Abstract
Whereas cell cycle arrest, apoptosis, and senescence are traditionally thought of as the major functions of the tumor suppressor p53, recent studies revealed two unique functions for this protein: p53 regulates cellular energy metabolism and antioxidant defense mechanisms. Here, we identify glutaminase 2 (GLS2) as a previously uncharacterized p53 target gene to mediate these two functions of the p53 protein. GLS2 encodes a mitochondrial glutaminase catalyzing the hydrolysis of glutamine to glutamate. p53 increases the GLS2 expression under both nonstressed and stressed conditions. GLS2 regulates cellular energy metabolism by increasing production of glutamate and α-ketoglutarate, which in turn results in enhanced mitochondrial respiration and ATP generation. Furthermore, GLS2 regulates antioxidant defense function in cells by increasing reduced glutathione (GSH) levels and decreasing ROS levels, which in turn protects cells from oxidative stress (e.g., H2O2)-induced apoptosis. Consistent with these functions of GLS2, the activation of p53 increases the levels of glutamate and α-ketoglutarate, mitochondrial respiration rate, and GSH levels and decreases reactive oxygen species (ROS) levels in cells. Furthermore, GLS2 expression is lost or greatly decreased in hepatocellular carcinomas and the overexpression of GLS2 greatly reduced tumor cell colony formation. These results demonstrated that as a unique p53 target gene, GLS2 is a mediator of p53’s role in energy metabolism and antioxidant defense, which can contribute to its role in tumor suppression.
Keywords: reactive oxygen species, oxidative phosphorylation
p53 mainly exerts its tumor suppression function through the transcriptional regulation of its target genes. In response to stress, p53 selectively regulates the expression of its target genes, which results in cell cycle arrest, apoptosis, or senescence (1, 2). Whereas these responses are traditionally thought of as the major functions of p53 in tumor prevention, recent studies revealed two unique functions for this protein: p53 regulates cellular energy metabolism and antioxidant defense mechanisms. Emerging evidence has shown that these two functions of p53 contribute greatly to p53’s role in tumor suppression (3–5).
The recent identification of SCO2 and TIGAR as two p53 target genes revealed a unique function of p53 in the regulation of energy metabolism and ATP generation pathways (3, 4). The SCO2 gene is a key regulator of the cytochrome c oxidase complex that is essential for mitochondrial respiration. TIGAR functions to lower fructose-2, 6,-bisphosphate levels and thus slows glycolysis and directs glucose to the pentose phosphate pathway. p53 induces SCO2 expression to enhance mitochondrial respiration and induces TIGAR expression to slow glycolysis. Loss of p53 results in decreased oxygen consumption and impaired mitochondrial respiration and promotes a switch to high glucose utilization in aerobic glycolysis in cells. In mice, p53 loss results in reduced endurance during physical exercise, suggesting a crucial role for p53 in ensuring efficient ATP production by aerobic respiration for prolonged exercise (3). Tumor cells primarily use glycolysis rather than the much more efficient aerobic mitochondrial respiration for their energy needs, a switch known as the Warburg effect (6, 7). As a hallmark of tumor cells, metabolic changes have been suggested to be a key contributor to tumorigenesis (6, 7). Considering the importance of p53 in tumor suppression, these findings suggest that p53 mutations could be a genetic change that contributes to the Warburg effect in tumors, which provides a unique mechanism for p53 in tumor suppression.
Organisms living in aerobic conditions are constantly subjected to reactive oxygen species (ROS). Increased ROS levels contribute to genetic instability and cancer initiation and progression (5, 8). Recent studies revealed a unique function for p53 in regulating cellular antioxidant defense mechanisms. p53 promotes the expression of several antioxidant proteins to decrease intracellular ROS levels, including sestrins, TIGAR, GPX1, and ALDH4 (5, 9 –11). The antioxidant role of p53 is important to reduce oxidative stress-induced DNA damage and mutations, which contributes greatly to the tumor suppression activity of p53 (5).
Here, we identify human glutaminase 2 (GLS2) as a unique p53 target gene to mediate the role of p53 in both cellular energy metabolism and antioxidant defense mechanisms. The GLS2 gene encodes a mitochondrial glutaminase that catalyzes the hydrolysis of glutamine to glutamate (12, 13). p53 increases GLS2 expression under both nonstressed and stressed conditions, which results in enhanced mitochondrial respiration and ATP generation and, furthermore, increased glutathione (GSH) levels and decreased ROS levels in cells. These results suggest that GLS2 is an important component in mediating the tumor-suppressive effects of p53.
Results
The Human GLS2 Gene Contains a p53 Consensus DNA-Binding Element.
