Abstract
The Mycobacterium tuberculosis genome encodes 11 serine/threonine protein kinases (STPKs) that are structurally related to eukaryotic kinases. To gain insight into the role of Ser/Thr phosphorylation in this major global pathogen, we used a phosphoproteomic approach to carry out an extensive analysis of protein phosphorylation in M. tuberculosis. We identified more than 500 phosphorylation events in 301 proteins that are involved in a broad range of functions. Bioinformatic analysis of quantitative in vitro kinase assays on peptides containing a subset of these phosphorylation sites revealed a dominant motif shared by six of the M. tuberculosis STPKs. Kinase assays on a second set of peptides incorporating targeted substitutions surrounding the phosphoacceptor validated this motif and identified additional residues preferred by individual kinases. Our data provide insight into processes regulated by STPKs in M. tuberculosis and create a resource for understanding how specific phosphorylation events modulate protein activity. The results further provide the potential to predict likely cognate STPKs for newly identified phosphoproteins.
Keywords: signal transduction, phosphorylation motif, phosphoproteomics
A key feature of all living cells is the ability to sense environmental signals and implement adaptive changes. These inputs propagate through complex signal transduction networks whose activity is often regulated by reversible protein phosphorylation. Although Ser/Thr/Tyr protein phosphorylation-based signaling in eukaryotes has been intensively studied, the extent to which this mechanism is used in prokaryotes has only recently begun to be appreciated (1). The number of protein kinases in prokaryotes varies widely. Although many bacteria have only a few or none of these enzymes, some cyanobacteria and streptomycetes have dozens of them (2). Bacteria that do possess Ser/Thr or Tyr kinases often have complex lifestyles and depend on these kinases to regulate critical processes, such as stress adaptation, development, and virulence (2).
Mycobacterium tuberculosis is an extraordinarily versatile pathogen that can exist in distinct states in the host, leading to asymptomatic latent tuberculosis (TB) infection in which bacteria are thought to be dormant, or active TB disease in which the organisms are actively replicating. To achieve these different physiologic states M. tuberculosis requires mechanisms to sense a wide range of signals from the host and to coordinately regulate multiple cellular processes. In most bacterial pathogens, the predominant phosphorylation-based signal transduction mechanism is the two-component system. The M. tuberculosis genome, however, encodes 11 Ser/Thr protein kinases (STPKs) and an equal number of two-component system sensor kinases, suggesting that these two phospho-based signaling systems are of comparable importance in this organism (3).
Knowledge of the substrates of each of the M. tuberculosis STPKs is essential for understanding their function; however, only a small number of kinase-substrate cognate pairs have been characterized to date. Examples include the essential kinases PknA and PknB, which regulate cell shape and cell wall synthesis via phosphorylation of the cell pole-localized protein Wag31 and the septum-associated penicillin-binding protein PbpA (4–6). A kinase that has been implicated in TB pathogenesis, PknG, phosphorylates the forkhead-associated (FHA) domain-containing protein GarA, which has been shown to regulate enzymes of central carbon and nitrogen metabolism in a phosphorylation state-specific manner (7–9).
Our current limited view of protein phosphorylation in M. tuberculosis mirrors the relatively sparse phosphorylation data in prokaryotic organisms more generally. To obtain a more comprehensive understanding of in vivo phosphorylation events in M. tuberculosis, we used a mass spectrometry-based approach to identify phosphorylation sites in M. tuberculosis proteins. These results provide the most extensive data on Ser/Thr phosphorylation currently available for any bacterium, more than doubling the currently known bacterial phosphoproteome, and provide insight into the range of functions regulated by Ser/Thr phosphorylation in M. tuberculosis. Bioinformatic analysis of these in vivo phosphorylations, and of data from in vitro kinase assays, enabled us to identify and validate a phosphorylation site motif shared by several kinases, leading to a model of STPK–substrate interaction. In addition to providing insights into Ser/Thr phosphorylation in M. tuberculosis, these data will serve as an important resource for further investigation of these signal transduction pathways in M. tuberculosis, and in prokaryotes more broadly.
Results
Identification of 301 Phosphoproteins in M. tuberculosis.
