Abstract
Mimicry among Heliconius butterflies provides a classic example of coevolution but unresolved relationships among mimetic subspecies have prevented examination of codiversification between species. We present amplified fragment length polymorphism and mtDNA datasets for the major comimetic races of Heliconius erato and H. melpomene. The AFLP data reveal unprecedented resolution, clustering samples by geography and race in both species. Our results show that, although H. erato and H. melpomene co-occur, mimic each other, and exhibit parallel shifts in color pattern, they experienced very different modes of diversification and geographic histories. Our results suggest that H. erato originated on the western side of South America whereas H. melpomene originated in the east. H. erato underwent rapid diversification and expansion with continued gene-flow following diversification, resulting in widely dispersed sister taxa. In contrast, H. melpomene underwent a slower pace of diversification with lower levels of gene flow, producing a stepwise directional expansion from west to east. Our results also suggest that each of the three main wing pattern phenotypes originated and/or was lost multiple times in each species. The rayed pattern is likely to be the ancestral phenotype in H. erato whereas postman or red patch is likely to be ancestral in H. melpomene. Finally, H. cydno and H. himera are monophyletic entities clearly nested within H. melpomene and H. erato, rather than being their respective sister species. Estimates of mtDNA divergence suggest a minimum age of 2.8 and 2.1 My for H. erato and H. melpomene, respectively, placing their origins in the late Pliocene.
Keywords: amplified fragment length polymorphism, Heliconius erato, Heliconius melpomene, Müllerian mimicry, mtDNA
Neotropical Heliconius butterflies are famous for extensive Müllerian mimicry, in which mutually protected species share the same warning pattern and thereby distribute the mortality associated with educating predators of their unpalatable chemical defenses. Although mimicry in Heliconius often involves other butterflies like ithomiines or even day-flying moths, the majority of mimetic relationships occur among species within the genus. Mimicry within Heliconius often involves pairs of comimetic species, with one member of each pair coming from each of two major clades, the pupal-mating and non–pupal-mating clade (1–6). Paradoxically, there is extreme color pattern variation among Heliconius species and among geographic subpopulations within species. The cause of this diversity remains enigmatic because, in their simplest forms, theories of warning coloration and Müllerian mimicry predict strong stabilizing selection and convergence of signals, as opposed to diversification (7–9).
The phenomenon of geographic variation in mimicry is well exemplified by the comimetic species pair Heliconius erato and Heliconius melpomene. Like other Heliconius comimics, one species (H. erato) belongs to the pupal-mating clade whereas the other (H. melpomene) belongs to the non–pupal-mating clade. These two species look nearly identical across their shared range of Central and South America, yet their wing patterns shift, in tandem, from location to location (4, 9, 10). The racial variation in H. erato and H. melpomene involves both major phenotypic shifts, such as the difference between rayed and postman patterns, as well as relatively minor variations, such as the subtle differences among rayed populations or among postman populations. Two primary hypotheses have been proposed to explain the coincident variation between H. erato and H. melpomene: (i) “Pleistocene refugia,” which posits that the species coradiated during periods of habitat fragmentation associated with Pleistocene glacial advances (4, 6, 9, 10), and (ii) “advergence,” which posits that H. erato radiated first and established the diversity of warning patterns that H. melpomene later evolved to match (11–13). Recent DNA sequence data have been used to support both hypotheses (12, 14–16), but poorly resolved relationships among racial phenotypes within each species have prevented the critical test for codiversification between the species.
Previous studies attempting to reconstruct the relationships among geographic races of H. erato and H. melpomene have focused on mitochondrial and nuclear DNA sequence data (12, 14, 15). These studies revealed broad geographic structuring of genetic variation in both species, but little resolution at the level of individual races. For instance, mtDNA phylogenies of both species grouped haplotypes into large biogeographic regions, such as east and west of the Andes mountains for H. erato (14, 15) and west of the Andes, Amazon, Guiana shield, and eastern Brazil for H. melpomene (15). Within these regions, however, there was no structuring, with individual haplotypes distributed among races and individual races containing multiple haplotypes. Similarly, gene trees for the nuclear genes Mannose phosphate isomerase and Triose phosphate isomerase revealed pronounced clustering of haplotypes into large biogeographic regions in H. melpomene but little population structure in H. erato (12). It is likely that a combination of recent diversification, large ancestral population sizes, and extensive ongoing hybridization among geographic races have served to inhibit or erase the strong signal of population genetic differentiation that would be required to resolve relationships at finer geographic scales based on one or few genes.
