation parameters, and typical SMRI images of polymer phantoms.
a) Single-pulse proton spectrum of the 20% PEO/PMMA blend. The molecular weights of PEO and PMMA were 3,400 and 35,000 respectively. The linewidth at half height of the polymer is around 1.9 kHz at room temperature. b) WASPI spectrum of a porcine bone specimen without projection gradients. The full width of the observed resonance at half height is around 1.8 kHz. c) Polymer phantom: T1 = 3.6 s, excitation angle = 12°. d) Porcine bone specimen: T1 = 3.4 s, excitation angle=13°. T1s were obtained by fitting data acquired with progressive-saturation sequences to Eq. [3]. e) The typical SMRI images of four polymer phantoms, whose densities were 1.171, 0.800, 0.557 and 0.402 g/cm3 respectively. f) The least squares fit of known polymer calibration phantom densities to their MRI intensities, r2= 0.99.
Fig. 2c and d show the proton T1 measurements of the 20% PEO/PMMA blend and porcine bone solid matrix. The proton T1 measurements were carried out using a series of progressive-saturation WASPI experiments. The projection gradient was applied only in the z-direction, resulting in a 1D WASPI to ensure that the signal arises from the components of interest while reducing the total experiment time. The signal was averaged over 128 scans, and the TR was varied from 0.1 s to 10 s. The measured signal amplitudes were fitted to the Ernst equation of T1 relaxation (22)
| [3] |
The fitting results showed that the proton T1 of the polymer blend and porcine bone matrix were ~3.6 s and ~3.4 s (Table 1), and the excitation angles β in each experiment were 12° and 13° respectively. Based on the T1s and excitation angles, a TR of 0.15 s was chosen in the following WASPI measurements of bone matrix.
Table 1.
1H spin-lattice relaxation times T1 and correction factors F for bone specimens and the 20% PEO/PMMA blend phantom at 4.7 T
| Porcine bone matrix | 20% PEO/PMMA | |
|---|---|---|
| T1, s | 3.4 | 3.6 |
| Excitation pulse β | 13° | 12° |
| TR, s | 0.15 | 0.15 |
| Correction factor F | 6.98 | 7.28 |
Fig. 2e shows the SMRI images of four polymer phantoms of various densities. The MRI intensity was proportional to the polymer density (Fig. 2f).
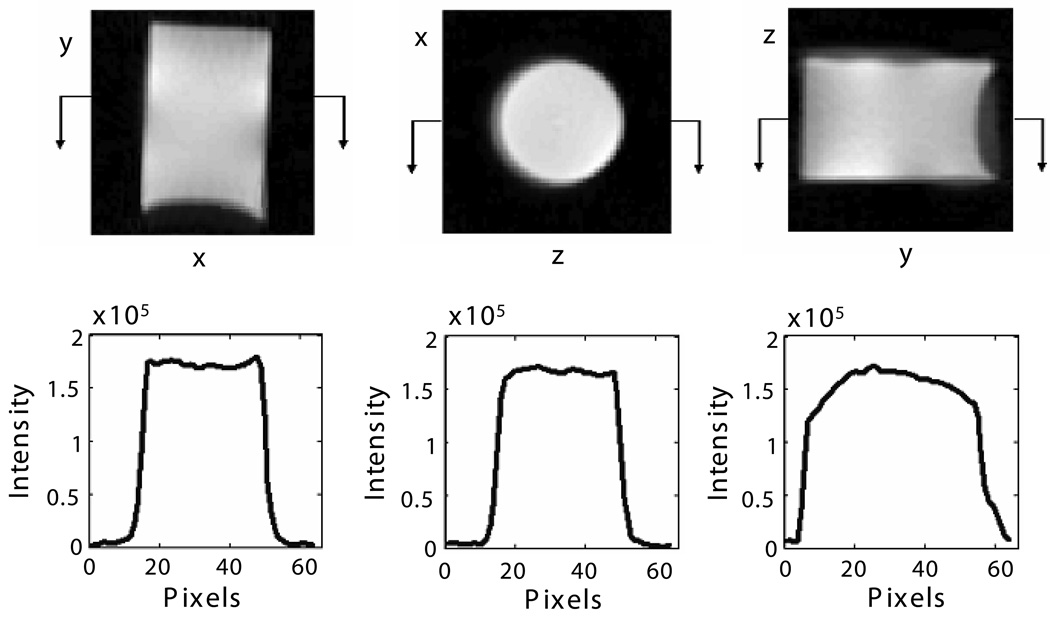
SMRI experiments with the same acquisition protocol as above were performed on short T2 (~1 ms) 10 mM aqueous MnSO4 filling the FOV in the specimen holder (Fig. 3). This is an alternative to B1 mapping, and is valid because the flip angles were small. The signal intensity over the x-z plane of the FOV where the bone and phantoms were located is homogenous, with variation smaller than 5%. The signal intensity over the longitudinal direction (y-x and z-y) showed that most of the FOV is as homogenous as in the transverse direction, but that there was a region in which the image intensity is about 20% stronger than the rest of the FOV due to the coil structure.