We used a proteomic approach to identify phosphoproteins and their phosphorylation sites in M. tuberculosis proteins (Fig. 1). To increase the number of phosphoproteins identified, protein extracts were prepared from M. tuberculosis H37Rv cultures (i) supplied with different carbon sources, (ii) grown to different growth stages, and (iii) exposed to stresses including NO, peroxide, and hypoxia. More than 150 samples were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) and phosphopeptides were identified using the ProteinPilot and Mascot algorithms with high stringency cutoffs.
Fig. 1.
Flow diagram of approach to Ser/Thr protein phosphorylation detection and phosphorylation site motif identification and verification in M. tuberculosis.
We identified 301 phosphoproteins containing at least 516 phosphorylation sites demonstrating that at least 7% of M. tuberculosis proteins are phosphorylated (Table S1). Of these phosphoproteins, more than 40% contained more than one phosphorylation site, with some proteins having as many as seven sites (Tables S1 and S2). Among the 301 phosphoproteins identified in this study are several of the previously defined M. tuberculosis phosphoproteins, including four STPKs, GarA, Rv1422, and FhaA (4, 8, 10). The MS/MS search algorithms that we used can identify the presence of a phosphorylation site with high specificity but cannot always determine the precise site phosphorylation within the peptide backbone. We therefore used the Ascore algorithm (11) to attempt to identify the specific phosphoacceptor residue within each phosphopeptide. Using this approach we identified 215 phosphoacceptor residues with high confidence.
Phosphorylation in M. tuberculosis was biased toward Thr compared with Ser (60%:40%), a striking departure from findings in eukaryotes, where Ser phosphorylation may account for 80–90% of total phosphorylation sites (12). Among other bacteria, Bacillus subtilis, Escherichia coli, and Pseudomonas species all show greater phosphorylation of Ser than Thr, whereas data from Lactococcus lactis showed 51% and 46% Thr and Ser phosphorylation, respectively (13–16).
To determine whether specific sequences are preferentially targeted for Ser/Thr phosphorylation, we used the motif-x algorithm (17) to search for sequence motifs surrounding the phosphoacceptor for the 215 well-localized phosphorylation sites. Four statistically significant motifs were identified, all with Thr as the phosphoacceptor (Fig. S1). This result indicates that the M. tuberculosis STPKs, in addition to preferentially phosphorylating Thr vs. Ser, target the phosphoacceptor in the context of specific sequences.
Ser/Thr Phosphorylation Regulates a Wide Range of Functions in M. tuberculosis.
The phosphoproteins identified in these experiments belong to all functional classes of proteins (18) (Fig. S2), with the largest numbers involved in cell wall/cell processes and intermediary metabolism/respiration. The proportion of phosphoproteins in each category is not significantly different from the distribution of all proteins in the M. tuberculosis H37Rv genome annotation (18). Closer inspection of these data, however, provides interesting insights regarding the regulation of cell physiology by STPKs in M. tuberculosis. Several chaperones, for example, are phosphorylated, suggesting extensive regulation of protein turnover by Ser/Thr phosphorylation (Table S1). Multiple phosphorylation events on transporter proteins and lipid metabolic enzymes suggest modulation of the interface between the cell and the extracellular environment by the STPKs. Many cell division proteins are phosphoproteins, indicating a key role for phosphorylation in regulating this process.
Phosphorylation of the STPKs themselves is of interest for understanding their regulation. We found that four STPKs (PknA, PknB, PknD, and PknG) were phosphorylated in vivo. In addition to activation loop phosphorylation, sites in the intracellular juxtamembrane region, which has been hypothesized to have a regulatory function, were identified for PknA, PknB, and PknD (19, 20). Some of these sites were previously shown to be phosphorylated in vitro (19–21). In contrast to the other protein kinases, we found that PknG is phosphorylated near its amino terminus, also consistent with previous in vitro results (8).
In Vitro Phosphorylation of Peptides Containing in Vivo Phosphorylation Sites.
We designed 336 13-mer biotinylated peptides corresponding to in vivo phosphorylation sites in M. tuberculosis proteins. The peptides were incubated with the recombinant kinase domain of nine STPKs (we were unable to produce active PknI and PknJ) in presence of [γ-33P] ATP. After binding peptides to streptavidin-coated plates, excess radiolabeled ATP was washed away and incorporated 33P signal was measured using a scintillation counter. Half of the peptides were phosphorylated 2-fold or more above background by at least one kinase in these experiments. Most peptides that were phosphorylated in vitro were phosphorylated by more than one kinase, and some were actively targeted by the majority of kinases (Table S3).