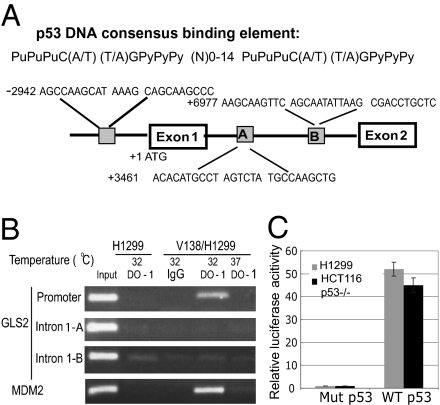
As a transcription factor, p53 binds to the p53 consensus DNA-binding elements in its target genes to regulate their transcription (14). By employing a p53 MH algorithm, a computer program scanning for potential p53 target genes by identifying the potential p53 consensus DNA-binding elements (15), we have recently indentified several unique p53 target genes (16–18). Employing this algorithm, the human GLS2 gene was identified as a potential p53 target gene containing three potential p53 consensus DNA-binding elements in its promoter region and intron 1 (Fig. 1A).
Fig. 1.
The human GLS2 gene contains a p53 DNA consensus binding element in the promoter region. (A) Putative p53 consensus binding elements in the human GLS2 gene predicted by the p53MH program. N, any nucleotide; Pu, purine; Py, pyramidine. (B) p53 binds to the p53 consensus binding element in the human GLS2 promoter region as detected by ChIP assays. The V138/H1299 cells were shifted to 32 °C for 24 h before assays. The p53-binding element in the MDM2 promoter serves as a positive control. DO-1: p53 antibody. (C) p53 activates luciferase activity of a reporter vector containing the p53-binding element in the GLS2 promoter region. The p53-null H1299 and HCT116 p53−/− cells were cotransfected with the luciferase reporter vectors and vectors expressing either wild-type (pRC p53) or mutant p53 protein (pRC 273H) 24 h before measuring luciferase activities.
GLS2 encodes a mitochondrial glutaminase (~65 kDa) that catalyzes the hydrolysis of glutamine to glutamate. GLS2 is highly expressed in postnatal liver and brain tissues and expressed at a low level in other tissues (12, 13). In cells, glutamate can be further converted into α-ketoglutarate, which is an important substrate for the citric acid cycle (TCA) to produce ATP in mitochondria. Furthermore, glutamate is a precursor of reduced GSH, one of the most important antioxidant molecules and a scavenger for ROS (8). Our finding that GLS2 could be a potential p53 target gene suggests that GLS2 may mediate p53’s function in the regulation of energy metabolism and antioxidant defense in cells.
To verify that p53 binds to these putative p53 consensus binding elements in the GLS2 gene in vivo, chromatin immunoprecipitation (ChIP) assays were performed. V138/H1299 cells are p53 null human lung H1299 cells stably transfected with a temperature-sensitive mutant p53 vector (alanine 138 to valine), which express the wild-type p53 protein at 32 °C and a mutant p53 protein at 37 °C (18). As shown in Fig. 1B, the DO-1 p53 antibody specifically pulled down the DNA fragment containing the potential p53-binding element in the GLS2 promoter but not the putative binding elements in intron 1 in V138/H1299 cells cultured at 32 °C, which expressed the wild-type p53 protein. In contrast, these chromatin fragments were not pulled down by DO-1 in either V138/H1299 cells cultured at 37 °C or H1299 cells cultured at 32 °C.
To investigate whether these putative p53 consensus binding elements confer p53-dependent transcriptional activity, DNA fragments containing one copy of the above putative DNA-binding elements were inserted into the promoter region of a pGL2 firefly luciferase reporter vector. The p53 null H1299 and colon HCT116 p53−/− cells were cotransfected with reporter vectors and vectors expressing either wild-type (pRC p53) or mutant p53 protein (pRC 273H). A pRL-SV40-TK vector expressing renilla luciferase was cotransfected as an internal control to normalize the transfection efficiency. Compared with the mutant p53, the expression of wild-type p53 protein in both cells greatly enhanced luciferase activities by 40- to 50-fold in the reporter vector containing the putative p53 DNA-binding element in the GLS2 promoter (Fig. 1C), but not in vectors containing two putative p53-binding elements in intron 1. These results together demonstrated that the human GLS2 gene contains a functional p53 DNA-binding element in its promoter region.
p53 Induces GLS2 Expression Under Both Stressed and Nonstressed Conditions.
