Abstract
Polyadenylation of RNA is a posttranscriptional modification that can play two somewhat opposite roles: stable polyadenylation of RNA encoded in the nuclear genomes of eukaryote cells contributes to nuclear export, translation initiation, and possibly transcript longevity as well. Conversely, transient polyadenylation targets RNA molecules to rapid exonucleolytic degradation. The latter role has been shown to take place in prokaryotes and organelles, as well as the nucleus of eukaryotic cells. Here we present evidence of hetero- and homopolymeric adenylation of truncated RNA molecules within the cytoplasm of human cells. RNAi-mediated silencing of the major RNA decay machinery of the cell resulted in the accumulation of these polyadenylated RNA fragments, indicating that they are degradation intermediates. Together, these results suggest that a mechanism of RNA decay, involving transient polyadenylation, is present in the cytoplasm of human cells.
Keywords: exosome, heteropolymeric tails, RNA polyadenylation
RNA polyadenylation occurs throughout the biological world. Stable poly(A) is associated with the mature 3′ end of most mRNAs encoded in the nuclear genome of eukaryotes. It is important for nuclear export and efficient translation initiation and may also contribute to transcript longevity (1). In turn, mRNA degradation in the cytoplasm usually initiates with deadenylation of the stable poly(A) tail, followed by either further degradation of the mRNA body by the exosome (3′–5′ pathway) or removal of the 5′ cap and subsequent 5′–3′ degradation by hXrn1 (exoribonuclease 1; 5′–3′ pathway) (2–4).
Unlike stable poly(A), transient poly(A) is not associated with the mature 3′ end of the transcript. RNA decay pathways that use transient poly(A) include a stage in which poly(A) or poly(A)-rich tails are added to the 3′ ends of truncated degradation intermediates and are believed to assist exoribonucleases in their rapid degradation. As the tail addition can occur after endonucleolytic cleavage of the RNA or repetitive adenylation and 3′-5′ digestion, it often appears to be at “internal” positions relative to the full RNA sequence (5, 6). The term “transient” is used because the short-lived tail is degraded along with the RNA fragment. Transient poly(A) was first disclosed in Escherichia coli and then in additional bacteria, organelles, archaea and, eventually, in yeast and human nuclei (4, 5, 7–9). In all systems in which truncated, polyadenylated RNA fragments were initially detected, they were later found to be correlated to poly(A)-assisted RNA decay (4, 7, 8).
In yeast nuclei, transient poly(A) was found to play a role in RNA quality control wherein polyadenylation, initiated by the TRAMP complex, targets incorrectly folded tRNA molecules to degradation by the nuclear exosome (10, 11). In human cells, cotranscriptionally cleaved 3′ regions of an introduced β-globin gene and a class of short, highly unstable RNAs, dubbed PROMPTs (promoter upstream transcripts), were found to be adenylated and accumulated when the exosome was down-regulated (12, 13). It has recently become clear that almost the entire genome is transcribed, which results in the production of antisense and noncoding RNAs (13–17). These cryptic unstable transcripts were found to undergo rapid poly(A)-assisted decay as well (10, 11, 14). However, although such nuclear mechanisms have been reported, poly(A)-assisted RNA decay has not been found in the cytoplasm of eukaryote cells.
Previously, we isolated truncated, poly(A)-tailed ribosomal RNA molecules from total RNA of human cells (18). Interestingly these tails were either of homopolymeric [poly(A)] or heteropolymeric [poly(A)-rich] nature. The heteropolymeric extensions were dominantly adenylated but contained other nucleotides as well and resembled the poly(A)-rich tails produced in bacteria, chloroplasts, and archaea (6, 7). However, neither the cellular location nor the function of these adenylation events was deciphered.
Here, we show the presence of truncated mRNAs and rRNAs, bearing homopolymeric or heteropolymeric tails, in the cytoplasm of human cells. RNAi-mediated silencing of 3′-5′ exoribonucleases and hXrn1 resulted in the accumulation of these molecules in the cytoplasm, strongly suggesting that poly(A)-assisted RNA decay occurs in the cytoplasm of human cells.
Results
Fragmented 28S rRNA Molecules Containing Homo- or Heteropolymeric Tails Are Present in Both the Cytoplasm and Nucleus of Human Cells.
In earlier work, we detected truncated 28S and 18S rRNAs containing homo- or heteropolymeric tails (18). It was plausible that these rRNA fragments are degradation intermediates of the nuclear process mentioned earlier, but a second option could not be ignored: transient, perhaps degradation-assisting, adenylation of RNA within the cytoplasm, coexisting with the stable poly(A) tails that characterize this cellular location. Hence, our first goal was to determine whether these tailed, truncated molecules are present in the nucleus, cytoplasm, or both.
