Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) plays an important role in the cellular response to stress and DNA damage. However, excessive activity of PARP-1 exacerbates brain injury via NAD+ depletion and energy failure. The purpose of this study was to determine if tagging single nucleotide polymorphisms (tSNPs) covering multiple regions of the PARP-1 gene are related to outcome after traumatic brain injury (TBI) in humans. DNA from 191 adult patients with severe TBI was assayed for four tSNPs corresponding to haplotype blocks spanning the PARP-1 gene. Categorization as favorable or poor outcome was based on Glasgow Outcome Scale (GOS) score assigned at 6 months. PARP-1 enzyme activity was indirectly evaluated by quantifying poly-ADP-ribose (PAR)-modified proteins in cerebrospinal fluid (CSF) using an enzyme-linked immunosorbent assay. In multiple logistic regression analysis controlling for age, initial Glasgow Coma Scale score, and gender, the AA genotype of SNP rs3219119 was an independent predictor of favorable neurologic outcome. This SNP tags a haplotype block spanning the automodification and catalytic domains of the PARP-1 gene. SNP rs2271347 correlated with CSF PAR-modified protein level. This SNP, which did not correlate with outcome, tags a haplotype block spanning the promoter region of the PARP-1 gene. We conclude that after severe TBI in humans, a PARP-1 polymorphism within the automodification-catalytic domain is associated with neurological outcome, while a polymorphism within the promoter region was associated with CSF PAR-modified protein level. These findings must be replicated in a prospective study before the relevance of PARP-1 polymorphisms after TBI can be established.
Key words: ADP-ribosyltransferase, traumatic brain injury, poly(ADP-ribose) polymerase, poly-ADP-ribosylation
Introduction
Poly(ADP-ribose) polymerase-1 (PARP-1), encoded by the PARP-1 gene, is a ubiquitous enzyme found in multiple cellular compartments, and it uses NAD+ as a substrate to add long-branching ADP-ribose chains to proteins in response to DNA damage (Ueda and Hayaishi, 1985; Virag and Szabo, 2002). These poly(ADP-ribose) (PAR)-modified proteins may include DNA repair proteins, transcription factors, and the PARP-1 enzyme itself (Virag and Szabo, 2002). Thus PARP-1 plays an important role in the cellular response to stress. However, since more than 200 molecules of NAD+ may be consumed during the poly-ADP-ribosylation of a single protein (Virag and Szabo, 2002), PARP-1 overactivation may lead to energy failure and cell death via NAD+ depletion, inhibition of electron transport, and ultimate reduction of ATP (Halmosi et al., 2001).
PARP-1 overactivation has been shown to exacerbate damage after experimental traumatic brain injury (TBI) (LaPlaca et al., 1999; Satchell et al., 2003; Whalen et al., 1999), and cerebral ischemia (Eliasson et al., 1997; Endres et al., 1998), and both genetic deletion of PARP-1 and PARP-1 inhibition have been shown to be beneficial in experimental trauma (Clark et al., 2007). Also, nuclear and/or mitochondrial PARP-1 activation have been shown to mediate apoptotic cell death via calpain activation and eventual translocation of apoptosis-inducing factor from the mitochondria to the nucleus (Du et al., 2003; Yu et al., 2002). Although there is a preponderance of experimental evidence that activated PARP-1 is deleterious after TBI by promoting energy failure and apoptosis, there is not a consensus (Nagayama et al., 2000). TBI results in more than 200,000 hospitalizations and 50,000 deaths annually in the United States alone (Thurman et al., 1999), and it is the leading cause of death and disability in young people (Myburgh et al., 2008). While the role of PARP-1 has been extensively studied in animal models of TBI, the influence of PARP-1 after TBI in humans has received limited attention (Fink et al., 2008).