Amplified fragment length polymorphisms (AFLPs) have been shown to provide phylogenetic resolution among recently and rapidly radiating groups in which sequence data have failed (17–22). The increased resolution of AFLPs is associated with their nuclear genome-wide distribution, which overcomes problems associated with locus-specific effects, and the large number of polymorphisms that can be easily characterized. Recent population genetic analyses of multiple Heliconius species based on AFLPs revealed pronounced genetic structure in both H. erato and H. melpomene over small spatial scales (13), suggesting that AFLPs should be effective at distinguishing closely related and geographically proximate races in each species.
Here, we use large AFLP and mtDNA datasets to infer relationships among the major comimetic races in H. erato and H. melpomene and determine whether they radiated in parallel across time and space. Specifically, we compare the resolution of both marker types and address the following questions: (i) Where did H. erato and H. melpomene each originate, and what explains their current biogeography? (ii) What was the ancestral wing pattern in each species? (iii) Do individual races constitute monophyletic groups? (iv) Are major wing pattern forms (such as the rayed or postman patterns) monophyletic in each species? Additionally, we infer the relationship between each species and its putative “sister” species, H. cydno for H. melpomene and H. himera for H. erato, and we also estimate the minimum age of each species group based on pairwise mtDNA sequence divergence. Our results illuminate the disparate histories behind these remarkable comimetic radiations and provide essential insights for future work focused on the molecular evolution of mimicry genes themselves.
Results
A list of samples and accompanying information is presented in Table S1. The AFLP and mtDNA phylogenies and Structure-based clustering results for H. erato and H. melpomene are presented in Figs. 1 and 2, respectively. For H. erato, 4,667 and 4,582 polymorphic AFLP loci were scored with and without the outgroups, respectively. For H. melpomene, 3,186 polymorphic AFLP loci were scored with outgroups included. In Structure analyses, both the H. erato and H. melpomene datasets had maximum log-likelihood values at seven clusters. For H. erato, each of the seven clusters formed a distinct group. For H. melpomene, one cluster (cluster 7 in Fig. 2C) did not form a distinct group. Clade construction indices (CCIs; a unique method presented here to quantify the degree of monophyly of a group, see Materials and Methods) show that AFLP data performed better than mtDNA data in clustering the samples by geographic location and/or race in both species (Table S2 and Fig. S1). In H. erato, 11 of the groupings were monophyletic in the AFLP tree whereas only two were in the mtDNA tree; in H. melpomene, 10 groupings were monophyletic in the AFLP tree whereas only four were in the mtDNA tree.
Fig. 1.
Phylogeography of H. erato. (A) Phylogeny of H. erato based on AFLP data (bootstrap values for key nodes are shown). “x” denotes a node not found in the bootstrap consensus. Dotted lines indicate relationships found in the bootstrap consensus and not in the original topology. mtDNA lineages (as noted in B), races, and wing patterns are indicated. The ingroup phylogeny shown is derived from AFLP data scored without outgroups. For the clade denoted by a red star, AFLP data were scored and analyzed separately. Gray boxes indicate all regions west of the Andes. Numbers in colored boxes are cross-referenced to C. (B) Maximum likelihood phylogeny of H. erato based on mtDNA sequences (bootstrap values above branches). Races are indicated as in A. Gray boxes indicate all regions west of the Andes. (C) Population structure inferred from AFLP data using Structure. The right panel shows separate analyses of groups 1, 4, and 5. Images of the races are shown, in which comimics are matched in left-to-right order with images in Fig. 2, with the exception of chestertonii (no H. melpomene comimic) and lativitta (H. melpomene comimic not included in this study).
Fig. 2.
Phylogeography of H. melpomene. (A) Phylogeny of H. melpomene based on AFLP data (bootstrap values for key nodes are shown). “x” denotes a node not found in the bootstrap consensus; the curly bracket indicates the alternative node found in the bootstrap consensus. mtDNA lineages (as noted in B), races and wing patterns are indicated. Gray boxes indicate all regions west of the Andes. Numbers in colored boxes are cross-referenced to C. (B) Maximum likelihood phylogeny of H. melpomene based on mtDNA sequences (bootstrap values above branches). Races are indicated as in A. Gray boxes indicate all regions west of the Andes. (C) Population structure inferred from AFLP data using Structure. The right panel shows individual analyses of groups 1, 3, and 4. Images of the races are shown, in which comimics are matched in left-to-right order with images in Fig. 1, with the exception of plesseni and vulcanus (corresponding H. erato comimics were not included in this study).