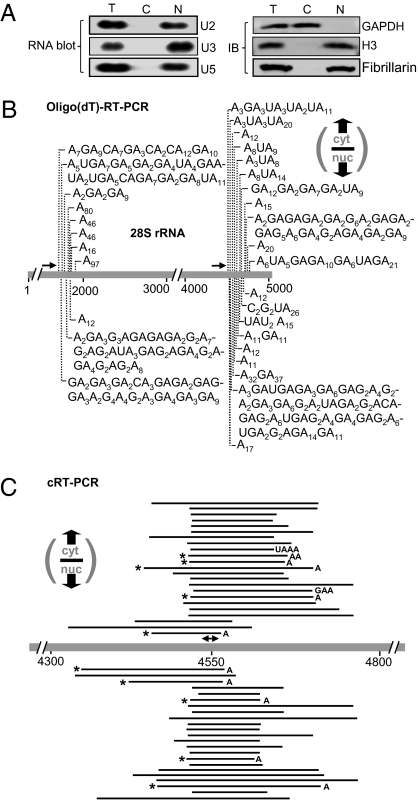
RNA and protein were obtained from cytoplasmic and nuclear fractions which were prepared from HeLa cells. A clear fraction-specific distribution of mature rRNA and pre-rRNA (and tRNA) was observed upon ethidium bromide staining of total, cytoplasmic, and nuclear RNA (Fig. S1). To assess purity of fractions, Northern blots and immunoblots were prepared, and Fig. 1A shows that the RNA and protein markers were detected exclusively in the corresponding fractions. Next, the presence of fragmented polyadenylated 28S rRNA was evaluated by oligo(dT) RT-PCR. The results disclosed 28S rRNA fragments, with either homo- or heteropolymeric extensions in both the nucleus and cytoplasm (Fig. 1B). To our knowledge, this is the first detection of internally adenylated RNA in the cytoplasm of human cells.
Fig. 1.
28S rRNA fragments with homo- or heteropolymeric poly(A) tails in the cytoplasm and nuclei of human cells. (A) Fractionation purity was determined by Immunoblots (IBs) and RNA blots using fraction-specific markers: C, cytoplasm: GAPDH. N, nucleus: histone H3 (H3), fibrillarin, and U2/U3/U5-snoRNAs. T, total cell RNA/proteins. (B) Oligo(dT) RT-PCR isolation of truncated, adenylated 28S rRNA molecules from cyt (above) and nuc (below) fractions. 28S rRNA is schematically presented with arrows indicating primer locations and vertical lines showing tail sites and content (Table S1). (C) cRT-PCR isolation of truncated 28S rRNA molecules from cyt (above) and nuc (below) fractions. Cases in which it is not known whether the “A” is encoded or added posttranscriptionally are marked with an asterisk (Table S2).
To evaluate the truncated molecules in both fractions by means unbiased to poly(A) extensions, circularized reverse transcription (cRT) was applied to the same region of 28S rRNA (Fig. 1C). Truncated molecules were successfully isolated from both the cytoplasmic and nuclear fractions. Although most fragments were tail-less, a small number had extensions of 1 to 4 nt; however, there are cases in which it is not clear whether the adenosine is part of the encoded sequence or was posttranscriptionally added. These molecules could reflect oligo(A) addition, or could be either at a stage before tail synthesis or amid 3′-5′ degradation. Therefore, as in many previous studies of transient adenylation, biased methods such as oligo(dT) RT-PCR are necessary to view long internal tails and assess their nt composition, which can often lead to the identity of the polyadenylating factor (7, 19).
Truncated, Intron-Less β-Actin Transcripts Harboring Homo- or Heteropolymeric Tails.
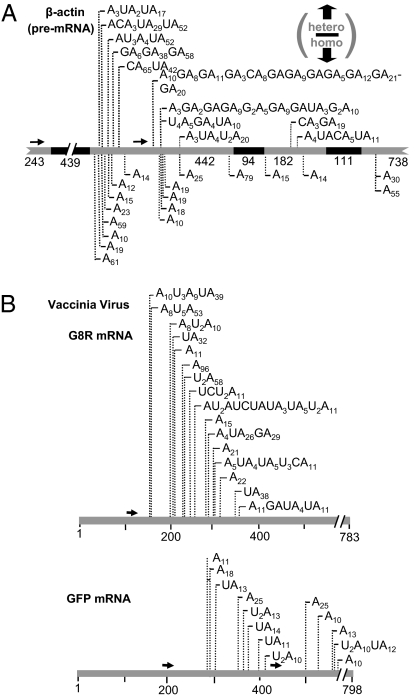
To check if, in addition to rRNA, mRNA can undergo this process, the presence of truncated adenylated β-actin mRNA molecules was analyzed using oligo(dT) RT-PCR. Gene-specific primers were applied; some of which targeted the mRNA sequence immediately upstream of introns, potentially allowing the isolation of both spliced and unspliced adenylated β-actin fragments. As shown in Fig. 2A, truncated adenylated β-actin fragments were isolated and similar to rRNA, both homo- and heteropolymeric tails were observed. Also, such molecules were obtained from both nuclear and cytoplasmic fractions (Table S1). No introns were present among the isolated sequences, disclosing that these are (partially, if not fully) spliced transcripts. These results show that like rRNA, mRNA can undergo transient adenylation.