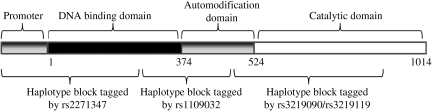
PARP-1 is the prototypical member of a large family of PARPs which are encoded by different genes and have a highly conserved catalytic domain (Ame et al., 2004). A schematic diagram of the mature protein product is displayed in Figure 1, and includes a 374-residue N-terminal zinc-finger DNA binding domain, a 150-residue central automodification domain, and a 490-residue C-terminal catalytic domain (Tao et al., 2008). The PARP-1 gene occupies a 47-kb segment on chromosome 1, and consists of 24 exons and 1162 codons. Humans with the heterozygous genotype of a single nucleotide polymorphism (SNP) of PARP-1 show delayed onset of Parkinson's disease (Infante et al., 2007), a condition in which oxidative stress contributes (Gandhi and Wood, 2005). PARP-1 polymorphisms were associated with the development of arthritis and nephritis in patients with systemic lupus erythematosus, a disease in which ineffective DNA repair is implicated (Hur et al., 2006). One study investigating genotype-phenotype relationships of PARP-1 polymorphisms found a reduction of in-vitro enzymatic activity with a SNP, resulting in an amino acid change of Val to Ala at codon 762 within the catalytic domain of the enzyme (Wang et al., 2007). Recently, a biomarker for PARP activity, the detection of PAR-modified proteins by enzyme-linked immunosorbent assay (ELISA), has been developed to indirectly quantify PARP activity in the cerebrospinal fluid (CSF) of TBI patients (Fink et al., 2008). There are currently no studies investigating genotype-phenotype relationships of PARP-1 polymorphisms and impact on outcome after TBI. Accordingly, we hypothesized that genetic variability caused by PARP-1 polymorphisms impacts neurological outcome and levels of PAR-modified proteins in CSF after TBI.
FIG. 1.
A schematic of the mature protein product, which includes a 374-residue N-terminal zinc-finger DNA binding domain, a 150-residue central automodification domain, and a 490-residue C-terminal catalytic domain.
Methods
Patient enrollment
This retrospective study was approved by the University of Pittsburgh Institutional Review Board, and included subjects admitted to the University of Pittsburgh Medical Center Neurointensive Care Unit from May 2000 to September 2006. Inclusion criteria were individuals aged 18–75 years who sustained severe TBI as defined by a Glasgow Coma Scale (GCS) score of < 9, and had an indwelling ventriculostomy catheter placed for intermittent CSF drainage and monitoring of intracranial pressure. Subjects were excluded from the study if they had a pre-existing neurologic deficit, penetrating TBI, or cardiac or respiratory arrest. Informed consent was obtained from the subjects' surrogate prior to data collection, and continued assent was obtained from the subject when possible. Data regarding demographics and specific characteristics of injury and treatment were collected from medical records. Samples of CSF collected from an indwelling ventriculostomy catheter and blood collected from an intravenous catheter were catalogued and stored at −80°C.
PARP-1 genotyping
Using HapMap Genome Browser Build 35 based on data available in June 2007, one tSNP was chosen to mark each of the four haplotype blocks of the PARP-1 gene, including sequences spanning 1 kb 5′ of the gene into the promoter region, through 1 kb past the most 3′ exon. The four tSNPs, rs1109032, rs3219090, rs3219119, and rs2271347, were chosen because they have a minor allele frequency of at least 20% in the population, and an r2 of at least 0.80.
DNA was extracted from CSF or blood samples using a commercially available kit (Qiagen, Valencia, CA). Genotyping was performed for each tSNP using TaqMan allele discrimination as previously described (Heid et al., 1996), using commercially available TaqMan assays (Applied Biosystems, Foster City, CA). Briefly, this approach uses dual-labeled hybridization probes, each containing one reporter dye (FAM, or carboxyfluorescein), whose fluorescence is cancelled by the quencher dye (TAMRA, or 6-carboxy-tetramethylrhodamine) while in close proximity prior to amplification. During DNA polymerization, the reporter is separated from the quencher, resulting in a quantifiable, allele-specific release of fluorescence. The sequence detector (ABI Prism; Applied Biosystems) measures the fluorescence spectra of all 96 wells continuously during PCR thermal cycling, which in turn determines the genotype.