Figure 3. Image slices and intensity profiles of MnSO4 solution in the specimen holder.
Upper (from left to right): Center slice of images in the y-x, x-z and z-y planes respectively. Lower: corresponding intensity profile of the images in the upper row.
Bone Specimens and Bone/Glass Mixtures
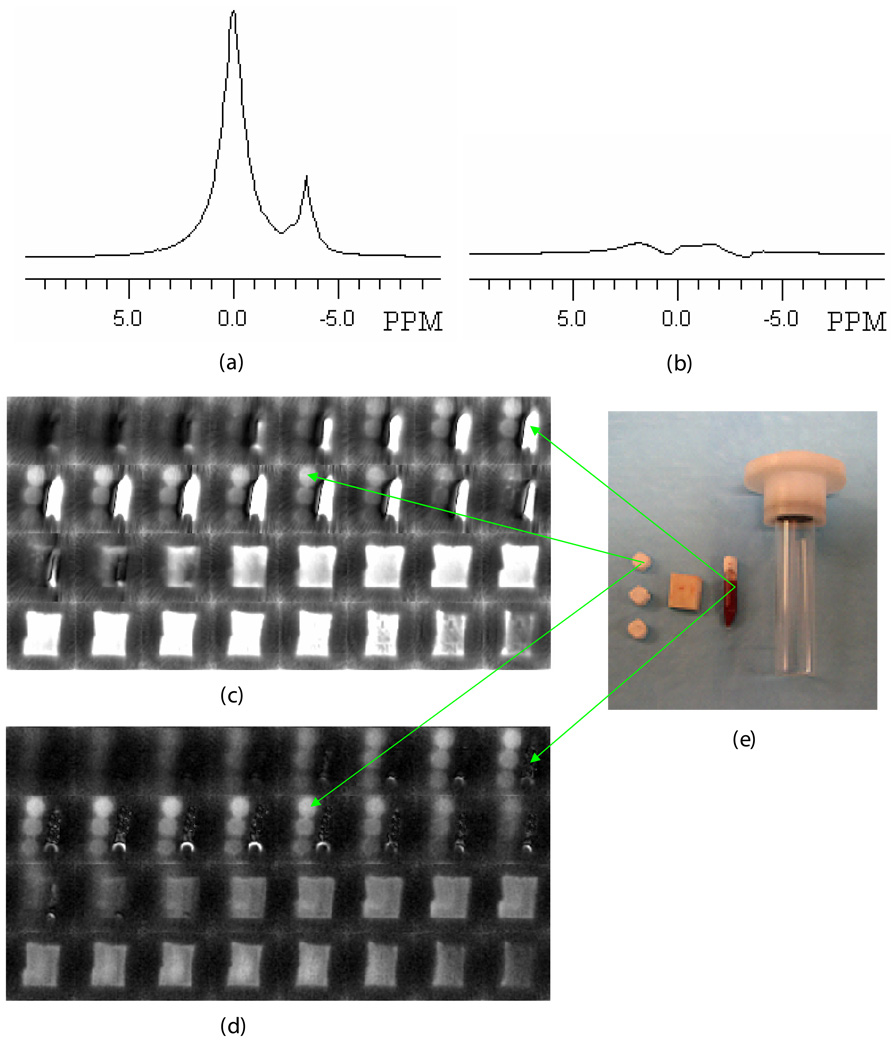
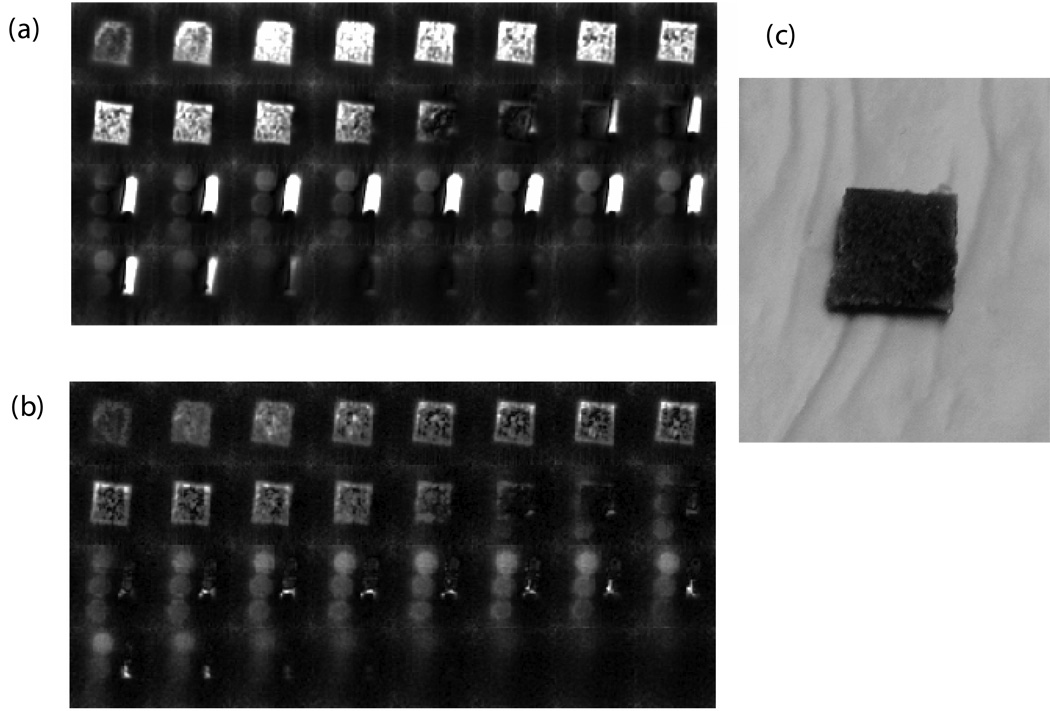
SMRI and WASPI experiments were performed on cortical bone specimens along with the calibration phantom and the marrow specimen. Fig. 4a shows the single-pulse proton spectrum of the marrow specimen; Fig. 4b shows the spectrum of the same marrow specimen acquired with the WASPI sequence without projection gradients. Under suppression, the water and fat signals in Fig 4b were less than 4% of their full values in Fig. 4a. Comparing SMRI images of the marrow specimen with its WASPI images, a similar degree of water and fat suppression was observed (Fig. 4c, d). It is interesting to point out that in SMRI (Fig. 4c), only the polymer pellet with highest density was clearly visualized, since the liquid state proton signal (water and fat) from bone marrow dominated the image. In contrast WASPI (Fig. 4d) enables all three polymer pellets to be observed with clear image boundaries, because the strong liquid signal has been suppressed. Similar degrees of water and fat suppression (monitored by the marrow specimen) were achieved in the trabecular bone specimens (Fig. 5a, b) and in the bone/glass mixtures.
Figure 4. Proton spectra, SMRI and WASPI images, and photograph of a cortical bone specimen, marrow specimen, and polymer phantoms.