Fig. 2.
(A) Spliced β-actin fragments with homo- or heteropolymeric poly(A) tails, isolated from human cells. Primary β-actin mRNA is schematically presented (introns in black, exons in gray). Lengths of each exon/intron are shown. Homo- and heteropolymeric tails are presented above and below the gene, respectively (Table S1). (B) Oligo(dT) RT-PCR isolation of truncated, adenylated VV RNA molecules [G8R (a viral mRNA) and GFP (integrated into the viral genome)] from HeLa cells 4 h after transfection (Table S3).
Truncated Vaccinia Virus RNAs Bearing Homo- or Heteropolymeric Tails Were Isolated from Infected Cells.
In addition to the fractionation purity assays (Fig. 1A), to further ensure that the RNA molecules obtained from cytoplasmic fractions as described earlier were not intermediates of a nuclear process that had “leaked” to the cytoplasm, either in vivo or during our experimental procedures, we adopted an additional approach: vaccinia virus (VV) belongs to the poxvirus family and contains a double-stranded DNA genome, encoding more than 200 proteins. Poxviruses are unique among most DNA viruses in that they do not enter the nucleus during their entire life cycle (20). To check if viral RNA is subjected to transient adenylation within the cytoplasm, RNA was purified from infected HeLa cells and analyzed for the presence of truncated, polyadenylated transcripts of genes encoded in the viral genome. Fig. 2B displays the results of the analysis of G8R mRNA, an intermediate viral gene and GFP mRNA, produced from a GFP gene inserted into the viral genome (21). Truncated transcripts originating from these genes, with either homo- or heteropolymeric tails, were detected. These results provide additional support that a mechanism involving transient RNA polyadenylation is present in the cytoplasm and applies not only to rRNA, but mRNA as well. Accordingly, the next step was to check if there is a link between this type of RNA adenylation and RNA decay, as in all previously studied systems in which such molecules were found.
Are the Truncated, Polyadenylated Transcripts Degradation Intermediates?
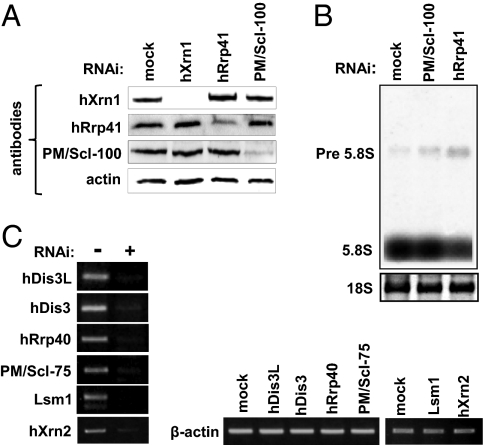
In all systems in which truncated, adenylated RNA molecules were detected, they were later shown to be intermediates in a poly(A)-assisted RNA decay pathway. We asked if the tails observed here, in the cytoplasm, are indeed telltale signs of such a process. If so, inhibition of exonucleolytic activity would result in their accumulation, as observed in bacteria, plant mitochondria, and the nuclei of yeast and human cells (12 –14, 22). To this end, we used RNAi to knock down (KD) hXrn1 and the exosome subunits. For the latter, PM/Scl-100, an exoribonuclease associated with the exosome and hRrp41, an exosome core protein, were targeted. Immunoblots disclosed significant KD of the targeted proteins (Fig. 3A). A reduction in exosome activity was validated by 5.8S rRNA precursor accumulation in both the PM/Scl-100 and hRrp41-silenced cells (Fig. 3B) (13, 23).
Fig. 3.
RNAi-mediated silencing of exosome subunits, hXrn1, hXrn2, and Lsm1. (A) Efficiency of silencing was measured via immunoblot with specific antibodies. (B) Loss of exosome activity was validated by the accumulation of the 5.8S rRNA precursor as detected by Northern blot. (C) KD efficiency of RNAi-silenced proteins for which antibodies were not available was monitored by RT-PCR. Immunoblots and RT-PCR were normalized with β-actin.