In contrast to the peptides phosphorylated by multiple kinases, we found 48 peptides that were phosphorylated by a single kinase. The peptides that are uniquely phosphorylated by a single kinase suggest several interesting STPK–substrate pairs. For example, PknD uniquely and strongly phosphorylates a peptide from the amino terminus of SecF, whereas PknH uniquely and strongly phosphorylates a peptide derived from the chaperone DnaJ1. These data suggest that PknD may regulate protein secretion and PknH may regulate protein turnover. These data thus predict candidate in vivo targets of individual STPKs and provide a basis for experimental investigation of their role in regulating cell physiology.
Identification of a Shared Phosphorylation Site Motif.
To identify sequence motifs among the peptides phosphorylated by the M. tuberculosis STPKs, we analyzed separately the peptides that were and were not phosphorylated in vitro, using motif-x (Figs. S3 and S4). We found motifs that were highly similar to those that we obtained in the analysis of well-localized in vivo phosphorylation sites (Fig. S1). Notably, the set of peptides that were phosphorylated in vitro had markedly different motifs from the peptides that were not (Figs. S3 and S4).
We next performed analyses to attempt to identify preferred phosphorylation site sequences for each kinase. We used two approaches: a threading algorithm that compared the frequency of residues at each position in sequences of highly phosphorylated peptides with the entire set of synthesized peptide sequences (SI Materials and Methods), and the motif-x algorithm (17) using the entire M. tuberculosis proteome as the background. We were able to identify significant motifs for the six most active kinases—PknA, PknB, PknD, PknE, PknF, and PknH—using motif-x and for five of these six (excluding PknH) with the threading algorithm (Fig. 2 and Figs. S5 and S6). For all of these kinases, major features of the preferred phosphorylation site motif include Thr as the phosphoacceptor and highly significant selection for hydrophobic residues at the +3 and +5 positions (3 and 5 residues carboxyl-terminal to the phosphoacceptor). At the +3 position, several large hydrophobic residues were highly selected, whereas at the +5 position Ile was the predominant residue (Fig. 2 and Fig. S6).
Fig. 2.
Phosphorylation site motif analysis. pLOGos generated using motif-x based on in vitro assays in which peptide phosphorylation was >3-fold above median are shown for PknA, PknB, PknD, PknE, PknF, and PknH. The pLOGos show the relative statistical significance (with respect to the M. tuberculosis proteomic background) of residues within 6 aa of the central Thr phosphorylation site. Residues above the midline are overrepresented, whereas those below the midline are underrepresented. The red horizontal line indicates the 0.01 significance level (after Bonferroni correction).
In addition to these major features at +3 and +5, we identified additional significantly overrepresented residues at other positions. Most prominent are the preferences of several kinases for acidic resides at positions N-terminal to the phosphoacceptor (−1 to −4) and for Pro or Arg at the +4 position (Fig. 2 and Fig. S6). The only motif identified with Ser as a phosphoacceptor had borderline significant preference by PknD for acidic residues at the −5 position (Fig. S5).
To verify that the major features of the phosphorylation motif identified in the peptide kinase assays are important for substrate recognition in the context of a full-length protein, we performed kinase assays with PknB, using WT and substituted forms of the M. tuberculosis protein GarA as the substrate. GarA has been shown to be phosphorylated on adjacent residues by PknB (Thr22) and PknG (Thr21) (8). The sequence surrounding the PknB-phosphorylated residue (VTVETTSVFRA, Thr22 in bold) contains the major features of the PknB phosphorylation motif, including the large hydrophobic residue (Phe) at +3 (Fig. 2) and an acidic residue (Glu) at −2. Substitution of Phe at +3 with Ala (F25A) resulted in markedly decreased phosphorylation of GarA by PknB, comparable to removal of the phosphoacceptor (T22A) (Fig. S7). Substitution of Glu at −2 (E20V) also severely decreased phosphorylation. In contrast, substitution of Thr21 (T21A) did not have a significant effect on PknB phosphorylation, confirming Thr22 as the PknB phosphoacceptor in these assays.