a) Single-pulse (SMRI excitation) proton NMR spectrum of the marrow specimen. Water and fat peaks were at 0.00 and −3.50 ppm, respectively. b) Water and fat suppressed (WASPI excitation) proton spectrum of the marrow specimen. The two peaks were reduced down to the baseline. c) SMRI image of the specimen (no water and fat suppression). The central 32 slices from the complete image data set of 64 slices are shown. d) WASPI image of the same specimen. e) Photograph (from right to left) of the specimen holder, marrow specimen, bone specimen, and three polymer pellets. The diameter of the cylindrical polymer pellets was 0.52 cm and thickness varied from 0.30 to 0.35 cm, with densities of 1.171, 0.800, and 0.557 g cm−3 respectively.
Figure 5. SMRI and WASPI images of a trabecular bone specimen, marrow specimen, and polymer phantoms and photograph of the trabecular bone specimen.
MRI imaging of bone specimens were taken along with the marrow specimen and three polymer pellets. a) SMRI (no water and fat suppression). b) WASPI. c) Photograph of the trabecular specimen.
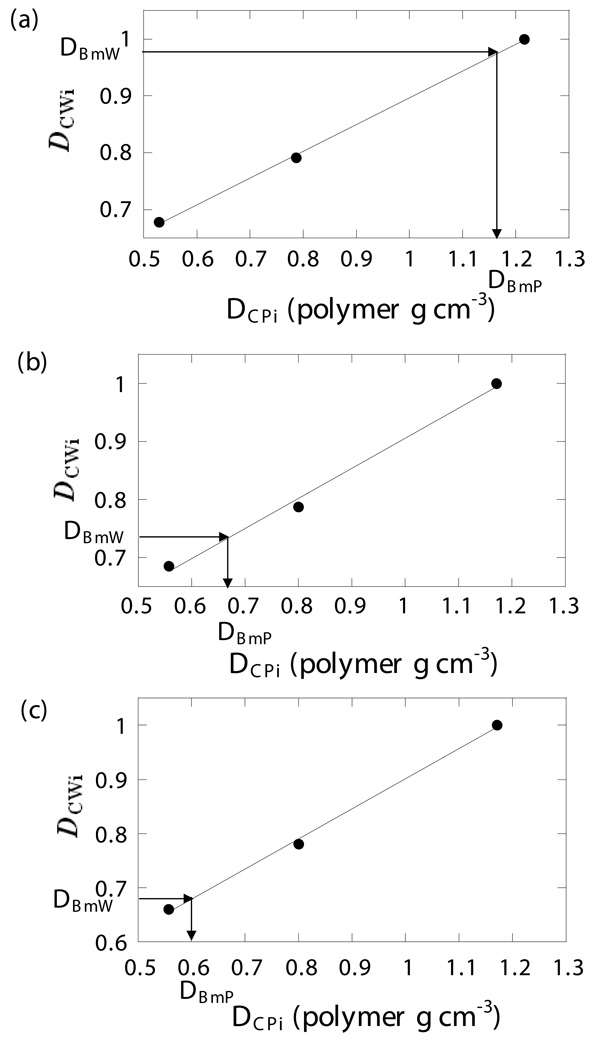
Using the data processing protocol described in the Methods section, the relationship between DCWi and DCPi was found by linear regression. The WASPI derived bone matrix density values DBmP (polymer g cm−3) for the cortical and trabecular bone specimens and the bone/sand mixtures were obtained from the DBmW values using the regression coefficients (Fig. 6). The results are listed in Table 2.