To investigate whether p53 regulates GLS2 transcription, the GLS2 mRNA levels were examined by real-time PCR in cells. First, two p53-inducible cell lines, V138/H1299 and LN-2024, were used. LN-2024 cells are p53 null human glioblastoma LN-Z308 cells stably transfected with a vector containing the wild-type p53 cDNA sequences under a tetracycline-regulated promoter, which express p53 protein in the presence of Doxycycline (Dox) (19). The expression of wild-type p53 protein greatly induced the GLS2 mRNA levels in both V138/H1299 cells cultured at 32 °C (by up to 35-fold) and LN-2024 cells treated with Dox (by up to 14-fold), respectively (Fig. 2A). As negative controls, GLS2 mRNA was not induced in H1299 cells cultured at 32 °C or LN-Z308 cells treated with Dox.
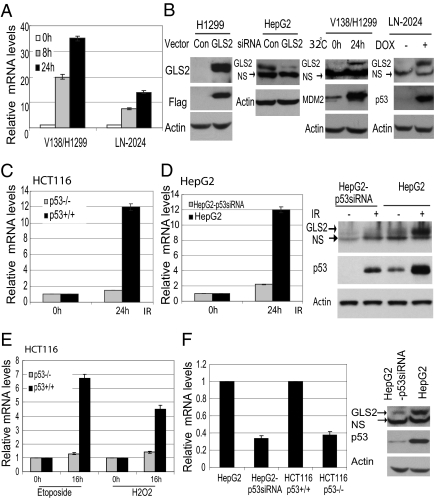
Fig. 2.
p53 increases GLS2 transcription under both stressed and nonstressed conditions. p53 induces GLS2 expression at mRNA (A) and protein levels (B) in V138/H1299 cells at 32 °C and in LN-2024 cells treated with Dox for different hours. The GLS2 mRNA levels were determined by real-time PCR and normalized with Actin. Exogenous GLS2 protein was detected using whole cell extracts from cells transfected with GLS2 expression vectors (B, panel 1). Endogenous GLS2 protein was detected using mitochondrial extracts (B, panels 2–4). GLS2 siRNA oligo was employed to knock down endogenous GLS2 expression in HepG2 cells (B, panel 2). NS: nonspecific band. (C and D) IR (10 Gy) induces GLS2 mRNA levels in a p53-dependent fashion in HCT116 and HepG2 cells. (E) Etoposide (20 μM) and H2O2 (150 μM) induce GLS2 mRNA levels in a p53-dependent fashion in HCT116 cells. (F) p53 regulates GLS2 basal expression levels.
To measure the GLS2 protein levels in cells, an antibody against the amino acids of the C terminus of GLS2 protein was produced. The specificity of the antibody was confirmed in H1299 cells transfected with GLS2 expression vectors with a C-terminal Flag tag (Fig. 2B , panel 1). The GLS2 antibody did not detect the endogenous GLS2 protein in the whole cell extracts because of the low levels of endogenous GLS2 in H1299 cells as measured by real-time PCR. To detect endogenous GLS2 protein, GLS2 was enriched by using mitochondrial extracts in the following experiments. The specificity of the antibody was further confirmed in HepG2 cells transfected with GLS2 siRNA oligo (Fig. 2B, panel 2). As shown in Fig. 2B (panels 3 and 4), p53 activation greatly induced the endogenous GLS2 protein levels in both V138/H1299 and LN-2024 cells.
Gamma-irradiation (IR) (10 Gy) was employed to treat cells as a stress signal to activate p53 in a pair of isogenic HCT116 p53+/+ and HCT116 p53−/− human colon cells. A clear induction of GLS2 mRNA was observed in HCT116 p53+/+ cells (~12-fold) but not in p53−/− cells (~1.4-fold) (Fig. 2C). Similar results were observed in a pair of isogenic human hepatocellular HepG2 (p53 wild type) and HepG2-p53siRNA cells, which were stably trasfected with a shRNA vector to knock down p53 in HepG2 cells (16) (Fig. 2D). In addition to IR, Etoposide and H2O2 (Fig. 2E) but not hypoxia or glucose starvation induced GLS2 mRNA levels in a p53-dependent fashion, suggesting that the p53 regulation of GLS2 depends on the nature of the stress that induces p53. These data demonstrated that GLS2 is a unique p53 target gene: p53 activation by various stress signals can induce GLS2 expression in various cells.
p53 not only induces GLS2 expression under stressed conditions, but also regulates the basal GLS2 levels under nonstressed conditions. Over 2.5- to 3-fold higher basal levels of GLS2 were observed in HCT116 p53+/+ cells than in p53−/− cells and in HepG2 cells than in HepG2-p53 siRNA cells (Fig. 2F). Similar results were observed in other cells: Knocking down p53 protein by transient transfection of siRNA led to the reduction of GLS2 mRNA levels by ~2- to 3-fold in cells, including lung H460 and brain HTB-15 cells. These results suggest an important role of p53 in maintaining the normal expression and function of GLS2 under nonstressed conditions.
GLS2 Increases Mitochondrial Oxidative Phosphorylation and ATP Generation.