Validation of the Dominant Motif and Identification of Specificity Determinants.
In addition to the phosphorylation motif shared by the six most active kinases, our analysis also suggested differences in the optimal substrate sequences for each kinase (Fig. 2 and Fig. S6). These include the specific hydrophobic residues preferred by each kinase at the +3 position, the position of acidic residues N-terminal to the phospho-Thr, and preference for Pro vs. Arg at +4. In addition, there was apparent selectivity by some kinases for residues at other positions, which did not reach statistical significance. To validate the motif and test these predictions, we chose a peptide (ITVAELTGEIPII) that was highly phosphorylated by the six most active kinases, and changed selected residues in a manner predicted to increase or decrease phosphorylation by some or all of the kinases. A set of 32 peptides was synthesized and incubated with the six kinases (Fig. 3).
Fig. 3.
In vitro phosphorylation of ideal (WT) and substituted peptides by the six kinases that share a substrate phosphorylation site motif. A peptide from Rv0497 that has features of the shared phosphorylation site motif was synthesized, together with 31 additional peptides incorporating substitutions predicted to enhance or diminish phosphorylation. The phosphoacceptor is shown in bold. Substituted residues are underlined. Phosphorylation of each peptide is expressed as a ratio relative to phosphorylation of the original peptide (WT). Substitutions that increase or decrease phosphorylation by 50% or more are shaded in green or red, respectively. Mean values of two independent experiments are shown.
Results of phosphorylation of these substituted peptides were mostly in agreement with predictions from the original in vitro phosphorylation data (Fig. 3). These experiments confirmed that large hydrophobic residues at +3 and +5 are dominant components of a common phosphorylation motif for PknA, PknB, PknD, PknE, PknF, and PknH. Acidic residues from −2 to −5 increase phosphorylation by most of these kinases, although generally they are not as important as the +3 and +5 positions. Surprisingly, Pro or Arg at +4 was required for optimal phosphorylation by all six kinases. It is particularly noteworthy that, as predicted from the kinase-specific motifs, Ser could not substitute for Thr as the phosphoacceptor for any kinase.
In addition to these shared components of the phosphorylation site motif, differences in kinase-specific preferences are evident in these data. For example, the importance of the acidic residues at the −2 to −5 positions, and whether Asp or Glu is optimal, varies among the different kinases. We also observed increased phosphorylation by PknB, PknE, PknF, and especially PknH when Val is at the +2 position. At the +4 position there was strong preference for Pro (PknA and PknB) vs. Arg (PknD, PknE, and PknH) among different STPKs. Also as predicted, Ile is preferred at position +5, although Leu can substitute without a significant decrease in phosphorylation by PknD, PknE, and PknH. Surprisingly, substitution of Gly for Ile at position +6 markedly impaired phosphorylation by PknA, PknB, and PknD. Combining the results of the initial in vitro phosphorylation with the refinement provided by the substituted peptides yields the shared motif XααααTX(X/V)ϕ(P/R)I (where α is an acidic residue and ϕ a large hydrophobic residue), with the potential for kinase-specific selectivity in the specific residues at positions that contribute to the motif.
Model of a PknB Peptide–Substrate Complex Suggests a Basis for the Phosphorylation Site Motif.
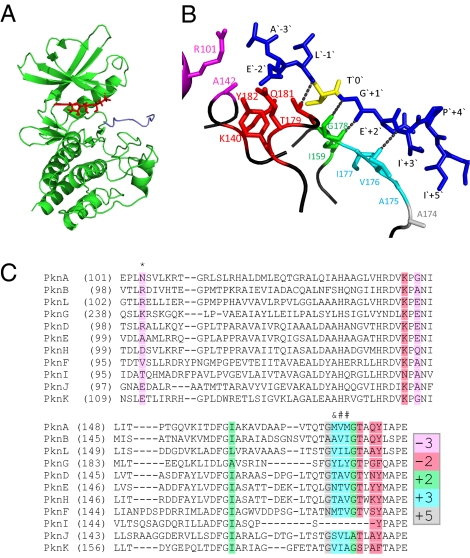
Several crystal structures of M. tuberculosis kinases have been reported; however, in none of these is the kinase cocrystallized with a substrate, nor do any of these structures include the activation loop. To search for structural features of M. tuberculosis STPK active sites that might explain substrate specificity, we generated a model PknB structure in complex with an optimal substrate peptide, using rabbit phosphorylase kinase (Phk) in complex with its substrate as a reference structure (Fig. 4A). The peptide substrate in this model contains residues from −3 to +5 around the phosphoacceptor Thr (AELTGEIPI).