H. erato.
The AFLP phylogeny of H. erato revealed substantial clustering by geography and, to a lesser extent, by race (Fig. 1A). The datasets obtained by scoring with and without outgroups produced topologies that differed in some of the relationships (Fig. S2 and Fig. 1A, respectively). In both trees, relationships among some of the clades were poorly supported by bootstrap analyses. Scoring without outgroups yielded a topology in which more groups were monophyletic (Fig. 1A). Based on this topology, there were 11 monophyletic groups: Costa Rica, French Guiana, Trinidad, Peru, “the isthmus” (i.e., Panama and Costa Rica), H. himera, hydara from Colombia, phyllis, chestertonii, etylus, and cyrbia, the latter four being the only monophyletic races of H. erato.
Structure results (Fig. 1C Left), which were largely consistent with the AFLP tree, revealed that H. himera, H. e. chestertonii, and H. e. cyrbia were strongly differentiated from other H. erato populations. This is also evident in the long branches leading to these taxa in the AFLP phylogram (Fig. 1A). Separate analyses of Structure clusters (Fig. 1C Right) revealed that large clusters could be further divided into subclusters that corresponded partially with a priori populations (i.e., races/geographic groups). The mtDNA phylogeny (Fig. 1B) showed much less clustering by race or geography in comparison with the AFLP phylogeny. The AFLP and mtDNA trees did, however, have two clades in common: H. e. etylus and E1, corresponding to the western clade of Brower (14).
H. melpomene.
The AFLP phylogeny of H. melpomene (Fig. 2A) also revealed strong clustering by geography, followed by race. Relationships among the clades in the H. melpomene tree were better supported compared with H. erato. Nine clades were evident in the AFLP phylogeny: H. melpomene from Costa Rica, H. cydno, H. melpomene from the isthmus, Peru, French Guiana, Trinidad, East Ecuador, cythera, and nanna. The latter two were the only monophyletic races of H. melpomene. Colombian specimens were the only group that did not emerge monophyletic, forming a paraphyletic grade.
As in H. erato, the Structure results (Fig. 2C Left) were largely consistent with the AFLP tree. Unlike H. erato, none of the H. melpomene groups showed strong genetic isolation, except perhaps H. m. nanna. Individual analyses of Structure clusters (Fig. 2C Right) revealed that large clusters resolved into subclusters that corresponded well to a priori populations. The mtDNA phylogeny (Fig. 2B) showed far less clustering by race or geography compared with the AFLP phylogeny. Clades in common between the AFLP and mtDNA trees were nanna from Brazil, Trinidadian specimens, and H. cydno. A notable phenomenon in the mtDNA phylogeny is the wide phylogenetic scattering of samples from Colombia, which are clustered in the AFLP phylogeny. The other AFLP clades show substantial clustering within the mtDNA tree, except the French Guiana AFLP clade, which appeared as two mtDNA clades (M2b and M2cii).
Discussion
Our results reveal, with greater resolution than previously described, the very different evolutionary histories experienced by H. erato and H. melpomene, species that display near-perfect convergence across a diversity of warning patterns today. Historically, the concordant color pattern variation between H. erato and H. melpomene was thought to be the result of parallel shifts that occurred in Pleistocene refugia (9, 10). In contrast, recent inferences of historical demography based on DNA sequence data suggest that populations of H. erato began expanding before those of H. melpomene (12). Our AFLP results, which reveal striking clustering by geography and racial phenotype, show that, whereas H. erato and H. melpomene co-occur, mimic one another, and shift color patterns in parallel today, they arrived at this diverse mimetic relationship starting from very different times, places, and phenotypes.
Although our AFLP data clearly provide improved population-level resolution relative to DNA sequence data, it is important to point out that there are potential limitations associated with using AFLPs for phylogenetic reconstruction (23, 24). Two primary issues are the degree of homoplasy in the data and whether distance measures are suitable for tree-building analyses (25). AFLPs are scored based on the presence or absence of DNA fragments of particular sizes. Homoplasy presents a greater problem when shared absences, as opposed to shared presences, are used in the calculation of distances because of the greater number of ways by which taxa/individuals can share absences. One way to reduce the influence of homoplasy is to use distance measures that rely only on shared presences, such as the Nei-Li distance measure employed in our neighbor-joining analyses. More significantly, studies have shown that phylogenetic information can indeed be gleaned from AFLP data (26), and a growing body of literature (e.g., refs. 27–29) attests to the ability of AFLP-derived phylogenies to corroborate other sources of data. Finally, it is encouraging that (i) the AFLP phylogenies of H. erato and H. melpomene show such strong geographic coherence, which would not be expected if the data were dominated by homoplasy; and (ii) our results from Structure analyses are largely consistent with those from the phylogenetic analyses of our AFLP data.