To investigate how GLS2 regulates energy metabolism, GLS2 was overexpressed by transfection of GLS2 expression vector with a C-terminal Flag tag or knocked down by siRNA oligo in cells. The exogenous GLS2 expression was confirmed at the mRNA level by real-time PCR and at the protein level by Western-blot assays (e.g., H1299 cells in Fig. 2B), respectively. Immunofluorescence staining was employed to ensure that the exogenous GLS2 protein expressed by the vector was localized in mitochondria (Fig. 3A).
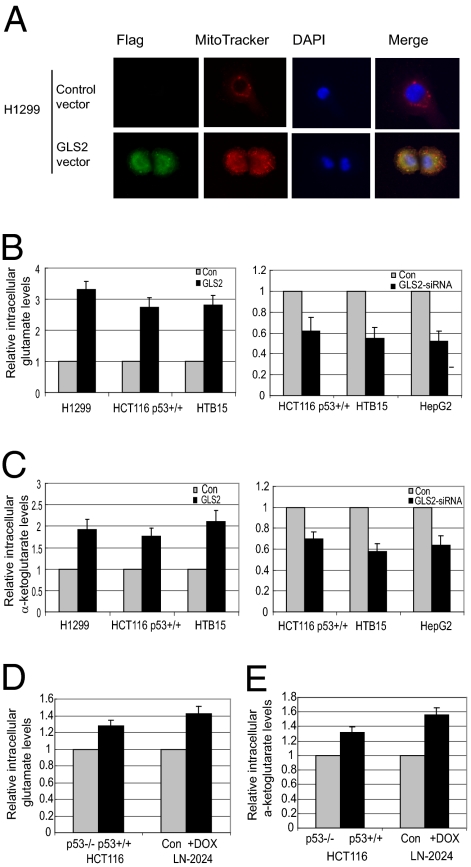
Fig. 3.
GLS2 expression increases the levels of intracellular glutamate and α-ketoglutarate. (A) The mitochondrial localization of exogenous GLS2 protein in cells transfected with GLS2 expression vectors. Mitochondria were stained with MitoTracker. (B) Exogenous GLS2 expression increases glutamate levels (Left), whereas GLS2 knockdown by siRNA (GLS2-siRNA) decreases glutamate levels in cells (Right) (P < 0.05). Glutamate levels were determined at 24 h after transfection. (C) (Left) Exogenous GLS2 expression increases α-ketoglutarate levels. (Right) GLS2 knockdown decreases α-ketoglutarate levels (P < 0.05). (D and E) p53 increases the levels of glutamate and α-ketoglutarate in cells (P < 0.05).
Consistent with the known biological function for GLS2 to catalyze the hydrolysis of glutamine to glutamate, exogenous GLS2 expression greatly increased the intracellular levels of glutamate in the H1299, HCT116 p53+/+, and HTB-15 cells (Fig. 3B Left) as compared with control cells transfected with empty vectors. GLS2 knockdown by siRNA oligo (>80% knockdown as detected by real-time PCR) clearly reduced the intracellular levels of glutamate in cells compared with control cells transfected with a scrambled siRNA oligo (Fig. 3B Right). Similar results were observed in other cells we tested. We further measured the α-ketoglutarate levels in cells. As shown in Fig. 3C, exogenous GLS2 expression increased the intracellular levels of α-ketoglutarate by ~2-fold in cells, whereas GLS2 knockdown clearly decreased the α-ketoglutarate levels. Significantly higher levels of glutamate and α-ketoglutarate were observed in HCT116 p53+/+ compared with HCT116 p53−/− cells and in LN-2024 cells after p53 protein expression (Fig. 3 D and E) (P < 0.05).
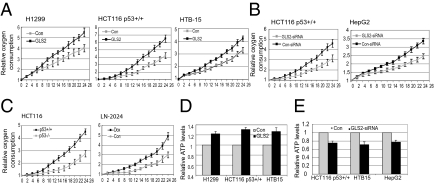
The role of GLS2 in mitochondrial respiration was further investigated. Exogenous GLS2 expression clearly increased the oxygen consumption and ATP levels, whereas GLS2 knockdown clearly decreased oxygen consumption and ATP levels (Fig. 4 A–E). Furthermore, glutamine depletion from media abolished the increased effect of GLS2 on ATP generation, suggesting that the enhanced glutamine metabolism is mainly responsible for the increase in ATP generation by GLS2 in cells. Interestingly, whereas p53 promoted mitochondrial respiration as demonstrated by the higher oxygen consumption in HCT116 p53+/+ cells than in p53−/− cells (Fig. 4C), no significant differences in ATP levels were observed between these two cells, suggesting that other metabolic pathways make up for the production of ATP in p53−/− cells. These results together demonstrate that GLS2 promotes the production of glutamate and α-ketoglutarate through its regulation of glutamine metabolism, which in turn promotes mitochondrial respiration and the TCA cycle, resulting in the efficient ATP gener-ation in cells.