Figure 6. Calibration curves and bone matrix density.
Calibration curves were created by linear regression of the physical densities DCPi of the three polymer calibration pellets to their WASPI densities DCWi. The polymer equivalent bone matrix densities DBmP (expressed in terms of polymer g cm−3) were obtained from the linear regression coefficients from their WASPI densities DBmW. a) Porcine cortical bone specimen 1. b) Porcine trabecular bone specimen 1. c) Bone/glass mixture 1.
Table 2.
Bone matrix densities of bone specimens and bone/glass mixtures measured by gravimetric analysis, amino acid analysis and WASPI.
| Specimen | Gravimetric Analysis DBmG (mass g cm−3) |
Amino Acid Analysis DBmA (protein g cm−3) |
WASPI DBmP (polymer g cm−3) |
|---|---|---|---|
| Cortical bone 1 | 0.543 | 0.507 | 1.167 |
| Cortical bone 2 | 0.416 | 0.346 | 0.856 |
| Cortical bone 3 | 0.583 | 0.487 | 1.323 |
| Trabecular bone 1 | 0.267 | 0.273 | 0.662 |
| Trabecular bone 2 | 0.253 | 0.195 | 0.549 |
| Trabecular bone 3 | 0.217 | 0.226 | 0.631 |
| Trabecular bone 4 | 0.270 | 0.236 | 0.595 |
| Trabecular bone 5 | 0.241 | 0.199 | 0.642 |
| Trabecular bone 6 | 0.345 | 0.366 | 0.669 |
| Trabecular bone 7 | 0.247 | 0.207 | 0.641 |
| Trabecular bone 8 | 0.334 | 0.293 | 0.754 |
| Bone/sand mixture 1 | 0.276 | 0.192 | 0.598 |
| Bone/sand mixture 2 | 0.249 | 0.180 | 0.537 |
| Bone/sand mixture 3 | 0.179 | 0.142 | 0.372 |
| Bone/sand mixture 4 | 0.166 | 0.107 | 0.332 |
| Bone/sand mixture 5 | 0.036 | 0.026 | 0.072 |
To assess the effect of repositioning and day to day instrument stability on the reproducibility of the WASPI measurement, three measurements of the same porcine cortical specimen were made, two on the same day, and the third on the following day. The measured values were 0.884, 0.874, and 0.891 polymer g cm−3 respectively, giving a mean value of 0.883 polymer g cm−3, and a relative standard deviation of 0.97%.
Gravimetric and amino acid analysis were performed on all cortical bone, trabecular bone specimens and bone/glass mixtures. The results are also listed in Table 2.
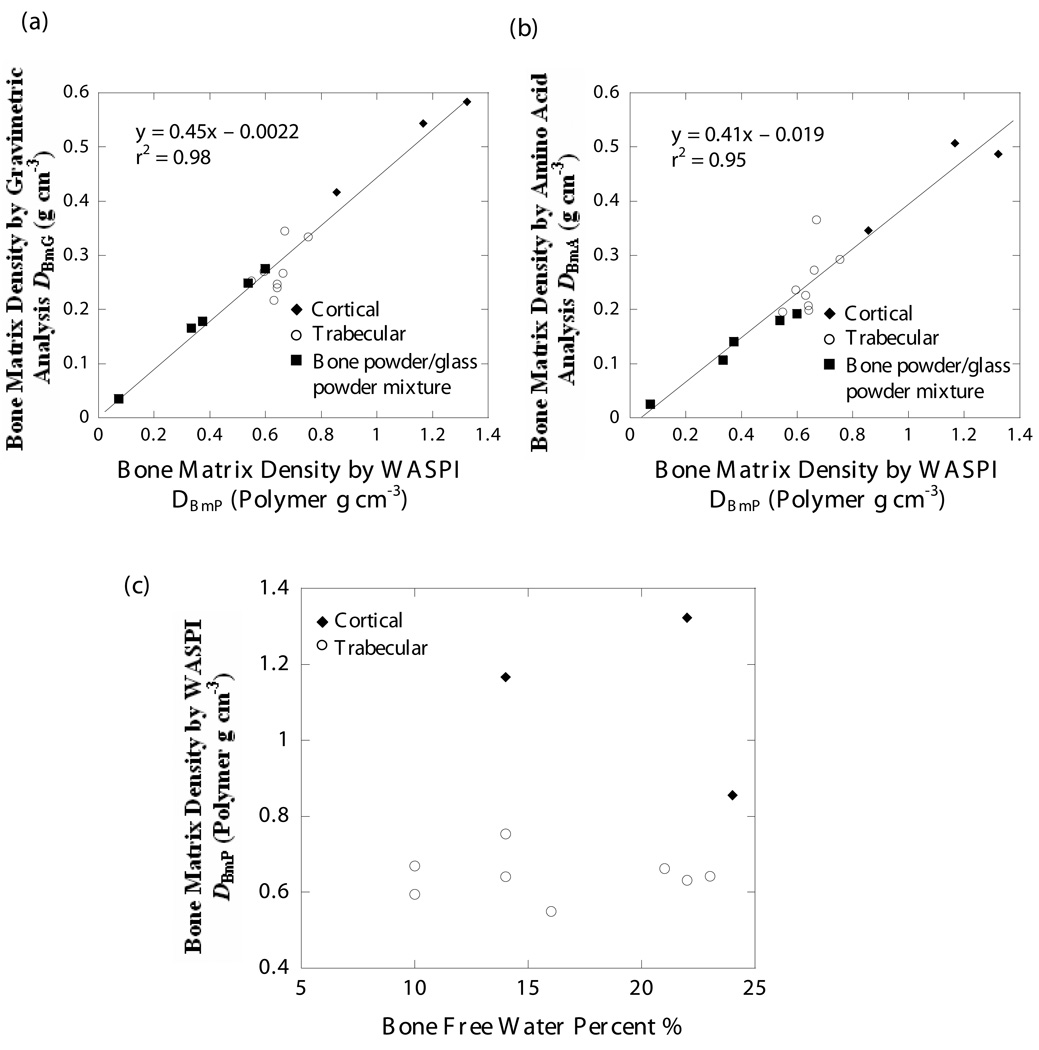
The WASPI measurements were strongly positively correlated with gravimetric analysis (r2 = 0.98) and amino acid analysis (r2 = 0.95). The conversion factor for WASPI derived density DBmP (expressed in units of polymer g cm−3) to bone matrix mass density DBmG (calibrated by gravimetric analysis and expressed in units of g cm−3) was 0.45 (Fig. 7a), while that for DBmP to bone matrix protein density DBmA (calibrated by amino acid analysis and expressed in units of g cm−3) was 0.41 (Fig. 7b). No relationship between DBmP and the free water content in bone was observed (Fig. 7c).
Figure 7. Correlation between WASPI and gravimetric analysis, WASPI and amino acid analysis, WASPI and free bone water content.
a) Correlation between bone matrix density derived by WASPI DBmP (polymer g cm−3) and by gravimetric analysis DBmG (g cm−3). The solid line is the result of a linear regression of data for the 3 cortical, 8 trabecular and 5 bone/glass mixture specimens, which yielded the following relationship: DBmG = 0.45 DBmP – 0.0022; r2 = 0.98. The standard errors of the slope and intercept were 0.03 and 0.02 respectively.