Western Origins for H. erato and Eastern Origins for H. melpomene.
The AFLP and mtDNA data presented here substantially modify our view of where the H. erato and H. melpomene radiations each originated. Brower (14) inferred from an mtDNA phylogeny based on parsimony that the basal clades within H. erato (H. himera and chestertonii) originated on the western side of the Andes, thus suggesting that H. erato began to diversify in this region and subsequently spread eastward into the Amazon basin. Our AFLP phylogeny indicates that H. chestertonii and H. himera are not basal lineages. Taken at face value, the tree shows that H. e. lativitta and H. e. etylus, both from Ecuador, occupy basal positions in the phylogeny, suggesting that H. erato may have originated in the western part of the continent but on the eastern, Amazonian slope of the Andes. Brower (15) also inferred that H. melpomene originated in the Guiana shield. Our mtDNA phylogeny (Fig. 2B), based on maximum likelihood analyses, places the French Guiana specimens as later-branching lineages and does not support a Guianan origin for H. melpomene. Additional phylogenetic analyses of our mtDNA sequences, with Brower's (15) sequences from Guiana included, placed them as members of lineage M2b (Fig. 2B), which is not a basal lineage. Nevertheless, the AFLP phylogeny shows a clear east-to-west axis, with the basal-most lineage occurring furthest east (in coastal Brazil) and progressively younger lineages branching off in a westward sequence (Fig. 3), suggesting an origin somewhere in the east. In addition, H. erato races in the west tend to be completely or nearly monophyletic whereas the same is true for H. melpomene races in the east, a pattern that is consistent with the geographic origins inferred here (SI Discussion).
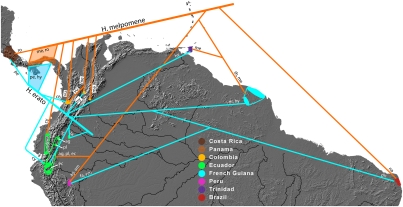
Fig. 3.
Sampling locations for specimens used in this study. AFLP phylogenies of H. erato and H. melpomene are superimposed. Races within each species are abbreviated to the first two letters of the race names. Note that “cy” stands for cythera in H. melpomene and for cyrbia for H. erato.
Rapid Expansion and Diversification with Gene Flow in H. erato, Sequential and Directional Radiation in H. melpomene with Less Gene Flow Following Diversification.
A notable difference between the AFLP phylograms of the two species is that clades within H. melpomene have longer subtending branches and/or better support compared with clades in H. erato. The nodes defining the terminal clades in the H. erato AFLP phylogram appear almost collapsed into the spine of the phylogeny, suggesting that these clades diverged from one another within a narrow window of time. These observations are consistent with rapid diversification and geographic expansion, coupled with continued gene flow following diversification. Consistent with this scenario is the absence of a distinct continental-scale geographic trend in the H. erato phylogeny and the observation of widely dispersed sister lineages (e.g., Peru with the eastern tip of Brazil). The exceptions are H. himera, H. e. chestertonii, and H. e. cyrbia, clades that are well defined by long branches. In contrast, the distinct east-to-west geographic trend in the H. melpomene AFLP phylogeny and the observation that sister lineages in H. melpomene tend to be from neighboring regions (French Guiana with Trinidad, Peru with Ecuador) points to a directional and stepwise geographic expansion. Also in contrast to H. erato, the individual tips within each clade in H. melpomene coalesce at points that are more distal from the backbone of the phylogeny, suggesting a slower tempo of diversification. The longer subtending branches and greater bootstrap support for H. melpomene clades may indicate historical bottlenecks and/or lower levels of gene flow following diversification. These inferences are supported by Flanagan et al. (12), who also showed that H. melpomene exhibits more phylogeographic structure than H. erato.
Divergent Ancestral Wing Patterns in H. erato and H. melpomene.