Fig. 4.
Model of PknB structure in a complex with an ideal peptide substrate. (A) The PknB kinase domain in complex with an ideal peptide substrate (AELTGEIPI) was modeled using x-ray crystal structures of the M. tuberculosis PknB kinase domain [Protein Data Bank (PDB) no. 1o6y] and of a Phk-peptide substrate complex (PDB no. 2phk). ATP is in red, and the peptide in blue. (B) Likely contacts between PknB active site residues and the peptide, within 4 Å, are shown. Colors of PknB residues correspond to highlighted amino acids shown in C. The peptide is in blue except for the phosphoacceptor Thr, which is yellow. Hydrogen atoms and the main chain atoms were omitted, except for hydrogen bond contacts. (C) Alignment of the kinase domains of all M. tuberculosis STPKs, highlighting residues that are predicted to be in close contact with the peptide. The numbers in the color code indicate the position in the peptide substrate with which the residues shaded in that color may interact. In addition to the interactions indicated by the color code, kinase residues labeled with “*” are predicted to interact with the −2 position, “#” with the +2 position, and “&” with +2 and +5 positions.
Although this model does not identify exact contacts of PknB with the peptide, it does suggest probable residues that contribute to substrate binding. As shown in Fig. 4B, the central part of the activation loop is in close contact with substrate residues around the phosphoacceptor, potentially forming several hydrogen bonds between the peptide backbone of the substrate (from −1 to +3) and the kinase active site (hydroxyl group from Thr179 and peptide bond of Val176 and Gly178). Several residues in this portion of the activation loop (positions 174–179) are highly conserved among the M. tuberculosis STPKs, suggesting that it may be a common binding site for substrate residues at the +2, +3, and +5 positions (Fig. 4C).
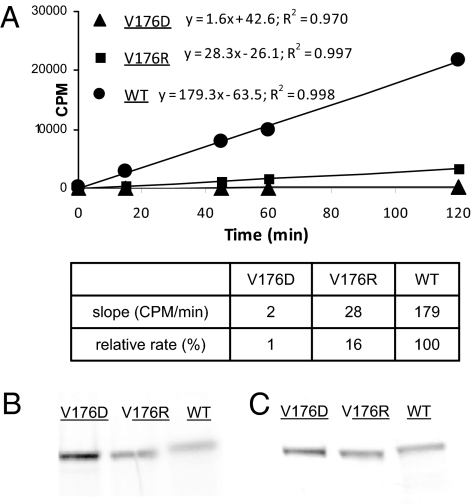
As an initial test of the model based on predicted interactions with the +3 hydrophobic residue in the optimal substrate motif, we investigated the effect of mutation of PknB at Val176 to either a negatively (V176D) or positively (V176R) charged residue (Fig. 5). These altered forms of PknB are catalytically functional on the basis of active autophosphorylation at levels comparable to WT PknB. Both substitutions resulted in markedly decreased phosphorylation of the “ideal peptide,” which contains Ile at the +3 position, compared with WT PknB, consistent with the prediction of the structural model.
Fig. 5.
Kinase activity of WT and substituted forms of PknB. (A) Val176 of PknB was mutated to Asp or Arg, and in vitro phosphorylation of an ideal peptide (peptide 1 in Fig. 3) by WT and substituted PknB proteins was performed and quantified by 33P incorporation at serial time points. (B) Autoradiography of WT and substituted PknB after incubation with γ-P33-ATP followed by SDS/PAGE. (C) GelCode Blue (Pierce) stained gel showing approximately equal amounts of protein loaded on the gel.