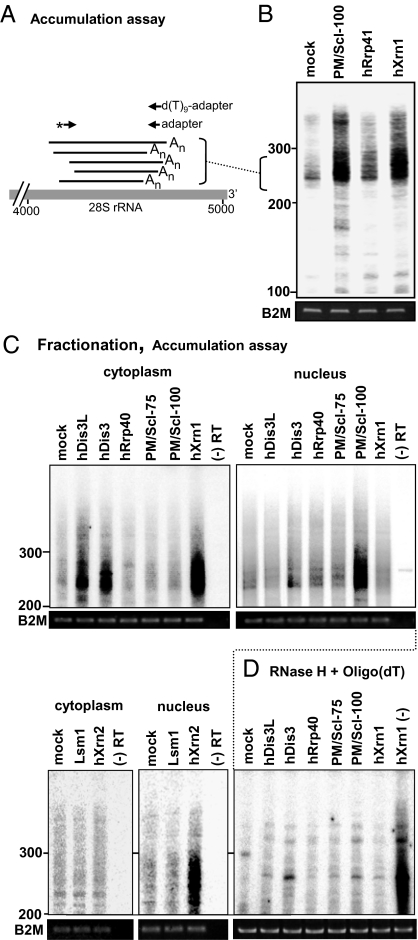
Degradation intermediates are nonabundant and rapidly degraded; hence it is beneficial to analyze highly expressed transcripts for such related molecules. Therefore, we chose to focus on the region of 28S rRNA from which adenylated fragments had been isolated by oligo(dT) RT-PCR, as described earlier. To visually evaluate fragmented, adenylated 28S rRNA accumulation, a radiolabeling assay was applied; loading amounts were normalized according to B2M mRNA via RT-PCR (Fig. 4A). Following oligo(dT) reverse transcription, the same primers with which the truncated molecules were previously isolated were used but this time, an extra PCR stage in which the products were labeled via a 5′ end [32P] 28S rRNA primer, was included. Fractionation on denaturing polyacrylamide gel, allowed sensitive assessment of accumulation of the labeled amplification products. As the truncated molecules were polyadenylated at varying sites along the 28S rRNA (Fig. 1B), and the extensions were of different lengths, accumulation did not result in a single band, but a smeared signal instead. Substantial accumulation of truncated 28S rRNA fragments was observed in the lanes representing the RNAi silencing of PM/Scl-100 and hXrn1 (Fig. 4B). Although PM/Scl-100 (Rrp6 in yeast) has been reported to exist in the cytoplasm, it is mainly found in the nucleus of human cells in association with the nuclear exosome (24, 25). Therefore, the corresponding signal in this lane is most likely the result of the accumulation of nucleus-localized degradation intermediates. In contrast, accumulation of truncated 28S rRNA fragments was much less drastic in the hRrp41-silenced cells.
Fig. 4.
Adenylated 28S rRNA degradation intermediates significantly accumulated in cells upon RNAi silencing of major exonucleases. (A) Schematic presentation of PCR-labeling assay to detect accumulation of adenylated, truncated 28S rRNA molecules after exonuclease KD. Briefly, following oligo(dT) RT of RNA from different KD cells, PCR with [32P]-labeled gene-specific primer (asterisk; same as used to isolate molecules as shown in Fig 1B) and adapter oligo was performed. (B) RNA purified from the mock and KD HeLa cells was subjected to this analysis to assess the accumulation of polyadenylated, fragmented 28S rRNA. The migration of DNA markers is shown to the left (nucleotide numbers). (C) Cytoplasmic or nuclear RNA from cells in which the indicated proteins were silenced by RNAi, was subjected to oligo(dT) RT-PCR labeling. A control, with reverse transcriptase omitted from the reaction, is shown in the far right lane of each panel. (D) To verify that the PCR products originated from polyadenylated transcripts, a control was performed in which the poly(A) tails of the cytoplasmic fractions were removed by incubation with oligo(dT) and RNase H before oligo(dT)-primed RT-PCR labeling. This resulted in reduction of amplification signals to background levels, as compared with the hXrn1 sample that was treated with RNase H but without oligo(dT) [lane labeled hXrn1 (-)].
hXrn1 plays a central role in 5′-3′ RNA degradation in the cytoplasm. Counter intuitively, its silencing resulted in significant accumulation of the polyadenylated molecules. A cytoplasmic localization of these transcripts is implied by that of hXrn1 but the obvious question is why the inhibition of a 5’-3’ exonuclease results in the accumulation of degradation intermediates with 3’ end poly(A) tails. This issue will be addressed in the Discussion.
Together, the information obtained using this assay suggests that the isolated polyadenylated 28S rRNA fragments are indeed degradation intermediates. To definitively determine cellular location, the analyses described here were repeated after fractionation of the nucleus and cytoplasm. Also, to further investigate whether the exosome core is involved in this process, several additional subunits and exoribonucleases were down-regulated.
RNAi Silencing of Exoribonucleases Confirmed That Poly(A)-Assisted RNA Decay Exists in the Cytoplasm of Human Cells.