Fig. 4.
GLS2 promotes oxygen consumption, mitochondrial respiration, and ATP generation. Cells were transfected with GLS2 expression vectors or GLS2 siRNA oligo. Cells were seeded in 96-well plates at 24 h after transfection and oxygen consumption was measured every 2 h after seeding (0 h). (A) Exogenous GLS2 expression promotes oxygen consumption. (B) GLS2 knockdown decreases oxygen consumption. (C) p53 increases oxygen consumption in cells. (D) Exogenous GLS2 expression increases ATP levels in cells (P < 0.05). (E) GLS2 knockdown decreases ATP levels in cells (P < 0.05). ATP levels were measured at 24 h after transfection.
GLS2 Expression Increases Cellular Antioxidant Function.
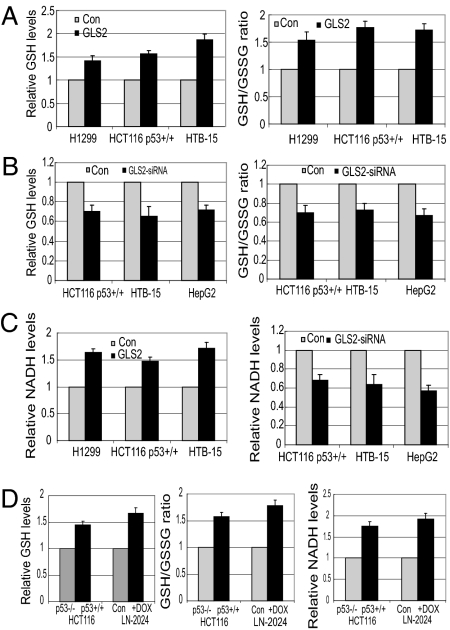
Glutamate is a precursor of GSH, a most important antioxidant molecule and a scavenger for ROS. As electrons are lost, the GSH becomes oxidized and forms oxidized glutathione (GSSG). GSH/GSSG balance reflects the redox state of cells (8). Interestingly, exogenous GLS2 expression significantly increased GSH levels and the GSH/GSSG ratio, whereas GLS2 knockdown decreased GSH levels and the GSH/GSSG ratio in cells (Fig. 5 A and B) (P < 0.05). Glutamine depletion from media abolished the increased effect of GLS2 on GSH levels and GSH/GSSG ratio, suggesting that the enhanced glutamine metabolism is mainly responsible for this effect. Our results further show that GLS2 expression regulates intracellular NADH production. GLS2 overexpression significantly increased the NADH levels, whereas GLS2 knockdown decreased NADH levels in cells (P < 0.05) (Fig. 5C). NADH is both the primary cofactor that drives reduction and oxidation reactions and an important antioxidant. A possible mechanism accounting for this is that GLS2 accelerates the TCA cycle, which in turn produces more NADH in cells. Consistent with these findings, significantly higher GSH levels, GSH/GSSG ratio, and NADH levels were observed in HCT116 p53+/+ as compared with HCT116 p53−/− cells and in LN2024 cells treated with Dox as compared with control LN2024 cells (Fig. 5D; P < 0.05).
Fig. 5.
GLS2 increases intracellular levels of GSH and NADH. Cells were transfected with GLS2 expression vector or GLS2 siRNA 24 h before assays. (A) Exogenous GLS2 expression increases GSH levels and relative GSH/GSSG ratio (P < 0.05). (B) GLS2 knockdown decreases GSH levels and GSH/GSSH ratio (P < 0.05). (C) Exogenous GLS2 expression increases NADH levels, and GLS2 knockdown decreases NADH levels (P < 0.05). (D) p53 increases GSH levels, GSH/GSSH ratio, and NADH levels (P < 0.05). LN-2024 cells were treated with Dox for 24 h.
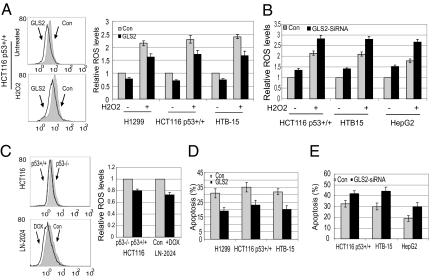
The ROS levels were measured in cells untreated (nonstressed) or treated with H2O2 to induce oxidative stress. Exogenous GLS2 expression clearly reduced intracellular ROS levels in both cells, whereas GLS2 knockdown by siRNA oligo resulted in a clear increase of ROS levels in both cells (Fig. 6 A and B). Significantly lower ROS levels were observed in HCT116 p53+/+ cells compared with HCT116 p53−/− cells and in LN-2024 cells treated with Dox compared with untreated cells (Fig. 6C).