In H. melpomene, the rayed pattern is restricted to a single clade in the AFLP phylogeny (white diamonds in Fig. 2A) whereas the postman and red patch patterns are more widely spread, suggesting that the rayed pattern is less likely to represent the ancestral wing pattern in H. melpomene. Brower (15) noted that the red patch pattern was basal in every mtDNA clade in which it occurs, but that is not the case in the AFLP tree (gray diamonds in Fig. 2A). However, the red patch pattern is basal in the AFLP clade that constitutes Structure cluster 1 (Colombia plus west of the Andes). The red patch pattern may be ancestral for the “French Guiana + Trinidad” AFLP clade. If the topology presented in Fig. 2A is correct, it is possible that the red patch is ancestral in the entire clade that is sister to “Brazil,” (i.e., everything minus H. m. nanna). Given that H. m. nanna sports the postman pattern and its sister may be ancestrally red patched, their most recent common ancestor could have been either.
At face value, the AFLP phylogeny suggests that the rayed pattern is ancestral in H. erato, as the two earliest branches (one individual of H. e. lativitta and a clade of H. e. etylus plus H. e. lativitta) have that pattern (Fig. 1A). However, these branching orders are not well supported, and Structure results indicate that all of the lativitta individuals share similar ancestry (Fig. 1C), which has much in common with a clade comprising French Guiana, Brazil, and Peru samples. If it is indeed true that the most recent common ancestor of H. erato originated in Amazonian Ecuador (as discussed earlier), it is likely that those ancestors sported the rayed wing pattern.
Multiple Origins of Similar Color Patterns in H. erato and H. melpomene.
Whereas other races appear clustered in the AFLP phylogeny, H. e. hydara is scattered roughly into three groups (Fig. 1A). Structure results also show three distinct groups of H. e. hydara. These findings indicate that the hydara form (“red patch” phenotype) likely evolved multiple times, as originally suggested by Brower (14). Its comimic, H. m. melpomene, is also widely scattered, falling into four groups in the AFLP phylogeny (Fig. 2A). Structure results show H. m. melpomene appearing in three groups (1, 4, and 5 in Fig. 2C). These findings support the idea that the H. m. melpomene pattern also may have evolved multiple times (15), and our results suggest it evolved at least thrice.
Whereas the red patch phenotypes in H. erato and H. melpomene are each geographically contiguous across the continent and display little pattern diversity, populations with the postman phenotype are geographically disjunct and those with the rayed phenotype display minor pattern differences. Therefore, populations of each form have been given a variety of subspecific names in each species. However, like the red patch phenotype, the AFLP tree topologies show that similarly patterned populations are not monophyletic in H. erato and H. melpomene, suggesting multiple origins and/or losses of both the postman and rayed phenotypes within each species.
Although our genome-wide AFLP data are consistent with multiple origins in each species, it is important to point out that hybridization among races could cause the evolutionary history of the wing pattern to become disconnected from the genome-wide background inferred from our AFLP data (SI Discussion). Eventually, comparing the interracial relationships identified here versus those inferred from mimicry genes themselves will provide the critical test of whether color pattern phenotypes have single or multiple origins in each species.
New Insights from the Matriline.
The mtDNA phylogenies of H. erato and H. melpomene reported here present a somewhat modified view from previous mtDNA analyses (14, 15), including the presence of additional clades, and different branching orders (SI Discussion). The differences stem from our larger ingroup sample size (88 vs. 52 for the H. erato complex and 83 vs. 42 for the H. melpomene complex) and our use of likelihood, as opposed to parsimony-based analyses. We also used a longer fragment of mtDNA with significantly greater coverage of COI (766 bp COI and 711 bp COII, vs. 126 bp COI and 598 bp COII).
Mean pairwise divergences among the three main H. erato clades, based on COI alone (766 bp), are as follows: H. himera - E1, 4.4%; H. himera - E2, 4.0%; E1 - E2: 4.3%; mean of approximately 4.2%. In H. melpomene, the mean pairwise divergence between M1 and M2 is 3.1%, or approximately 73% that in H. erato. Applying a rate of 1.5% uncorrected pairwise divergence per My (30) yields an estimated minimum age of 2.8 My for H. erato and 2.1 My for H. melpomene, placing the estimated minimum ages of both species groups in the later Pliocene. These estimates are older than previous ones (14, 15) because we used a different divergence rate, and because we relied only on COI. We focus only on COI because it has been found to exhibit the most clock-like rate among commonly used mtDNA markers (31). For H. melpomene, the estimate is much older compared to that inferred by Beltran et al. (32), who suggested a Pleistocene origin (1.5 My) based on the mtDNA divergence between H. melpomene and H. cydno. However, they concede that one might be paraphyletic to the other, and indeed, this study shows H. cydno mtDNA lineages to be nested within H. melpomene, as corroborated by the AFLP data.