Measurement of PAR-modified proteins
The CSF samples used for this assay were collected by intermittent drainage as part of standard of care. PAR-modified proteins were quantified from CSF samples obtained approximately 24 h after injury, the time when CSF PAR-modified proteins peak after TBI in children, using ELISA as previously described (Fink et al., 2008). Briefly, a standard was produced by preparing a reaction mixture consisting of a known concentration of the PARP-1 substrate histone-1, active PARP-1 enzyme, NAD+, and nicked DNA (Trevigen, Gaithersburg, MD), and incubating for 1 h at 37°C. Assuming nearly 100% completion of the reaction, serial dilutions of the PAR-modified histone mixture were used to generate standard curves. PAR-modified proteins in duplicate CSF samples and PAR-modified histone standards were diluted 1:5 with coating buffer solution (Alpha Diagnostic International Inc., San Antonio, TX), were placed into 96-well high-binding plates (Corning Inc., Corning, NY), and incubated at 4°C overnight. A 1:1000 dilution of rabbit polyclonal antibody against PAR (Trevigen) was then added to each well and incubated at room temperature for 60 min. Then a 1:2000 dilution of a secondary anti-rabbit antibody conjugated to horseradish peroxidase (Alpha Diagnostic) was added to each well and incubated at room temperature for 30 min. A colorimetric detection system was used and absorbance was determined at 450 nm. CSF PAR-modified protein concentrations were calculated using the curve generated from PAR-modified histone standards.
Measures of outcome
Functional outcome determined as GOS assigned at 6 months was dichotomized into unfavorable outcome (GOS 1–3: death, persistent vegetative state, or severe disability), and favorable outcome (GOS 4–5: moderate disability or good recovery). The GOS assigned at 6 months was used, since data were incomplete at later time points. All assessments were performed face-to-face if possible, or by phone interview by a neuropsychological technician supervised by a neuropsychiatric attending or neurosurgery attending, all of whom were blinded to PARP-1 genotype and CSF PAR-modified protein level.
Statistical analysis
Demographics and initial GCS score for each genotype for each tSNP were compared using ANOVA. Relationships between genotype and outcome were tested using the chi-square test for linear trends. Based this analysis, genotypes for each SNP were dichotomized for multivariate logistic analysis. Multivariate analysis was also used to test whether age, GCS, gender, outcome, or mortality were independent contributors to CSF PAR levels.
Results
Patient demographics
A total of 191 subjects were included in the analysis. Table 1 shows patient demographic data. The age range of the subjects was 16–73 years, with median 31.5 years, and 79% of the subjects were male. The median GCS score assigned upon arrival to the emergency department was 5. Overall mortality was 32.5%, and the percentage of subjects with poor outcome at 12 months was 61%. The population was 94.1% Caucasian and 4.3% African-American. Table 2 outlines the association between the genotype of the tSNPs for the four haplotype blocks, and age, initial GCS, and gender. Importantly, there were no statistically significant differences between genotype groups and any of these factors, all of which are known to influence outcome in TBI. The genotype results showed each tSNP to be in Hardy-Weinberg equilibrium. In some cases, the minor allele frequencies were lower than expected in our population, compared to published data from HapMap, leading to small numbers in some of the groups. For example, only 12 (6%) of subjects had the AA genotype of rs2271347, and only 10 (5%) had the TT genotype of rs1109032; one of the selection criteria for each tSNP was an expected minor allele frequency of approximately 20%.
Table 1.
Patient Demographics
| Age (y) | 34.2 ± 14.7 a |
| Initial Glasgow Coma Scale score | 5 [3–13] a |
| Male (%) | 79 |
| Mortality (%) | 32.5 |
| Poor outcome (%) | 61 |
| Caucasian (%) | 94.1 |
| African-American (%) | 4.3 |
| Asian/Pacific Islander (%) | 1.8 |
Data are mean ± standard deviation or median [range].
Table 2.