Fig. 6.
GLS2 decreases ROS levels in cells and protects cells from H2O2-induced apoptosis. Cells were transfected with GLS2 expression vectors or siRNA oligo for 24 h and then treated with H2O2 (400 μM) for 4 h before measuring ROS or for 24 h before measuring apoptosis. (A) Exogenous GLS2 expression decreases ROS levels in cells (P < 0.05). (B) GLS2 knockdown increases ROS levels in cells (P < 0.05). (C) p53 decreases ROS levels in cells (P < 0.05). LN-2024 cells were treated with Dox for 24 h. (D) GLS2 overexpression protects cells from H2O2-induced apoptosis (P < 0.05). (E) GLS2 knockdown sensitizes cells to H2O2-induced apoptosis (P < 0.05).
To further investigate whether GLS2 can protect cells from oxidative stress-induced apoptosis, cells with exogenous GLS2 expression or GLS2 knockdown were treated with H2O2 and apoptosis was measured. Exogenous GLS2 expression significantly reduced H2O2-induced apoptosis, whereas the GLS2 knockdown by siRNA oligo significantly sensitized cells to H2O2-induced apoptosis (P < 0.05) (Fig. 6 D and E). These results together demonstrate an important role of GLS2 in antioxidant defense in cells, suggesting that as a unique p53 target gene, GLS2 contributes greatly to p53’s role in antioxidant defense.
Loss of GLS2 Expression in Liver Tumors.
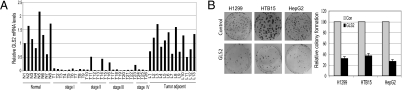
Considering the critical role of p53 and its pathway in tumor suppression and the importance of maintaining proper energy metabolism and antioxidant defense function of cells in tumor suppression, our finding that GLS2 is a unique p53 target gene that mediates a portion of p53’s functions in energy metabolism and antioxidant defense suggests a potential role of GLS2 in tumor suppression. It is known that GLS2 is highly expressed in adult liver tissues. Our data show that the mRNA expression of GLS2 is almost absent or significantly decreased in all 26 specimens from hepatocellular carcinoma (HCC) at different stages that we analyzed (Origene Technologies) compared with adjacent normal liver tissues (n = 8) or tissues with cirrhosis, fatty changes, or chronic hepatitis (n = 13) as measured by real-time PCR. The expression levels of GLS2 mRNA were significantly lower in HCC compared to normal adjacent liver tissue (P < 0.0001), but not significantly different from normal liver tissues and liver tissues with cirrhosis, fatty changes, or chronic hepatitis (P = 0.23) (Fig. 7A). These results suggest that loss of GLS2 expression could be a specific event and tumor biomarker for HCC, which may contribute to liver tumorigenesis. It is well documented that ~40–50% of HCC contains DNA mutations in the p53 gene. Our data show that almost all of these HCC tissues we studied have significantly lower GLS2 expression, which suggests that there should be some other factors leading to the down-regulation of GLS2 expression in HCC in addition to p53 mutations.
Fig. 7.
Loss of GLS2 expression in hepatocellular carcinomas and the inhibition of colony formation by GLS2 in tumor cells. (A) Loss or significant decrease of mRNA expression of GLS2 in human hepatocellular carcinomas. HCC, hepatocellular carcinoma; N, normal liver. Tumor adjacent tissues: L1–5, tissues with cirrhosis; L6–10, tissues with fatty changes; L11–13, tissues with chronic hepatitis. The GLS2 expression was detected by real-time PCR and normalized with Actin. GLS2 levels of N1 are designated as 1. Samples were provided by Origene Techonologies. P < 0.0001 (normal vs. tumor), P < 0.0001 (tumor adjacent tissues vs. tumor), and P = 0.2367 (normal vs. tumor adjacent tissue). (B) Exogenous GLS2 expression reduces cell colony formation in tumor cells, including H1299, HepG2, and HTB15 cells (P < 0.05).
GLS2 Overexpression Reduces Tumor Cell Colony Formation Abilities.
To further investigate the potential role of GLS2 in tumor suppression, tumor cells, including H1299, HepG2, and HTB15 cells, were transfected with GLS2 expression vectors, and the cell colony formation ability was examined under the selection of G418. Compared with control cells transfected with empty vectors, tumor cells transfected with GLS2 expression vectors show significantly decreased colony formation ability (up to 3-to 4-fold, P < 0.05) (Fig. 7B). These data suggest that GLS2 may play a role in tumor suppression.