To confirm that degradation of transiently adenylated RNA occurs in the cytoplasm of human cells, the analysis was repeated after nuclear and cytoplasmic fractions were prepared. Two additional exosome core components, hRrp40 and PM/Scl-75, and two exoribonucleases, hDis3 and hDis3L, were knocked down. hDis3 is a homologue of the yeast Dis3 protein; a hydrolytic 3′-5′ exoribonuclease with additional endonuclease activity that is associated with the exosome in both the nucleus and cytoplasm in yeast. This protein serves as the only catalytic subunit in the cytoplasmic yeast exosome (25, 26). While this work was in progress, an additional Dis3 homologue in human cells, hDis3L, was discovered, and unlike hDis3, was found to be strongly associated with the exosome core and to localize to the cytoplasm. Therefore, it is the more likely Dis3 orthologue in human cells and its silencing was included (27). Two additional enzymes were silenced: hXrn2 and Lsm1. The former is a major, nucleus-localized 5′-3′ exonuclease and so its KD could be anticipated to cause accumulation in the nucleus but not in the cytoplasm (4). The Lsm proteins play a role similar to the bacterial Hfq proteins, involved in bacterial mRNA decay via tail addition. Lsm1 is associated with hXrn1, decapping enzymes (Dcp1/Dcp2), and mRNA decay in the cytoplasm. Also, it was shown to be directly involved in histone mRNA oligo(U)-mediated decay. Thus, the effect of its RNAi silencing was evaluated as well (28). RT-PCR quantification, for lack of appropriate antibodies, showed significant reduction in the mRNA levels of the silenced proteins (Fig. 3C).
No accumulation was observed in the cytoplasm of cells silenced of the exosome components, hRrp40 or PM/Scl-75, and only minor accumulation could be observed in their nuclear fractions (Fig. 4C). In contrast, KD of either hDis3 or hDis3L resulted in distinct cytoplasmic accumulation of truncated 28S rRNA fragments, suggesting that these two proteins are involved in the degradation process. As anticipated, hXrn1 and PM/Scl-100 silencing caused substantial accumulation in the cytoplasmic and nuclear fraction, respectively, and KD of hXrn2 resulted in accumulation only in the nuclear fraction. Silencing of Lsm1 did not result in accumulation within either fraction. To be sure that the labeled products observed in the gel were indeed obtained by the amplification of polyadenylated 28S rRNA molecules and not artifacts of nonspecific reverse transcription, RNA purified from the cytoplasm was incubated with RNase H and oligo(dT) before the oligo(dT)-primed reverse transcription (Fig. 4D). This way, all poly(A) tails would be removed and no cDNA would be produced during the subsequent oligo(dT)-primed RT. Indeed, this treatment reduced the observed amplified signals to the background level. It is especially evident when comparing the result obtained for RNA purified from hXrn1-silenced cells that was either incubated with RNase H alone or with RNase H together with oligo(dT) (Fig. 4D).
Together, these results confirm that rRNA can be transiently adenylated, not only in the nucleus of human cells, but in the cytoplasm as well, and this is linked to rRNA decay. In the cytoplasm, the involved exoribonucleases include hDis3L, hDis3, and hXrn1, as the adenylated degradation intermediates accumulate upon their down-regulation.
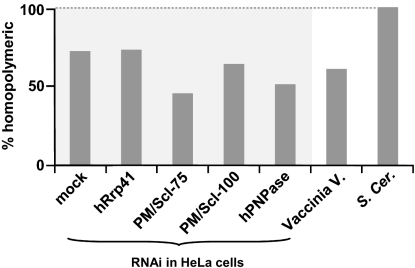
Exosome or hPNPase Silencing Does Not Cause a Decrease in the Percentage of Hetero-Adenylated Clones.
At this stage, we attempted to identify the enzyme responsible for the addition of the tails, in particular those of heteropolymeric nature. PNPase and the archaeal exosome are responsible for both hetero-tailing and degrading bacterial, organellar, and archaeal RNA (7). The structurally similar eukaryotic exosome was initially a candidate for the hetero- activity observed here and if it were responsible for producing the hetero-tails, a shift in favor of the homo- extensions would be expected to occur upon its KD. However, the exosome was reported to lack phosphorolytic activity (25) and in agreement with this, we found that the percentage of homo- tails within the total amount of isolated and sequenced clones did not increase upon KD of exosome subunits, hRrp41, PM/Scl-100, and PM/Scl-75 (Fig. 5).
Fig. 5.
Percentage of homopolymerically tailed molecules isolated from HeLa cells with RNAi silencing of exosome components or hPNPase, cells infected with VV, and yeast. Percentage of homopolymerically tailed molecules among the total number of sequenced molecules isolated from HeLa cells did not increase upon RNAi silencing of: hRrp41, PM/Scl-75, PM/Scl-100, or hPNPase (28S rRNA was analyzed). In VV-infected cells, the viral G8R RNA was analyzed, and in S. cerevisiae, 25S rRNA, CYH2 and Actin1 (Tables S3, S4, and S5).