The model indicates that Pro at the +4 position in the PknB substrate motif serves to position the +3 and +5 residues for binding. Arg, a preferred residue at this position for some kinases, would serve a similar purpose, but might also have contacts with the active site in kinases that prefer this residue. This model also locates the basic residues Arg101 and Lys140 of PknB close to the acidic residues present at the −2 to −4 positions of the substrate (Fig. 4 B and C). Lys140 is conserved in all M. tuberculosis STPKs except PknI, whereas Arg101 is less conserved (Fig. 4C).
Discussion
The presence of 11 STPK genes in the M. tuberculosis genome suggests that Ser/Thr phosphorylation is an important mechanism for signal transduction in this organism but that the total phosphoproteome is likely to be substantially smaller than that of most eukaryotes. By searching for phosphoproteins in M. tuberculosis grown under several conditions, we identified 516 phosphorylation events, occurring in 301 M. tuberculosis proteins, several-fold more than any prior study in prokaryotes. On the basis of our identification of some but not all previously described phosphoproteins, however, there are likely many phosphoproteins that have not been identified.
Comparing our results to studies of Ser/Thr phosphorylation in other bacteria shows interesting similarities. Of 41 phosphoproteins identified in the soil actinomycete Corynebacterium glutamicum (22), for example, almost one third are also phosphorylated in M. tuberculosis. This finding suggests that the M. tuberculosis phosphoproteome identified here may provide insight into proteins targeted by STPKs in other Actinomycetes, and possibly in other bacteria. Phosphorylation of ribosomal proteins has been found in Pseudomonas putida (16), L. lactis (13), and E. coli (15). We found that five ribosomal and a ribosome-associated protein are phosphorylated in M. tuberculosis, indicating regulation of translation by Ser/Thr phosphorylation in this pathogen. Together with phosphorylation of proteins involved in DNA and RNA turnover, and in essential metabolic pathways, these data suggest that Ser/Thr phosphorylation provides a mechanism for the coordinate regulation of M. tuberculosis physiology during infection.
We previously determined that PknA and PknB both preferentially phosphorylate peptides that have a large hydrophobic residue at the +3 position (4). Here, motif extraction from quantitative phosphorylation of more than 300 peptides, followed by validation using peptides with targeted substitutions, allowed us to define an extensive motif shared by these two kinases and by PknD, PknE, PknF, and PknH. The conservation of the dominant components of the motif among these six kinases, although initially surprising, is consistent with the similarity of the predicted substrate binding sites of these STPKs (Fig. 4). Beyond the shared components of the motif, this approach provided insight into potential differences in the sequences that each of these kinases optimally target. In contrast, the peptides phosphorylated by PknG and PknK do not match this common motif, consistent with their kinase domains being markedly dissimilar to those of the other STPKs (Fig. S8).
This common motif has at least two potential implications for substrate targeting by M. tuberculosis STPKs. One is that multiple kinases may target a single protein, which has been shown previously for at least two M. tuberculosis STPK substrates, GarA and Rv1422 (4, 23). The other implication is that other factors must contribute to in vivo substrate specificity of the individual STPKs. These factors are likely to include coordinated expression and colocalization of kinase and substrate, and protein–protein interactions both between the substrate and a cognate STPK, and in many cases with other members of larger protein complexes. In addition, less dominant kinase-specific phosphorylation site sequence preferences may be important determinants of specificity. Although the shared motif of the six STPKs limits direct mapping of individual substrates to a cognate kinase, these secondary motif characteristics should allow prediction in some cases of the STPK(s) most likely to phosphorylate newly identified M. tuberculosis phosphoproteins, and potentially to predict de novo protein substrates of individual STPKs.
The importance of the +3 position in the common motif, which is typically a critical position for FHA recognition (24), suggests that phospho-dependent interactions of STPK substrates with FHA domain proteins may also contribute to specific downstream signaling mediated by these kinases. In support of this idea, the GarA FHA domain was previously shown to prefer hydrophobic residues at the +3 position relative to phospho-Thr (25). As noted above, the sequence adjacent to the Thr22 residue in GarA that is phosphorylated by PknB, and that is also bound by its FHA domain, matches remarkably well the preferred phosphorylation site motif of PknB, including the hydrophobic residue (Phe) at +3.