Discussion
The recent identification of SCO2 and TIGAR as two p53 target genes revealed p53’s role in enhancing mitochondrial respiration and inhibiting glycolysis (3, 4). Here, we identified GLS2 as a unique p53 target gene involved in energy metabolism. The induction of GLS2 increases intracellular levels of glutamate and α-ketoglutarate and leads to the enhanced oxygen consumption, mitochondrial respiration, and ATP generation in cells. Through the regulation of GLS2, p53 increases the reliance upon glutamine metabolism, the production of glutamate and α-ketoglutarate, and mitochondrial respiration. Compared with p53-deficient cells, p53 wild-type cells have higher intracellular levels of glutamate and α-ketoglutarate, higher oxygen consumption, and higher rates of mitochondrial respiration, which are consistent with the effects of increased GLS2 expression in cells. These results demonstrated that the regulation of GLS2 and glutamine metabolism is a unique mechanism for p53 to regulate energy metabolism: restoring cells to a greater dependence upon oxidative phosphorylation.
p53 has been reported to modulate up cellular antioxidant defense mechanisms, especially under conditions of no stress or low stress, and this antioxidant activity plays an important role in the overall tumor suppressor function of p53 (5). Here, p53 promotes the expression of several antioxidant proteins that function to lower ROS levels, including sestrins, TIGAR, GPX1, and ALDH4. Sestrins are a family of proteins required for regeneration of peroxiredoxins, the major reductants of endogenously produced peroxides in cells (9). TIGAR diverts glucose through the pentose phosphate pathway to lower ROS levels (4). ALDH4 is a mitochondrial NAD+-dependent enzyme that catalyzes the proline degradation pathway to lower ROS levels (11). GPX1 is a primary antioxidant enzyme that scavenges hydrogen peroxide or organic hydroperoxides in cells (10). The identification of GLS2 as a p53-regulated gene that functions to lower ROS levels provides further direct evidence to support the important role of p53 in antioxidant defense in cells. GLS2 increases GSH levels and reduces ROS levels in cells through increasing the levels of glutamate, a precursor of GSH. Furthermore, GLS2 increases the NADH levels. These effects contribute to GLS2’s function in lowering ROS levels in cells and thus protect cells from oxidative stress (H2O2)-induced cell death. These findings presented here not only provide further strong evidence that p53 plays an important role in cellular antioxidant defense, but also provide a unique mechanism for p53 in the regulation of antioxidant defense mechanisms through regulating glutamine metabolism and GSH production.
Our results that GLS2 levels were significantly decreased in HCC and that overexpression of GLS2 in tumor cells, including HCC cells, significantly reduced tumor cell colony formation strongly suggest a potential role of GLS2 in tumor cell growth suppression. It is known that GLS2 is expressed only in postnatal liver but not in fetal liver (12). Fetal liver, like cancer cells, use glycolysis to generate ATP and precursors for cell division. GLS2 promotes oxidative phosphorylation and the efficient use of glucose to produce ATP. Therefore, loss of GLS2 expression in HCC suggests that liver tumor cells return to a fetal-like metabolic phenotype after neoplastic transformation. Consistent with our findings, it was recently reported that GLS2 expression is lost in many brain tumors, including highly malignant glioblastomas and anaplastic astrocytomas, and restoration of GLS2 expression in glioblastoma cells inhibited tumor cell proliferation and migration (20). Brain is one of several tissues where GLS2 is highly expressed. These data together strongly suggest a potential role of GLS2 in tumor suppression. Interestingly, the GLS1 gene located in 2q32 shares a considerable degree of sequence similarity with GLS2 and encodes a kidney-type glutaminase isoform (~70 kDa). The oncogene Myc was reported to increase GLS1 expression and promote tumor cell proliferation in human P-493 B lymphoma and PC3 prostate cancer cells (21). It appears that GLS1 and GLS2 may have contrasting roles in tumorigenesis. Several possible mechanisms may account for this: (i) The expression of GLS1 and GLS2 is regulated by different mechanisms in cells under different circumstances. For example, p53 induces the expression of GLS2 but not GLS1 as we observed in various cells. Myc induces the expression of GLS1 but not GLS2 (21). (ii) GLS1 and GLS2 have different kinetic, immunologic, and molecular characteristics. GLS1 is activated by high phosphate levels and strongly inhibited by the end-product glutamate, whereas GLS2 is activated by low phosphate levels and not inhibited by glutamate (13). Therefore, these two glutaminase isoforms may have different impacts upon the fine regulation of energy metabolism and antioxidant defense. (iii) GLS2 has been reported to interact with other proteins through the PDZ domain in its C terminus (22). GLS2 may exert its anti-tumorigenesis effect through its interaction with other PDZ domain-containing proteins; and further, (iv) GLS2 overexpression in cells results in altered expression of genes, suggesting that GLS2 may act indirectly or directly as a transcription factor (20). Further studies are required to address the quite different roles and mechanisms for GLS1 and GLS2 in tumorigenesis.