H. cydno Is Not Sister to H. melpomene, and H. himera Is Not Sister to H. erato.
The relationship between H. melpomene and H. cydno has been the subject of much speculation (12, 32, 33), and early mtDNA studies indicated a sister species relationship (15). The AFLP phylogeny clearly places H. cydno as monophyletic and deeply nested within H. melpomene, indicating that these two entities are not sister taxa, as suspected by Beltrán et al. (33). Similarly, H. himera is clearly monophyletic and nested within H. erato, and thus the two cannot be considered sister species. The long branches that support H. cydno, H. himera, and H. e. chestertonii are consistent with peripatric events as suggested by Flanagan et al. (12).
Conclusions
The evolutionary processes resulting in widespread geographic variation between the comimetic species pair H. erato and H. melpomene have been the topic of substantial speculation (7–12, 14–16). Our analyses of these comimetic radiations incorporated both AFLP data and a large segment of mtDNA for the same set of specimens, enabling direct comparisons between the two data types. The results reveal striking clustering by geography and racial phenotype based on AFLP data, far more than is apparent in DNA sequence data. The fine-scale resolution provided by the AFLP data permits detailed reconstruction of the histories of the two comimics. Their vastly divergent trajectories reject the hypothesis of coradiation and our present age estimates additionally reject Pleistocene radiations in both species. Our evidence for independent radiations, combined with different estimated ages for the two species, suggest that the concordant variation between H. erato and H. melpomene has been the result of one-sided advergence, with H. erato radiating first and establishing the color pattern template for the future radiation of H. melpomene. The intriguing question then becomes: how did H. erato diversify in the first place? A brief discussion of the possible roles of natural and sexual selection as well as genetic drift is given in the SI Discussion. Additional research is required to understand the balance among these evolutionary forces in driving the early diversification of H. erato.
Materials and Methods
Sampling.
Specimens were wild-caught between 1994 and 2007, avoiding locations known to be hybrid zones. Between two and 10 specimens per race per geographic location were included. For the investigation of H. erato, 85 specimens of H. erato, three specimens of H. himera, and two specimens of each of the outgroups H. hecalesia and H. clysonymus were used (Table S1). For the investigation of H. melpomene, 78 specimens of H. melpomene, five specimens of H. cydno, and the outgroups H. atthis, H. hecale, and H. ismenius were used (Table S1).
Molecular Data.
DNA were extracted using a Qiagen DNeasy Kit. A 1,602-bp fragment of mtDNA spanning COI (766 bp), tRNALEU, and COII (711 bp) was amplified by PCR and sequenced using published (32) primers and methods. Outgroup sequences were downloaded from GenBank. Sequences were edited and aligned in Sequencher 4.9 (Gene Codes). AFLPs were generated using the Applied Biosystems Plant Mapping Kit and scored using Applied Biosystems Genemapper software. Additional details regarding AFLP primers, controls, and scoring parameters are available in SI Materials and Methods.
Phylogenetic and Population Genetic Analyses.
For the AFLP markers, phylogenetic analyses were conducted using neighbor joining with Nei-Li distances (34) in Paup*4.0b10 (35). Maximum likelihood phylogenetic analyses were performed on the mtDNA sequences using PHYML, under the GTR+I+G model, which was selected by the Akaike Information Criterion (36) as implemented in ModelTest 3.7 (37). Support for all phylogenetic analyses was assessed with 2,000 bootstrap pseudoreplicates. The model-based clustering method implemented in Structure, version 2.2 (38, 39), was used to search for population structure in both H. erato and H. melpomene. Additional details regarding Structure analyses are available in SI Materials and Methods.
CCI.
No metric currently exists to quantify the degree to which the monophyly of a given group is disrupted on a given topology. Therefore, we developed the Clade Construction Index (CCI) to compare the degree of monophyly of each race/geographic population between the mtDNA and AFLP trees. The CCI is the number of lineages that must be removed in order for a given group to become monophyletic; thus, a CCI of 0 defines a monophyletic group. The CCI is a simple measure of the topological violation of monophyly and does not consider the relative branch lengths or node support of the lineages that have to be removed in order for that group to achieve monophyly (although these could be incorporated).