b) Correlation between bone matrix density derived by WASPI DBmP (polymer g cm−3) and by amino acid analysis DBmA (g cm−3). The solid line is the result of a linear regression of data for the 3 cortical, 8 trabecular and 5 bone/sand mixture specimens, which yielded the following relationship: DBmA = 0.41 DBmP – 0.019; r2 = 0.95. The standard errors of the slope and intercept were 0.04 and 0.03 respectively.
c) Relationship between bone matrix density DBmP derived by WASPI (polymer g cm−3) and free water percent in the bone specimens. The water content was obtained by weight loss on drying in the gravimetric analysis. The points are the results of 3 cortical and 8 trabecular bone specimens. There is no significant correlation between WASPI bone matrix density and free water content in the bone specimens.
DISCUSSION
The role of polymer phantom pellets is to serve as stable intensity reference standards, and relate bone matrix signal intensity to bone matrix mass density in units of polymer g cm−3, which can then be converted to true bone matrix mass or protein density in units of g cm−3 as described in the Methods section.
The intrinsic resolution of WASPI images is determined by the resonance linewidth of the proton signal and the applied gradient strength. Having similar resonance linewidths of the polymer blend and solid bone matrix ensures that the images of both substances measured under the same conditions have the same intrinsic resolution. Similarity of the T2s ensures that the same fraction of phantom and matrix signals contribute to image formation, hence reducing measurement errors. More specifically, if the T2 of the calibration phantom were to be much longer than that of the solid bone matrix, as in the case of liquid media phantoms, its signal would be suppressed due to the suppression effect of WASPI on long T2 signals (8). On the other hand, if T2 of the phantom were to be too short compared with that of solid bone matrix, the phantom would not be visualized in the image.
As demonstrated in this study, the PEO/PMMA blend is a good candidate for the calibration phantom for bone matrix density measurement. PEO/PMMA is a miscible blend system. The blends are fully amorphous when the PEO content is below 20–30 weight percent (23–26). In this study, we chose 20% PEO/PMMA blends, although we believe that as long as the blends have PEO content less than 30% and are amorphous, they should work well. Since PEO and PMMA are miscible, spin diffusion between the PEO and PMMA forces all protons in the specimen to have a common T1 and T2, and the wide-line spectrum exhibits a single resonance of linewidth intermediate between the linewidths of the pure components. Thus the linewidth of the blend can be adjusted by changing the PEO molecular weight, giving us freedom to adjust the calibration phantom to the linewidth of the solid bone matrix.