Demographics of PARP-1 Polymorphisms
| rs2271347 Genotype | AA | AG | GG | p |
|---|---|---|---|---|
| n (freq %) | 12 (6) | 65 (35) | 111 (59) | |
| Age | 29.0 ± 14.5 | 33.3 ± 14.3 | 34.7 ± 14.9 | 0.41 |
| Initial Glasgow Coma Scale score | 5.1 ± 1.6 | 5.3 ± 1.7 | 5.5 ± 1.9 | 0.63 |
| Sex (% male) | 75 | 77 | 80 | 0.41 |
|
rs1109032 Genotype
|
AA |
AT |
TT |
P |
| n (freq %) | 129 (68) | 52 (27) | 10 (5) | |
| Age | 34.6 ± 15.5 | 33.0 ± 12.9 | 36.3 ± 15.4 | 0.73 |
| Initial Glasgow Coma Scale score | 5.3 ± 1.7 | 5.4 ± 2.2 | 6.0 ± 2.2 | 0.54 |
| Sex (% male) | 79 | 85 | 50 | 0.73 |
| rs3219090 Genotype | ||||
| n (freq %) | 24 (13) | 79 (42) | 84 (45) | |
| Age | 37.5 ± 13.6 | 33.1 ± 14.0 | 34.6 ± 15.7 | 0.44 |
| Initial Glasgow Coma Scale score | 5.5 ± 2.0 | 5.4 ± 1.9 | 5.4 ± 1.8 | 0.95 |
| Sex (% male) | 71 | 84 | 77 | 0.44 |
| rs3219119 Genotype | ||||
| n (freq %) | 83 (45) | 77 (41) | 26 (14) | |
| Age | 34.1 ± 15.5 | 33.9 ± 14.7 | 36.8 ± 13.3 | 0.68 |
| Initial Glasgow Coma Scale score | 5.5 ± 1.7 | 5.4 ± 2.0 | 5.4 ± 2.0 | 0.94 |
| Sex (% male) | 75 | 82 | 73 | 0.68 |
Data are mean ± standard deviation or median [range].
PARP-1, poly(ADP-ribose) polymerase-1.
PARP-1 genotype and outcome
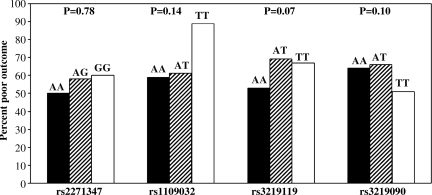
Figure 2 shows the relationship between genotype for each of the tSNPs and percentage of poor outcome, using chi-square analysis for linear trends. From this analysis, multivariate models were developed based on dichotomized genotypes, to determine whether individual polymorphisms were associated with clinical outcome. In multiple logistic regression controlling for age, initial GCS, and gender, the AA genotype of rs3219119 was an independent predictor of favorable neurologic outcome, as was the TT genotype of rs3219090 (Table 3).
FIG. 2.
Relationships between genotype for each of the tagging single nucleotide polymorphisms and percentage of poor outcome, using chi-square analysis for linear trends.
Table 3.
Associations between Genotype and Outcome
| Genotype | Odds ratio for poor outcome | p |
|---|---|---|
| rs2271347GG | 0.96 [0.51–1.82] | 0.78 |
| rs1109032AA | 0.19 [0.02–1.65] | 0.13 |
| rs3219090TT | 0.49 [0.26–0.93] | 0.03 |
| rs3219119AA | 0.46 [0.24–0.89] | 0.02 |
CSF PAR-modified protein level
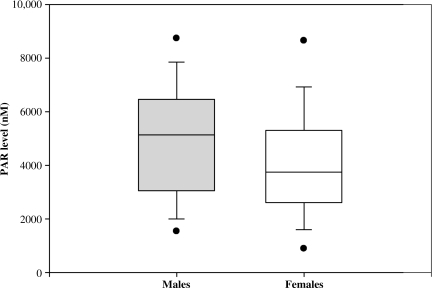
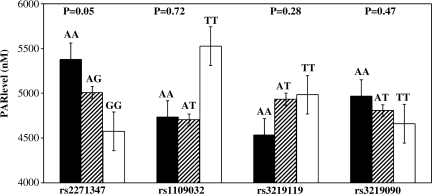
The mean CSF PAR level for all patients was 4748 ±2167 nM. PAR level was not associated with GOS (4642 ± 2231 versus 4912 ± 2072 nM, poor versus favorable outcome, respectively). Table 4 shows a multivariate analysis investigating the clinical factors associated with a PAR level higher than the median value. Among subjects with a PAR level below the median, 64% had poor outcome, compared to 58% in those with a PAR level above the median (OR 0.86 [0.42–1.76], p = 0.67). Age, initial GCS score, and mortality were also not associated with PAR level. However, males were 2.62 times more likely to have a PAR level above the median than were females (95% CI 1.09, 6.27; p = 0.03). The association between PAR level and gender is shown in Figure 3. Relationships between tSNP genotypes and PAR levels are shown in Figure 4. Using multivariate analysis controlling for gender, rs2271347 was associated with PAR level (AA 5378 ± 1917, AG 5008 ± 2196, and GG 4574 ± 2172 nM; p = 0.05), suggesting a genotype-phenotype relationship. The two tSNPs associated with outcome, rs3219119 and rs3219090, were not associated with PAR level.