PNPase is present in human cells (hPNPase) and phosphorolytically active (29). Although localized to the mitochondrial intermembrane space (IMS) (30), we considered the possibility that hPNPase, similar to other IMS proteins such as cytochrome C and endonuclease G (31), could be liberated from this compartment and enter the cytoplasm (and/or nucleus), where it could polymerize hetero- tails. However, no increase in homo- tail rate was observed in cells with constant hPNPase shRNA silencing either (32).
Discussion
Poly(A)-Assisted RNA Decay Occurs in the Cytoplasm of Human Cells.
A nuclear RNA surveillance mechanism involving polyadenylation and rapid degradation of nonfunctional, antisense, and noncoding RNAs has been described in yeast and human cells (4, 9, 33 –36). We initially assumed that the truncated, polyadenylated rRNA molecules that we had previously detected in human cells (18) are solely related to this process, but in this study we found that transient adenylation can occur in the cytoplasm as well, as such molecules were detected in cytoplasmic fractions. However, in yeast, unstable transcripts derived from intergenic regions (i.e., nuclear) were reported to be stabilized upon inhibition of Xrn1 and Dcp1 (i.e., cytoplasmic). In such a case, export from the nucleus would be entailed (35). To ensure this was not the case in our work, besides assays that demonstrated fractionation purity, both at protein and RNA levels, additional cytoplasmic localization evidence was obtained by analyzing the RNA of VV, a virus that spends its entire life cycle in the cytoplasm. Analysis of the β-actin transcript was applied as well, demonstrating that endogenous mRNA can undergo this process.
If indeed the observed molecules are degradation intermediates of poly(A)-assisted RNA decay, we expected to observe their accumulation upon exoribonuclease down-regulation. Silencing of three of the nine exosome core proteins caused only slight accumulation of adenylated rRNA in the nucleus and no noticeable accumulation in the cytoplasm. PM/Scl-100 silencing resulted in accumulation only in the nuclear fraction and therefore, although reported to partially reside in the cytoplasm of human cells, PM/Scl-100 does not seem to be involved in this cytoplasmic process (24, 37, 38). Conversely, RNAi silencing of the two Dis3 (Rrp44) homologues, hDis3 and hDis3L, resulted in cytoplasmic accumulation of truncated adenylated rRNA. The fact that silencing of hDis3, the previously identified yeast Dis3 orthologue (38), had this effect, suggests that this Dis3 family member, although most likely not associated with the exosome, acts as a 3′-5′ exonuclease. hDis3L, however, is most likely associated with the cytoplasmic exosome core (27), and the accumulation of truncated adenylated rRNA in the cytoplasm upon its silencing suggests either exosome involvement in this process (despite no such effect when silencing core subunits) or a level of exosome-independent activity of hDis3L. In previous studies, although substantially knocked down by RNAi (validated by assessment of 5.8S rRNA processing), not all exosome subunits that were silenced caused a substantial effect on AU-rich element (ARE)–mediated decay, and a similar situation could occur in this case (23). hXrn2 KD led to accumulation within the nucleus but not in the cytoplasm, in correlation with its cellular localization.
Lsm1 silencing did not result in the accumulation of adenylated rRNA fragments. This result is somewhat surprising, as Lsm1 is known to play a central role in mRNA degradation following stable poly(A) tail shortening . Also, recent work disclosed a novel oligo(U)-mediated histone mRNA decay pathway with Lsm1 as an integral component (28). There, its silencing greatly stabilized histone mRNA (i.e., hampered its degradation), as did that of Xrn1 and Dcp2. Here, its apparent uninvolvement in poly(A)-mediated rRNA decay could be interpreted as the following: Lsm1 is known to recruit the decapping enzymes and allow Xrn1 5′-3′ degradation after the cap is removed. However, rRNA (uncapped) was the focus of the assay presented here. The poly(A)-mediated decay of rRNA may differ from that of mRNA (capped). First, Lsm1 could well be involved in cap removal of mRNA, allowing hXrn1 activity. Second, hXrn1 may not be involved in mRNA decay. Third, in any case, in previous work, we traced truncated adenylated 28S rRNA molecules to an endonucleolytically cleaved region of the 28S transcript (18). Multiple endonucleolytic cleavage events of rRNA or mRNA before transient tail addition, as in other poly(A)-mediated RNA decay pathways, could provide bare 5′ (and 3′) ends, thereby bypassing the need to decap the molecule (18). More studies are needed to characterize any differences between poly(A)-mediated rRNA and mRNA decay.
Why Did Silencing of hXrn1 Result in the Accumulation of Truncated Transcripts, Adenylated at the 3′ End?