The similarity of the T1s of the solid bone matrix and the calibration material is also important, since the Ernst angle is used in WASPI measurements to optimize the SNR. The proton T1 of the blend used here is very close to that of the solid bone matrix as measured in this study, hence no T1 correction is needed. To prove this, the correction factor F under the experimental conditions can be calculated as
| [4] |
With T1 of the polymer = 3.6 s, T1 of the solid bone matrix = 3.4 s, β = 13°, 12° for polymer and bone experiments respectively, and TR = 0.15 s, the F values of the bone specimen and polymer phantom are very close, the difference being about 4% (Table 1).
Compensation for spatial B1 variation using a uniform fluid phantom should suffice in this study, since the coil used had a relatively uniform B1 (Fig. 3). For larger B1 variation, the RF field must be mapped and the correction factor computed for the known spatial variation in flip angle and the known relaxation parameters.
The correlations between the WASPI derived densities DCWi and the physical densities DCPi of the calibration phantoms were excellent (Fig. 6). The simple cylindrical shape of the polymer pellets ensured the accuracy of the measurement of their volumes, hence the accuracy of the density. The utilization of clean and dry silicon dioxide to dilute the polymer pellets guarantees that the observed proton signal arises solely from the polymer powder (finely divided materials such as silica gel can have huge surfaces that adsorb water which contributes a broad line signal). The WASPI scans of cortical bone and trabecular bone specimens showed that the polymer phantom density range from 0.56 g cm−3–1.17 g cm−3 is large enough to cover the measurement of both types of bone.
The conversion factor for WASPI data to amino acid analysis data is slightly lower than the conversion factor for WASPI data to gravimetric analysis data, as it is expected to be. The bone matrix density measured by amino acid analysis was found to be about 91% of that measured by gravimetric analysis. This difference arises because the two analyses measure different substances, although both have been used as measures of bone matrix. WASPI measures the proton signal arising from the solid bone matrix as defined in the Introduction section, which includes a portion of collagen protons, tightly bound water, and other motionally restricted molecules in bone matrix, but does not include fluid (spectrally resolvable) water and fat. The gravimetric analysis measures the components of bone matrix pyrolyzed at 600 °C, which includes the collagen, noncollagenous proteins, cellular material, and tightly bound water that can’t be completely removed under these conditions, but does not include extractable fat. Though removal of all fat is difficult, most fat should be removed by the solvent extraction and therefore is not included in the material vaporized by pyrolysis in gravimetric analysis. Amino acid analysis only measures the total protein in the bone specimen, and does not include tightly bound water or protein-bound lipid or glycosyl residues. Therefore it is reasonable that bone matrix density measured by amino acid analysis is lower than that measured by gravimetric analysis because the matrix is not entirely protein. In light of these considerations, the conversion factor to be used for an in vivo study would depend on whether the bone matrix mass density or the bone matrix protein density is desired.
The free water and fat content in the bone specimens obtained in the way described in the Methods section are listed in Table 3. The fat content in the trabecular specimens varied in the range of 21–38%, while there was little fat in the cortical specimens (around 1%), which justified skipping the solvent extraction of the bone/sand mixtures. The free water content in bone ranged from 10 to 24%. It is interesting to observe that there is no obvious relationship of any kind (r2 = 0.05) between the WASPI derived bone matrix density in polymer mass density units, DBmP, and the free water content in bone (Table 2, 3, Fig. 7c). This and the linear correlations of WASPI measurements with gravimetric analysis and amino acid analysis of bone specimens are consistent with our assignment of the WASPI signal primarily to solid bone matrix as a composite substance (with most of the signal probably arising from organic molecules), rather than to water.
Table 3.
Free water and fat content of porcine bone specimens, obtained by gravimetric analysis.
| Specimen | Water (weight fraction) | Fat (weight fraction) |
|---|---|---|
| Trabecular bone 1 | 0.21 | 0.28 |
| Trabecular bone 2 | 0.16 | 0.36 |
| Trabecular bone 3 | 0.22 | 0.32 |
| Trabecular bone 4 | 0.10 | 0.37 |
| Trabecular bone 5 | 0.23 | 0.29 |
| Trabecular bone 6 | 0.10 | 0.21 |
| Trabecular bone 7 | 0.14 | 0.38 |
| Trabecular bone 8 | 0.14 | 0.26 |
| Cortical bone 1 | 0.14 | 0.01 |
| Cortical bone 2 | 0.24 | 0.01 |
| Cortical bone 3 | 0.22 | 0.01 |
CONCLUSIONS
The utilization of a properly designed polymer phantom as a mass density reference to enable quantitative WASPI bone matrix density measurements has been demonstrated. In general, this type of calibration phantom can be used to convert the MRI intensity of a solid material exhibiting proton resonance linewidths of the order of a few thousand Hz to the true mass density of the solid in g cm−3. The solid proton signal might arise from a pure homogeneous solid sample, or from solid constituents in a material such as bone containing both fluid and solid constituents. By doing so, the WASPI data can be converted to true three-dimensional bone matrix substance density. If this method proves to be successful in live subjects, it would have potential clinical application in determining the contribution of osteoporosis and/or osteomalacia to a deficit of bone density.