Table 4.
Clinical Factors Associated with High PAR Level
| OR for high PAR level | p | |
|---|---|---|
| Age | 0.99 [0.24–2.95] | 0.40 |
| Initial Glasgow Coma Scale score | 0.97 [0.91–1.04] | 0.42 |
| Male gender | 2.62 [1.09–6.27] | 0.03 |
| Outcome | 0.86 [0.42–1.76] | 0.67 |
| Mortality | 1.099 [0.51–2.354] | 0.81 |
The odds ratio [OR] for having a PAR level higher than the median is represented.
PAR, poly-ADP-ribose.
FIG. 3.
Box and whisker plot depicting relationship between gender and PAR level, with median, quartiles, and outliers (PAR, poly-ADP-ribose).
FIG. 4.
Relationships between tSNP genotypes and PAR levels, using multivariate analysis controlling for patient gender (tSNP, tagging single nucleotide polymorphisms; PAR, poly-ADP-ribose).
Discussion
Here we show that PARP-1 polymorphisms within the automodification-catalytic domain are associated with neurological outcome after TBI, whereas a PARP-1 polymorphism within the promoter region is associated with CSF PAR-modified protein levels. The latter may represent a genotype-phenotype relationship between PARP-1 polymorphism within the promoter region and enzyme activity. The approach was to tag each PARP-1 gene haplotype block, or segment of DNA inherited together, with a single nucleotide polymorphism, thereby attempting to account for all of the variability in the gene. Using data from HapMap Genome Browser Build 35 in July 2007, one tSNP was chosen for each of the four haplotype blocks, including sequences spanning 1 kb 5′ into the promoter region, through 1 kb past the most 3′ exon. However, our genotyping data showed that two of the tSNPs, rs3219119 and rs3219090, were very closely associated, and therefore were not independent of one another as previously thought. Of the nine possible genotype combinations for these two tSNPs, 97% of subjects had one of three genotypes, whereas if these tSNPs were not in linkage disequilibrium, a less skewed distribution would be expected. Evaluation of updated HapMap data from Build 36 demonstrates that these two tSNPs are indeed correlated, validating our genotyping data to some degree. This association between the tSNPs rs3219119 and rs3219090 led to a gap in the coverage of the PARP-1 gene in sequences 3′ to exon 20. This includes the 129 transcribed codons from exons 21–24, as well as introns 21–24, and therefore a portion of the catalytic domain of the mature protein. This represents a limitation in the study, since it is plausible that variations in sequences coding for areas of the catalytic domain of this important DNA repair enzyme may impact function, enzymatic activity, and possibly outcome.
The haplotype block tagged by rs3219119 and rs3219090 includes sequences from intron 7 through exon 20, which code for residues in the automodification domain and part of the catalytic domain. These tSNPs were associated with outcome, as the AA genotype of rs3219119 and the TT genotype of rs3219090 were independent predictors of favorable outcome. However, these tSNPs were not associated with CSF PAR-modified protein level.
The haplotype block tagged by rs2271347 includes sequences 5′ to the promoter region through exon 1, which includes part of the DNA binding domain. This tSNP was associated with CSF PAR-modified protein level, a reflection of PARP-1 activity (Du et al., 2003; Satchell et al., 2003; Yu et al., 2002), consistent with the possibility that this polymorphism influences the amount of PARP-1 enzyme. However, this tSNP was not associated with outcome.