If the tails observed here provide the same function as in all known cases of transient adenylation, to assist in 3′-5′ degradation, why did inhibition of hXrn1’s 5′-3′ activity result in the accumulation of these 3′ tailed molecules? Perhaps this case resembles ARE-mediated mRNA decay in the sense that, although AREs are present in the 3′ UTR, Xrn1 is required to fully degrade the substrates (23). Also, oligo(U)-mediated histone mRNA degradation was shown to include degradation from both the 5′ and 3′ ends of the transcript (28). Moreover, if endonucleolytic cleavage events were to precede poly(A) tail addition, hXrn1 would be a likely candidate to degrade the distal cleavage products, starting from their unprotected 5′ ends.
Interestingly, recent studies have questioned the necessity of polyadenylation of nuclear exosome substrates by Trf4/5 of the TRAMP complex, as a polyadenylation-defective form of Trf4 (Trf4-DADA) can activate exosomal degradation of RNA substrates and rescue the lethality of Trf4/5 double mutants (14, 39). This suggests that the general addition of poly(A) tails to nuclear degradation intermediates may only be necessary to promote the degradation of certain structured RNAs. If cytoplasmic poly(A)-assisted RNA decay is similar in this manner, more weight would be placed on the 5′-3′ pathway; hence, hXrn1’s involvement.
Which Enzyme Produces the Homo- and Heteropolymeric Tails?
Neither hPNPase nor exosome silencing caused an increase in the percentage of homo-tailed molecules within the total amount isolated and sequenced. Theoretically, this could indicate two alternatives: (i) neither enzyme produces the heteropolymeric tails or (ii) one of the enzymes produces both homo- and heteropolymeric tails. The latter scenario is highly unanticipated as, traditionally, polyadenylating enzymes in such pathways are known to synthesize a single type of tail, as far as nucleotide composition. Furthermore, the exosome's reported lack of phosphorolytic activity and “inconvenient” IMS localization of hPNPase agree with their lack of involvement. Possible candidates, especially for producing the observed A/U-rich heteropolymeric tails, could be among the seven noncanonical poly(A) polymerases recently identified in human cells, as nc-PAPs have been shown to add either As or Us (28, 40, 41). However, many of the isolated heteropolymeric sequences are A/G-rich and could possibly be produced by a different factor. Initial results obtained by RNAi silencing of the seven human ncPAPs are yet inconclusive in terms of their possible involvement in the transient tail addition (Fig. S2 and Table S6). Degenerative or overlapping activities may necessitate combinatorial RNAi KD of these proteins in future analysis. Interestingly, only homopolymeric tails were found in Saccharomyces cerevisiae (Fig. 5), suggesting that the component responsible for heteropolymeric tails is not present in S. cerevisiae. Clear identification of the polyadenylating elements involved will assist in understanding the apparently complex model that governs this pathway.
In summary, in this work, we revealed transient homo- and heteropolymeric adenylation of rRNA and mRNA fragments within the cytoplasm of human cells. The adenylated rRNA fragments accumulated upon KD of RNA degradation machinery, showing that they are intermediates in an RNA decay process that involves transient poly(A). Although much remains to be done to decipher the specific functionality of the two tail types, the enzyme(s) that produce them, and the precise mechanism of decay, we have identified a number of exonucleases involved in their degradation.
Materials and Methods
Cells.
HeLa and HEp-2 cells were grown as a monolayer at 37 °C, 5% CO2, in DMEM (Sigma) supplemented with 10% FCS, 2 mM L-glutamine, and penicillin–streptomycin (32). Growing and infection of cells with recombinant viruses containing GFP was performed as described (21).
Transient siRNA Application.
Cells were transfected with 400 pmol siRNA duplex (Table S7) using Lipofectamine 2000 (Invitrogen). After 24 h, cells were detached with trypsin and replated, followed 24 h later by a second transfection with 800 pmol siRNA. Seventy-two hours after the initial transfection, total RNA was harvested with MasterPure kit (Epicentre) or total protein was harvested with an SDS-lysis buffer (1% SDS, 10% glycerol, and 0.1 M DTT).
Cell Fractionation.
Nuclear and cytoplasmic fractions were obtained from HeLa cells as previously described (24). Briefly, detached cells were washed in PBS solution, resuspended in lysis buffer, incubated on ice for 10 min, and centrifuged for 10 min at 3,000 × g. The supernatant (cytoplasmic fraction) was transferred to a new tube and the pellet (nuclear fraction) was resuspended in lysis buffer and lysed via sonication. Following centrifugation at 12,000 × g for 10 min at 4 °C, RNA was purified from both fractions by the MasterPure kit (Epicenter).
Random-Primed RT-PCR and Northern Blots.
Random-primed RT was performed on 3 μg total RNA using AffinityScript RT (Stratagene) with 500 ng random primers (Amersham Biosciences) and normalized with β-actin by PCR. RNAi silencing efficiency was measured via semiquantitative RT-PCR using random primed cDNA. RNA blot was performed as described previously (32). The [32P] uniformly labeled antisense riboprobe (beginning at +1 of the mature 5.8S rRNA and ending 48 nt downstream of its 3′ end) was used. The Northern blot to validate cell fractionation was probed with a [32P]-labeled U2, U3, or U5 snoRNA oligonucleotide (Table S7).