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01-EB004012 (National Institute of Biomedical Imaging and Bioengineering), R01-AG014701 (National Institute of Aging), S10-RR16811 and P41-RR14075 (NIH Division of Research Resources); the Peabody Foundation; the MIND Institute; and the Athinoula A. Martinos Center for Biomedical Imaging. We thank Dr. Brian D. Snyder, and Ara Nazarian, of the Department of Orthopaedic Surgery and the Orthopaedic Biomechanics Laboratory, Beth Israel Deaconess Medical Center, Boston, who provided considerable assistance in the pycnometer measurements of bone specimen volumes.
Grant Sponsors: NIBIB-NIH, NIA-NIH
LIST OF SYMBOLS
- B1
Upper case italic letter B, subscript numeral one
- F
Upper case italic letter f
- M
Upper case italic letter M
- M0
Upper case italic letter M, subscript numeral zero
- DBmA
Upper case italic letter D, subscript upper case letter B, subscript lower case letter m, subscript upper case letter A
- DBmG
Upper case italic letter D, subscript upper case letter B, subscript lower case letter m, subscript upper case letter G
- DBmP
Upper case italic letter D, subscript upper case letter B, subscript lower case letter m, subscript upper case letter P
- DBmW
Upper case italic letter D, subscript upper case letter B, subscript lower case letter m, subscript upper case letter W
- DCPi
Upper case italic letter D, subscript upper case letter C, subscript upper case letter P, subscript lower case letter i
- DCWi
Upper case italic letter D, subscript upper case letter C, subscript upper case letter W, subscript lower case letter i
- MBmW
Upper case italic letter M, subscript upper case letter B, subscript lower case letter m, subscript upper case W
- MCWi
Upper case italic letter M, subscript upper case letter C, subscript upper case W, subscript lower case letter i
- T1
Upper case italic letter T, subscript lower case 1
- T2
Upper case italic letter T, subscript lower case 2
- T2*
Upper case italic letter T, subscript lower case 2, lower case asteroid
- VBt
Upper case italic letter V, subscript upper case letter B, subscript lower case letter t
- VCi
Upper case italic letter D, subscript upper case letter C, subscript lower case letter i
- x
Lower case letter x
- y
Lower case letter y
- z
Lower case letter z
- β
Greek lower case italic beta
REFERENCES
- 1.Basha B, Rao DS, Han Z-H, Parfitt AM. Osteomalacia due to vitamin D depletion: a neglected consequence of intestinal malabsortion. Am J Med. 2000;108:296–300. doi: 10.1016/s0002-9343(99)00460-x. [DOI] [PubMed] [Google Scholar]
- 2.Sartoris DJ. Quantitative bone mineral analysis. In: Resnick D, editor. Bone and Joint Imaging. 2nd ed. W.B. Saunders Company; 1996. p. 154. [Google Scholar]
- 3.Bolotin HH. DXA in vivo BMD methodology: An erroneous and misleading research and clinical gauge of bone mineral status, bone fragility, and bone remodeling. Bone. 2007;41:138–154. doi: 10.1016/j.bone.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Parfitt AM, Qiu S, Rao DS. The mineralization index-A new approach to the histomorphometric appraisal of osteomalacia. Bone. 2004;35:320–325. doi: 10.1016/j.bone.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y, Ackerman JL, Chesler DA, Li J, Neer RM, Wang J, Glimcher MJ. Evaluation of bone mineral density using three dimensional solid state phosphorus-31 NMR projection imaging. Calcif Tissue Int. 1998;62:512–518. doi: 10.1007/s002239900471. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, Chesler DA, Glimcher ML, Garrido L, Wang J, Jiang HJ, Ackerman JL. Multinuclear solid state three dimensional MRI of bone and synthetic calcium phosphates. Proc Nat Acad Sci USA. 1999;96:1574–1578. doi: 10.1073/pnas.96.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y, Ackerman JL, Chesler DA, Wang J, Glimcher MJ. In vivo solid state 31P MRI of human tibia at 1.5 T. Abstracts, Seventh Scientific Meeting, International Society of Magnetic Resonance in Medicine; May 22–28, 1999; Philadelphia, PA, USA. p. 313. [Google Scholar]