While speculative, it is possible that polymorphism within the automodification and catalytic domains alters PARP-1 enzyme efficiency, but not activity. For example, PARP-1 corresponding to each polymorphism may place different numbers of ADP-ribose per modified protein, yielding different amounts of NAD+ consumed, or may have different affinities for various PAR protein targets. The ELISA used in this study measures the number of proteins modified by PARP, but not the number of ADP-ribose moieties attached to each protein, or the specific proteins modified. This could be addressed using ex-vivo studies and the individual PARP-1 enzymes. In an ex-vivo experiment, we have shown that PAR-modification of electron transport chain proteins affects oxygen consumption (Lai et al., 2008).
A limitation of this study is that measuring PAR-modified proteins in CSF represents an indirect reflection of PARP-1 activity. Further, it assumes that the amount and type of PAR-modified proteins in CSF reflect the amount and type of PAR-modified proteins within cells. Furthermore, since the course of oxidative stress after TBI is time-dependent (Deng et al., 2007), and may begin as early as 3 h post-injury (Ansari et al., 2008), the timing of CSF sample collection is very important. Samples were collected at approximately 1 day post-injury, based on our pediatric study that showed that CSF PAR-modified protein levels peaked at 24 h after injury (Fink et al., 2008). However, we recognize that small differences in timing among the samples could lead to differences in CSF PAR levels.
In the present study, males with severe TBI were more likely to have a higher CSF PAR-modified protein level than females. Our previous study in children with TBI also showed a similar sex difference in patients with TBI (Fink et al., 2008). Consistent results were found in an in-vitro experimental study, in which cultured primary cortical neurons from male rats were more sensitive to peroxynitrite, a potent PARP-1 activator, than neurons from females (Du et al., 2004). While this suggests that sex-dependent differences in PARP-1 are at least in part independent of circulating sex hormones, the powerful influence of sex hormones cannot be discounted. In our study, women over 45 tended to have higher CSF PAR levels than those under 45 (4691 ± 2388 [n = 9] versus 3723 ± 1736 [n = 20] nM; p = 0.23). This is consistent with a study in which ovariectomized female mice were less protected from PARP-1 activation than normal females, which may be explained by estrogen anchoring PARP-1 to estrogen receptor-α, inhibiting its DNA binding (Mabley et al., 2005). Our study was not powered to detect a difference in PAR levels between pre- and post-menopausal females, but this warrants further investigation.
A final caveat pertains to genetic polymorphism studies in general. To date, dozens of polymorphisms have been implicated in influencing outcome after TBI, most notoriously apolipoprotein E (Diaz-Arrastia and Baxter, 2006). Despite promising results, many of these studies have been difficult to replicate, particularly those with relatively small sample sizes (Ioannidis et al., 2003). Furthermore, elucidating functional links between genetic variation and phenotypic traits, not to mention outcome, remains challenging (Frazer et al., 2009). Our data are an example of this, as the genotypic and phenotypic results could be interpreted as conflicting.
We have shown that after severe TBI in humans, different PARP-1 polymorphisms are associated with neurological outcome and indirect measures of enzyme activity. Given the powerful effects of manipulation of this enzyme in experimental brain injury, this pathway may be an important therapeutic target in human TBI, but may be dependent on both PARP-1 genotype and patient gender. These findings must be replicated in a prospective study before the relevance of PARP-1 polymorphisms after TBI can be established.
Acknowledgments
The authors would like to thank the National Institute of Child Health and Human Development (T32 HD 40686), the National Institute of Neurological Disorders and Stroke (NS30318), and the National Institute of Nursing Research (R01NR008424).
Author Disclosure Statement
No competing financial interests exist.