In summary, we identified glutaminase 2 as a unique p53 target gene. GLS2 increases mitochondrial respiration and ATP generation. Furthermore, GLS2 increases cellular levels of GSH and NADH and decreases ROS levels in cells. Our results demonstrate that GLS2 is an important component in mediating these two unique functions of p53 in the regulation of energy metabolism and antioxidant defense.
Materials and Methods
Luciferase Activity Assay and ChIP Assay.
pGL2 luciferase reporter (Promega) was used to construct luciferase reporter containing putative p53-binding elements in the GLS2 gene. Luciferase activity assays were performed as previously described (16, 23). ChIP assays were performed as previously described (16, 23).
Western-Blot Analysis and Immunofluorescence Staining.
Rabbit polyclonal antibody to GLS2 was raised against a 15-aa peptide corresponding to the GLS2 human protein (PFAKDRWGNIP LDDC) (Genescript). For detection of endogenous GLS2 protein by Western blot, mitochondria were isolated and mitochondria extracts were used. Exogenous GLS2 protein was stained with anti-Flag (M2; Sigma). For MitoTracker staining, cells were incubated in MitoTracker for 30 min before fixation.
Cellular Apoptosis and Cell Colony Formation Assays.
Apoptosis was measured in a flow cytometry assay as previously described (24). Colony formation assays were performed as previously described (25).
Measurement of Parameters of Energy Metabolism.
Glutamate levels were measured by using the Amplex Red Glutamine Acid/Glutamate oxidase assay kit (Invitrogen). α-Ketoglutarate levels were measured by using the α-ketoglutarate assay kit (Biovision). ATP levels were measured by using the ATP Bioluminescence assay kit (Roche). ROS levels were measured by dihydrorhodamine 123 staining in a flow cytometry assay as described (4). Oxygen consumption in cells was measured by using the BD Oxygen Biosensor System.
See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant CA143204-01 (to Z.F.), by National Institutes of Health Grant P01 CA 87497 (to A.L.), and by a University of Medicine and Dentistry of New Jersey Foundation grant (to Z.F.). C.Z. was supported by a New Jersey Cancer Center Research postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001006107/DCSupplemental.
See Commentary on page 7117.
References
- 1.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Hu W, Feng Z. The P53 pathway: What questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 3.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 4.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 5.Sablina AA, et al. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensaad K, Vousden KH. p53: New roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 8.D'Autreaux B, Toledano MB. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 9.Budanov AV, et al. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 10.Tan M, et al. Transcriptional activation of the human glutathione peroxidase promoter by p53. J Biol Chem. 1999;274:12061–12066. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- 11.Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49:134–140. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 12.Perez-Gomez C, et al. Genomic organization and transcriptional analysis of the human l-glutaminase gene. Biochem J. 2003;370:771–784. doi: 10.1042/BJ20021445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos-Sandoval JA, et al. Expression of functional human glutaminase in baculovirus system: Affinity purification, kinetic and molecular characterization. Int J Biochem Cell Biol. 2007;39:765–773. doi: 10.1016/j.biocel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 14.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 15.Hoh J, et al. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 17.Feng Z, et al. p53 tumor suppressor protein regulates the levels of huntingtin gene expression. Oncogene. 2006;25:1–7. doi: 10.1038/sj.onc.1209021. [DOI] [PubMed] [Google Scholar]
- 18.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albertoni M, et al. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene. 2002;21:4212–4219. doi: 10.1038/sj.onc.1205610. [DOI] [PubMed] [Google Scholar]
- 20.Szeliga M, et al. Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia. 2009;57:1014–1023. doi: 10.1002/glia.20825. [DOI] [PubMed] [Google Scholar]
- 21.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olalla L, et al. Expression of the scaffolding PDZ protein glutaminase-interacting protein in mammalian brain. J Neurosci Res. 2008;86:281–292. doi: 10.1002/jnr.21505. [DOI] [PubMed] [Google Scholar]
- 23.Feng Z, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 24.Feng Z, et al. Declining p53 function in the aging process: A possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci USA. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu W, Feng Z, Tang MS. Chromium(VI) enhances (+/−)-anti-7beta,8alpha-dihydroxy-9alpha,10alpha-epoxy-7,8,9,10-tetrahydro benzo[a]pyrene-induced cytoto-xicity and mutagenicity in mammalian cells through its inhibitory effect on nucleotide excision repair. Biochemistry. 2004;43:14282–14289. doi: 10.1021/bi048560o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.