- 8.Wu Y, Ackerman JL, Chesler DA, Graham L, Wang Y, Glimcher MJ. Density of organic matrix of native mineralized bone measured by water and fat suppressed proton projection MRI. Magn Reson Med. 2003;50:59–68. doi: 10.1002/mrm.10512. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y, Dai G, Ackerman JL, Hrovat MI, Glimcher MJ, Snyder BD, Nazarian A, Chesler DA. Water and fat suppression projection MRI (WASPI) of rat femur bone. Magn Reson Med. 2007;57:554–567. doi: 10.1002/mrm.21174. [DOI] [PubMed] [Google Scholar]
- 10.Tyler DJ, Robson MD, Henkelman RM, Young IR, Bydder GM. Magnetic resonance imaging with ultrashort TE (UTE) pulse sequences: Technical considerations. J Magn Reson Imaging. 2007;25:279–289. doi: 10.1002/jmri.20851. [DOI] [PubMed] [Google Scholar]
- 11.Techawiboonwong A, Song HK, Wehrli FW. In vivo MRI of submillisecond T(2) species with two-dimensional and three-dimensional radial sequences and applications to the measurement of cortical bone water. NMR Biomed. 2008;21:59–70. doi: 10.1002/nbm.1179. [DOI] [PubMed] [Google Scholar]
- 12.Idiyatullin D, Corum C, McIntosh A, Moeller S, Garwood M. Direct MRI of human teeth by swift. Abstracts, Joint Annual Meeting, International Society of Magnetic Resonance in Medicine, and European Society for Magnetic Resonance in Medicine and Biology; May 19–25, 2007; Berlin, Germany. p. 383. [Google Scholar]
- 13.Yanasak N, Allison J. Use of capillaries in the construction of an MRI phantom for the assessment of diffusion tensor imaging: demonstration of performance. Magn Reson Imaging. 2006;24:1349–1361. doi: 10.1016/j.mri.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Roe JE, Prentice WE, Hornak JP. A multipurpose MRI phantom based on a reverse micelle solution. Magn Reson Med. 1996;35:136–141. doi: 10.1002/mrm.1910350118. [DOI] [PubMed] [Google Scholar]
- 15.Mazzara GP, Briggs RW, Wu Z, Steinback BG. Use of a modified polysaccharide gel in developing a realistic breast phantom for MRI. Magn Reson Imaging. 1996;14:639–648. doi: 10.1016/0730-725x(96)00054-9. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Kanazawa Y, Okumura M, Taninaka A, Yokawa T, Shinohara H. Lanthanoid endohedral metallofullerenols for MRI contrast agents. J Am Chem Soc. 2003;125:4391–4397. doi: 10.1021/ja027555+. [DOI] [PubMed] [Google Scholar]
- 17.Ruminski J, Bobek-Billewicz B. Parametric imaging in dynamic susceptibility contrast MRI-phantom and in vivo studies; Conf Proc IEEE Eng Med Biol Soc; 2004. pp. 1104–1107. [DOI] [PubMed] [Google Scholar]
- 18.Allen MJ, MacRenaris KW, Venkatasubramanian PN, Meade TJ. Cellular delivery of MRI contrast agents. Chem Biol. 2004;11:301–307. doi: 10.1016/j.chembiol.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Lutz NW, Schultz E. Phantom substances for quantitative evaluation of MRT images. III. Effect of various protein concentrations on MRT intensity values. Digitale Bilddiagnostik. 1987;7:1–4. [PubMed] [Google Scholar]
- 20.Brix G, Schad LR, Lorenz WJ. Evaluation of proton density by magnetic resonance imaging: phantom experiments and analysis of multiple component proton transverse relaxation. Phys Med Biol. 1990;35:53–66. doi: 10.1088/0031-9155/35/1/006. [DOI] [PubMed] [Google Scholar]
- 21.Hobe J, Faber S, Stammberger T, Reiser M, Englmeier K-H, Eckstein F. A technique for 3D in vivo quantification of proton density and magnetization transfer coefficients of knee joint cartilage. Osteoarthritis Cartilage. 2000;8:426–433. doi: 10.1053/joca.1999.0318. [DOI] [PubMed] [Google Scholar]
- 22.Ernst RR, Bodenhausen G, Wokaun A. Principles of nuclear magnetic resonance in one and two dimensions. Oxford: Clarendon Press; 1987. p. 124. [Google Scholar]
- 23.Cao H, Lin G, Jones AA. Deuterium solid echo line shape study of poly(ethylene oxide) dynamics in a blend with poly(methyl methacrylate) J Polym Sci B: Polym Phys. 2005;43:2433–2444. [Google Scholar]
- 24.Lartigue C, Guillermo A, CohenAddad JP. Proton NMR investigation of the local dynamics of PEO in PEO/PMMA blends. J Polym Sci B: Polym Phys. 1997;35:1095–1105. [Google Scholar]
- 25.Guillermo A, Lartigue C, CohenAddad JP. PEO temporary network in PEO/PMMA blends: NMR approach. Macromolecules. 1998;3:769–775. [Google Scholar]
- 26.Lutz TR, He Y, Ediger MD, Cao H, Lin G, Jones AA. Rapid poly(ethylene oxide) segmental dynamics in blends with Poly(methyl methacrylate) Macromolecules. 2003;36:1724–1730. [Google Scholar]