References
- Ame J.C. Spenlehauer C. de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Ansari M.A. Roberts K.N. Scheff S.W. A time course of contusion-induced oxidative stress and synaptic proteins in cortex in a rat model of TBI. J. Neurotrauma. 2008;25:513–526. doi: 10.1089/neu.2007.0451. [DOI] [PubMed] [Google Scholar]
- Clark R.S. Vagni V.A. Nathaniel P.D. Jenkins L.W. Dixon C.E. Szabo C. Local administration of the poly(ADP-ribose) polymerase inhibitor INO-1001 prevents NAD+ depletion and improves water maze performance after traumatic brain injury in mice. J. Neurotrauma. 2007;24:1399–1405. doi: 10.1089/neu.2007.0305. [DOI] [PubMed] [Google Scholar]
- Deng Y. Thompson B.M. Gao X. Hall E.D. Temporal relationship of peroxynitrite-induced oxidative damage, calpain-mediated cytoskeletal degradation and neurodegeneration after traumatic brain injury. Exp. Neurol. 2007;205:154–165. doi: 10.1016/j.expneurol.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Arrastia R. Baxter V.K. Genetic factors in outcome after traumatic brain injury: what the human genome project can teach us about brain trauma. J. Head Trauma Rehabil. 2006;21:361–374. doi: 10.1097/00001199-200607000-00007. [DOI] [PubMed] [Google Scholar]
- Du L. Bayir H. Lai Y. Zhang X. Kochanek P.M. Watkins S.C. Graham S.H. Clark R.S. Innate gender-based proclivity in response to cytotoxicity and programmed cell death pathway. J. Biol. Chem. 2004;279:38563–38570. doi: 10.1074/jbc.M405461200. [DOI] [PubMed] [Google Scholar]
- Du L. Zhang X. Han Y.Y. Burke N.A. Kochanek P.M. Watkins S.C. Graham S.H. Carcillo J.A. Szabo C. Clark R.S. Intra-mitochondrial poly(ADP-ribosylation) contributes to NAD+ depletion and cell death induced by oxidative stress. J. Biol. Chem. 2003;278:18426–18433. doi: 10.1074/jbc.M301295200. [DOI] [PubMed] [Google Scholar]
- Eliasson M.J. Sampei K. Mandir A.S. Hurn P.D. Traystman R.J. Bao J. Pieper A. Wang Z.Q. Dawson T.M. Snyder S.H. Dawson V.L. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat. Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- Endres M. Scott G.S. Salzman A.L. Kun E. Moskowitz M.A. Szabo C. Protective effects of 5-iodo-6-amino-1,2-benzopyrone, an inhibitor of poly(ADP-ribose) synthetase against peroxynitrite-induced glial damage and stroke development. Eur. J. Pharmacol. 1998;351:377–382. doi: 10.1016/s0014-2999(98)00381-1. [DOI] [PubMed] [Google Scholar]
- Fink E.L. Lai Y. Zhang X. Janesko-Feldman K. Adelson P.D. Szabo C. Berger R.P. Sarnaik A.A. Kochanek P.M. Clark R.S. Quantification of poly(ADP-ribose)-modified proteins in cerebrospinal fluid from infants and children after traumatic brain injury. J. Cereb. Blood Flow Metab. 2008;28:1523–1529. doi: 10.1038/jcbfm.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A. Murray S.S. Schork N.J. Topol E.J. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Gandhi S. Wood N.W. Molecular pathogenesis of Parkinson's disease. Hum. Mol. Genet. 2005;14(Spec No2):2749–2755. doi: 10.1093/hmg/ddi308. [DOI] [PubMed] [Google Scholar]
- Halmosi R. Berente Z. Osz E. Toth K. Literati-Nagy P. Sumegi B. Effect of poly(ADP-ribose) polymerase inhibitors on the ischemia-reperfusion-induced oxidative cell damage and mitochondrial metabolism in Langendorff heart perfusion system. Mol. Pharmacol. 2001;59:1497–1505. doi: 10.1124/mol.59.6.1497. [DOI] [PubMed] [Google Scholar]
- Heid C.A. Stevens J. Livak K.J. Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hur J.W. Sung Y.K. Shin H.D. Park B.L. Cheong H.S. Bae S.C. Poly(ADP-ribose) polymerase (PARP) polymorphisms associated with nephritis and arthritis in systemic lupus erythematosus. Rheumatology (Oxford) 2006;45:711–717. doi: 10.1093/rheumatology/kei262. [DOI] [PubMed] [Google Scholar]