Supplementary Material
Acknowledgments
We thank C. Jiggins for samples, N. Chamberlain for assistance in the laboratory, C. S. Walker for assistance with maps, N. Pierce for advice, and J. Mallet and three anonymous reviewers for comments on the manuscript. This work was funded by Coordenadoria de Aperfeicoamento de Pessoal de Nivel Superior (P.A.d.M.), Fundação de Amparo à Pesquisa do Estado de Rio Grande do Norte/Conselho Nacional de Desenvolvimento Científico e Tecnológico Research Grant PPP/2007 (to M.Z.C.), Ministério do Meio Ambiente (Brazil) collection permit 10894-1 (to M.Z.C.), National Science Foundation (W.O.M.), and National Institutes of Health/National Institute of General Medical Sciences Grant GM068763 (to M.R.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The mtDNA sequences reported in this paper have been deposited in the GenBank database (accession nos. GU330020–GU330187).
This article contains supporting information online at www.pnas.org/cgi/content/full/0911572107/DCSupplemental.
References
- 1.Brower AVZ. Phylogeny of Heliconius butterflies inferred from mitochondrial DNA sequences (Lepidoptera: Nymphalidae) Mol Phylogenet Evol. 1994;3:159–174. doi: 10.1006/mpev.1994.1018. [DOI] [PubMed] [Google Scholar]
- 2.Brower AVZ, Egan MG. Cladistic analysis of Heliconius butterflies and relatives (Nymphalidae: Heliconiiti): A revised phylogenetic position for Eueides based on sequences from mtDNA and a nuclear gene. Proc R Soc Lond B Biol Sci. 1997;264:969–977. [Google Scholar]
- 3.Brown KS. The Biology of Heliconius and Related Genera. Annu Rev Entomol. 1981;26:427–456. [Google Scholar]
- 4.Sheppard PM, Turner JRG, Brown KS, Benson WW, Singer MC. Genetics and the evolution of Muellerian mimicry in Heliconius butterflies. Philos Trans R Soc Lond B Biol Sci. 1985;308:433–613. [Google Scholar]
- 5.Turner JRG. Studies of Müllerian mimicry and its evolution in burnet moths and heliconid butterflies. In: Creed ER, editor. Ecological Genetics and Evolution. Oxford, UK: Blackwell; 1971. pp. 224–260. [Google Scholar]
- 6.Turner JRG. Adaptive radiation and convergence in subdivisions of butterfly genus Heliconius (Lepidoptera: Nymphalidae) Zool J Linn Soc. 1976;58:297–308. [Google Scholar]
- 7.Joron M, Mallet JLB. Diversity in mimicry: Paradox or paradigm? Trends Ecol Evol. 1998;13:461–466. doi: 10.1016/s0169-5347(98)01483-9. [DOI] [PubMed] [Google Scholar]
- 8.Mallet J, Joron M. Evolution of diversity in warning color and mimicry: Polymorphisms, shifting balance, and speciation. Annu Rev Ecol Syst. 1999;30:201–233. [Google Scholar]
- 9.Turner JRG, Mallet JLB. Did forest islands drive the diversity of warningly coloured butterflies? Biotic drift and the shifting balance. Philos Trans R Soc Lond B Biol Sci. 1996;351:835–845. [Google Scholar]
- 10.Brown KS, Sheppard PM, Turner JRG. Quaternary refugia in tropical America: Evidence from race formation in Heliconius butterflies. Proc R Soc Lond B Biol Sci. 1974;187:369–378. [Google Scholar]
- 11.Eltringham H. On specific and mimetic relationships in the genus Heliconius. Trans Entomol Soc Lond. 1916;1916:101–148. [Google Scholar]
- 12.Flanagan NS, et al. Historical demography of Mullerian mimicry in the neotropical Heliconius butterflies. Proc Natl Acad Sci USA. 2004;101:9704–9709. doi: 10.1073/pnas.0306243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kronforst MR, Gilbert LE. The population genetics of mimetic diversity in Heliconius butterflies. Proc R Soc Lond B Biol Sci. 2008;275:493–500. doi: 10.1098/rspb.2007.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brower AVZ. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brower AVZ. Parallel race formation and the evolution of mimicry in Heliconius butterflies: A phylogenetic hypothesis from mitochondrial DNA sequences. Evolution. 1996;50:195–221. doi: 10.1111/j.1558-5646.1996.tb04486.x. [DOI] [PubMed] [Google Scholar]
- 16.Mallet J, Jiggins CD, McMillan W. Evolution: Mimicry meets the mitochondrion. Curr Biol. 1996;6:937–940. doi: 10.1016/s0960-9822(02)00631-0. [DOI] [PubMed] [Google Scholar]