Western Blot Analysis.
Proteins were analyzed by 8% to 10% SDS/PAGE gel, transferred to a nitrocellulose membrane, blotted with appropriate antibody (Table S7), and normalized with actin rabbit polyclonal antibody (Santa Cruz Biotechnology).
Oligo(dT) RT-PCR Isolation and Sequencing of Truncated Polyadenylated RNA.
Briefly, cDNA was generated with dT10-adapter. PCR was applied to cDNA with a gene-specific primer and adapter followed by T/A cloning and sequencing (19).
PCR Labeling Assays.
For all reactions RNA amounts were measured with ND-1000 NanoDrop and cDNA was normalized according to B2M. The normalized oligo(dT)-primed cDNA was then subjected to 25 PCR cycles with a 28S rRNA-specific forward primer coupled with adapter oligo. Products were used as templates in a second round of PCR-labeling using the same 28S rRNA primer, which was 5′-end [32P]-labeled by PNK and γ-[32P]ATP, and the adapter. The second round lasted five cycles followed by 10 min extension at 72 °C. PCR-labeled products were resolved by 6% denaturing PAGE and autoradiography. EtBr-stained DNA and [32P]-labeled RNA molecules were run on the gels as length markers.
Supplementary Material
Acknowledgments
We thank Dr. Wen Chang (Academia Sinica, Taiwan) for help with the VV experiments and Dr. Mark Christensen (Georgetown College, Georgetown, Kentucky) for providing us with the fibrillarin antibody. This work was supported by grants from the Israel Science Foundation, the Niedersachsen Foundation, and The Israel Ministry of Science (Israel-Taiwan Collaboration Program).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910621107/DCSupplemental.
References
- 1.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 2.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 3.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 4.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slomovic S, Portnoy V, Liveanu V, Schuster G. RNA polyadenylation in prokaryotes and organelles; different tails tell different tales. Crit Rev Plant Sci. 2006;25:65–77. [Google Scholar]
- 7.Slomovic S, Portnoy V, Yehudai-Resheff S, Bronshtein E, Schuster G. Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases. Biochim Biophys Acta. 2008;1779:247–255. doi: 10.1016/j.bbagrm.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Wilusz CJ, Wilusz J. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev. 2008;22:1–7. doi: 10.1101/gad.1634508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanácová S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 12.West S, Gromak N, Norbury CJ, Proudfoot NJ. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell. 2006;21:437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 14.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 15.Lykke-Andersen S, Jensen TH. CUT it out: silencing of noise in the transcriptome. Nat Struct Mol Biol. 2006;13:860–861. doi: 10.1038/nsmb1006-860. [DOI] [PubMed] [Google Scholar]
- 16.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 18.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slomovic S, Portnoy V, Schuster G. Detection and characterization of polyadenylated RNA in Eukarya, Bacteria, Archaea, and organelles. Methods Enzymol. 2008;447:501–520. doi: 10.1016/S0076-6879(08)02224-6. [DOI] [PubMed] [Google Scholar]
- 20.Broyles SS. Vaccinia virus transcription. J Gen Virol. 2003;84:2293–2303. doi: 10.1099/vir.0.18942-0. [DOI] [PubMed] [Google Scholar]
- 21.Hsiao JC, et al. A poxvirus host range protein, CP77, binds to a cellular protein, HMG20A, and regulates its dissociation from the vaccinia virus genome in CHO-K1 cells. J Virol. 2006;80:7714–7728. doi: 10.1128/JVI.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kushner SR. mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- 23.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. J Cell Sci. 2009;122:1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- 26.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 27.Staals R, Bronkhorst W, Schilders G, Raijmakers R, Pruijn G. A novel Dis3-like ribonuclease associated with human exosome complexes. 2009 doi: 10.1038/emboj.2010.122. RNA meeting 2009 abstract 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Portnoy V, Palnizky G, Yehudai-Resheff S, Glaser F, Schuster G. Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts. RNA. 2008;14:297–309. doi: 10.1261/rna.698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HW, Koehler CM, Teitell MA. Human polynucleotide phosphorylase: location matters. Trends Cell Biol. 2007;17:600–608. doi: 10.1016/j.tcb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem. 2005;94:1078–1087. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- 32.Slomovic S, Schuster G. Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA. 2008;14:310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neil H, et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 34.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudfoot N, Gullerova M. Gene silencing CUTs both ways. Cell. 2007;131:649–651. doi: 10.1016/j.cell.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 38.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 39.San Paolo S, et al. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 2009;5:e1000555. doi: 10.1371/journal.pgen.1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.