- Infante J. Sanchez-Juan P. Mateo I. Rodriguez-Rodriguez E. Sanchez-Quintana C. Llorca J. Fontalba A. Terrazas J. Oterino A. Berciano J. Combarros O. Poly (ADP-ribose) polymerase-1 (PARP-1) genetic variants are protective against Parkinson's disease. J. Neurol. Sci. 2007;256:68–70. doi: 10.1016/j.jns.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Ioannidis J.P.A. Trikalinos T.A. Ntzani E.E. Contopoulos-Ionnidis D.G. Genetic associations in large versus small studies: an empirical assessment. Lancet. 2003;361:567–571. doi: 10.1016/S0140-6736(03)12516-0. [DOI] [PubMed] [Google Scholar]
- Lai Y. Chen Y. Watkins S.C. Nathaniel P.D. Guo F. Kochanek P.M. Jenkins L.W. Szabo C. Clark R.S. Identification of poly-ADP-ribosylated mitochondrial proteins after traumatic brain injury. J. Neurochem. 2008;104:1700–1711. doi: 10.1111/j.1471-4159.2007.05114.x. [DOI] [PubMed] [Google Scholar]
- LaPlaca M.C. Raghupathi R. Verma A. Pieper A.A. Saatman K.E. Snyder S.H. McIntosh T.K. Temporal patterns of poly(ADP-ribose) polymerase activation in the cortex following experimental brain injury in the rat. J. Neurochem. 1999;73:205–213. doi: 10.1046/j.1471-4159.1999.0730205.x. [DOI] [PubMed] [Google Scholar]
- Mabley J.G. Horvath E.M. Murthy K.G. Zsengeller Z. Vaslin A. Benko R. Kollai M. Szabo C. Gender differences in the endotoxin-induced inflammatory and vascular responses: potential role of poly(ADP-ribose) polymerase activation. J. Pharmacol. Exp. Ther. 2005;315:812–820. doi: 10.1124/jpet.105.090480. [DOI] [PubMed] [Google Scholar]
- Myburgh J.A. Cooper D.J. Finfer S.R. Venkatesh B. Jones D. Higgins A. Bishop N. Higlett T. Epidemiology and 12-month outcomes from traumatic brain injury in Australia and New Zealand. J. Trauma. 2008;64:854–862. doi: 10.1097/TA.0b013e3180340e77. [DOI] [PubMed] [Google Scholar]
- Nagayama T. Simon R.P. Chen D. Henshall D.C. Pei W. Stetler R.A. Chen J. Activation of poly(ADP-ribose) polymerase in the rat hippocampus may contribute to cellular recovery following sublethal transient global ischemia. J. Neurochem. 2000;74:1636–1645. doi: 10.1046/j.1471-4159.2000.0741636.x. [DOI] [PubMed] [Google Scholar]
- Satchell M.A. Zhang X. Kochanek P.M. Dixon C.E. Jenkins L.W. Melick J. Szabo C. Clark R.S. A dual role for poly-ADP-ribosylation in spatial memory acquisition after traumatic brain injury in mice involving NAD+ depletion and ribosylation of 14-3-3gamma. J. Neurochem. 2003;85:697–708. doi: 10.1046/j.1471-4159.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- Tao Z. Gao P. Hoffman D.W. Liu H.W. Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry. 2008;47:5804–5813. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- Thurman D.J. Alverson C. Dunn K.A. Guerrero J. Sniezek J.E. Traumatic brain injury in the United States: A public health perspective. J. Head Trauma Rehabil. 1999;14:602–615. doi: 10.1097/00001199-199912000-00009. [DOI] [PubMed] [Google Scholar]
- Ueda K. Hayaishi O. ADP-ribosylation. Annu. Rev. Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- Virag L. Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- Wang X.G. Wang Z.Q. Tong W.M. Shen Y. PARP1 Val762Ala polymorphism reduces enzymatic activity. Biochem. Biophys. Res. Commun. 2007;354:122–126. doi: 10.1016/j.bbrc.2006.12.162. [DOI] [PubMed] [Google Scholar]
- Whalen M.J. Clark R.S. Dixon C.E. Robichaud P. Marion D.W. Vagni V. Graham S.H. Virag L. Hasko G. Stachlewitz R. Szabo C. Kochanek P.M. Reduction of cognitive and motor deficits after traumatic brain injury in mice deficient in poly(ADP-ribose) polymerase. J. Cereb. Blood Flow Metab. 1999;19:835–842. doi: 10.1097/00004647-199908000-00002. [DOI] [PubMed] [Google Scholar]
- Yu S.W. Wang H. Poitras M.F. Coombs C. Bowers W.J. Federoff H.J. Poirier G.G. Dawson T.M. Dawson V.L. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]