- 17.Albertson RC, Markert JA, Danley PD, Kocher TD. Phylogeny of a rapidly evolving clade: The cichlid fishes of Lake Malawi, East Africa. Proc Natl Acad Sci USA. 1999;96:5107–5110. doi: 10.1073/pnas.96.9.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beardsley PM, Yen A, Olmstead RG. AFLP phylogeny of Mimulus section Erythranthe and the evolution of hummingbird pollination. Evolution. 2003;57:1397–1410. doi: 10.1111/j.0014-3820.2003.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 19.Koblmuller S, et al. Reticulate phylogeny of gastropod-shell-breeding cichlids from Lake Tanganyika - the result of repeated introgressive hybridization. BMC Evol Biol. 2007;7:7. doi: 10.1186/1471-2148-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendelson TC, Shaw KL. Rapid speciation in an arthropod. Nature. 2005;433:375–376. doi: 10.1038/433375a. [DOI] [PubMed] [Google Scholar]
- 21.Rudh A, Rogell B, Hoglund J. Non-gradual variation in colour morphs of the strawberry poison frog Dendrobates pumilio: Genetic and geographical isolation suggest a role for selection in maintaining polymorphism. Mol Ecol. 2007;16:4284–4294. doi: 10.1111/j.1365-294X.2007.03479.x. [DOI] [PubMed] [Google Scholar]
- 22.Savage WK, Mullen SP. A single origin of Batesian mimicry among hybridizing populations of admiral butterflies (Limenitis arthemis) rejects an evolutionary reversion to the ancestral phenotype. Proc R Soc Lond B Biol Sci. 2009;276:2557–2565. doi: 10.1098/rspb.2009.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollingsworth PM, Ennos RA. Neighbour joining trees, dominant markers and population genetic structure. Heredity. 2004;92:490–498. doi: 10.1038/sj.hdy.6800445. [DOI] [PubMed] [Google Scholar]
- 24.Kosman E, Leonard KJ. Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Mol Ecol. 2005;14:415–424. doi: 10.1111/j.1365-294X.2005.02416.x. [DOI] [PubMed] [Google Scholar]
- 25.Meudt HM, Clarke AC. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007;12:106–117. doi: 10.1016/j.tplants.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Koopman WJM. Phylogenetic signal in AFLP data sets. Syst Biol. 2005;54:197–217. doi: 10.1080/10635150590924181. [DOI] [PubMed] [Google Scholar]
- 27.Kingston SE, Adams LD, Rosel PE. Testing mitochondrial sequences and anonymous nuclear markers for phylogeny reconstruction in a rapidly radiating group: Molecular systematics of the Delphininae (Cetacea: Odontoceti: Delphinidae) BMC Evol Biol. 2009;9:19. doi: 10.1186/1471-2148-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meudt HM, Lockhart PJ, Bryant D. Species delimitation and phylogeny of a New Zealand plant species radiation. BMC Evol Biol. 2009;9:17. doi: 10.1186/1471-2148-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi R, Watanabe K, Nishida M, Hori M. Evolution of feeding specialization in Tanganyikan scale-eating cichlids: A molecular phylogenetic approach. BMC Evol Biol. 2007;7:11. doi: 10.1186/1471-2148-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quek SP, Davies SJ, Itino T, Pierce NE. Codiversification in an ant-plant mutualism: Stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae) Evolution. 2004;58:554–570. [PubMed] [Google Scholar]
- 31.Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 32.Beltran M, et al. Phylogenetic discordance at the species boundary: Comparative gene genealogies among rapidly radiating Heliconius butterflies. Mol Biol Evol. 2002;19:2176–2190. doi: 10.1093/oxfordjournals.molbev.a004042. [DOI] [PubMed] [Google Scholar]
- 33.Beltran M, Jiggins CD, Brower AVZ, Bermingham E, Mallet J. Do pollen feeding, pupal-mating and larval gregariousness have a single origin in Heliconius butterflies? Inferences from multilocus DNA sequence data. Biol J Linn Soc. 2007;92:221–239. [Google Scholar]
- 34.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 1999. [Google Scholar]
- 36.Akaike H. A new look at statistical model identification. IEEE Trans Automat Control. 1974;19:716–723. [Google Scholar]
- 37.Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 38.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.