In summary, we have identified extensive Ser/Thr phosphorylation of M. tuberculosis proteins, demonstrating that the STPKs of this organism regulate a broad range of cellular processes and indicating that this regulatory mechanism is likely to be important for the virulence of this major global pathogen. We defined common and specific components of a preferred substrate motif shared by six STPKs. These data provide insight into signal transduction in prokaryotes, and provide a resource for investigation of the regulation of M. tuberculosis physiology and pathogenesis by Ser/Thr phosphorylation.
Materials and Methods
M. tuberculosis Cultures, Protein/Peptide Preparation, and Analysis.
M. tuberculosis H37Rv was grown in broth culture under several conditions (NO stress, oxidative stress, hypoxia, and glucose or acetate as a carbon source) and harvested at different growth stages. Proteins were extracted using TRIzol, separated by SDS/PAGE, reduced, alkylated, and trypsin-digested, followed by phosphopeptide enrichment (see SI Materials and Methods). A total of 152 samples were run on a Thermo Finnigan LTQ instrument. Phosphopeptides were identified using the ProteinPilot 2.0 (Applied Biosystems) and Mascot (Matrix Science) software. Spectra were analyzed with the Ascore algorithm to attempt to unambiguously localize the phosphoacceptor in each tryptic phosphopeptide (11).
Recombinant Protein Kinase Expression and in Vitro Kinase Assays.
Kinase domains of all 11 STPK genes were cloned from M. tuberculosis genomic DNA and expressed in E. coli as GST-tagged proteins or as His-tagged proteins. Active PknG, PknI, and PknJ were not successfully obtained using this approach; therefore, full-length proteins were expressed in Mycobacterium smegmatis.
A total of 336 biotinylated 13-mer peptides containing phosphorylation sites identified from M. tuberculosis protein lysates were synthesized as crude product (BioTides; JPT Peptide Technologies). Kinase assays were performed in 50 μL containing the following components: recombinant kinase (10–200 ng/μL), ATP (10 μM), [γ-33P] ATP (EasyTide; PerkinElmer) (0.5 μCi), and peptide substrate (5 μM) in 50 mM Mops (pH 7.4), 10 mM MgCl2, and 10 mM MnCl2. The reaction was incubated at least 4 h at 30 °C, stopped, and transferred to streptavidin-coated FlashPlates (Perkin-Elmer). After overnight incubation, plates were washed and read in a TopCount instrument (Perkin-Elmer). For motif validation, 32 peptides were synthesized and purified to at least 70% purity, and kinase assays were performed as described above, except that incubation times were shorter (1.5–2 h).
Mutagenesis of PknB and GarA and Phosphorylation Analysis.
The WT GST-tagged PknB kinase domain was mutated using the QuikChange kit (Stratagene), expressed in E. coli, and purified using the B-PER GST kit (Pierce Biotechnology). GarA was cloned into the pENTR vector (Invitrogen), mutated using QuikChange (Stratagene), and transferred to pDEST17 (Invitrogen) to express His-tagged proteins in E. coli. HisPur (Pierce Biotechnology) was used for purification of WT and mutant GarA proteins.
PknB mutants were analyzed for ideal peptide phosphorylation (peptide 1 in Fig. 3) using FlashPlates as described above. Kinase assays in the presence of [γ-33P] ATP were performed to test autophosphorylation of PknB mutants and phosphorylation of GarA. Reactions were run on SDS/PAGE, and radioactive signal was detected and quantified using a Storm phosphorimager (GE Healthcare).
Phosphorylation Site Motif Analysis.
Two complementary approaches were used to identify preferred phosphorylation site motifs, an internal working version of the motif-x algorithm (17), and a threading algorithm. Motif-x was used to analyze in vivo phosphorylated sequences, and sequences of peptides that were phosphorylated in vitro at least 3-fold over the median for all peptides in two replicates (SI Materials and Methods). Probability log-based logos (pLOGos) were generated for all phosphorylated and nonphosphrylated peptides, and for the peptides phosphorylated by each kinase. All analyses were performed using the M. tuberculosis proteome as background. Residues that had values over the 0.01 significance level (after Bonferroni correction) were deemed statistically significant and used by the motif-x algorithm to fix motif positions in Fig. 2 and Figs. S1, S3, and S4.
For the threading algorithm, peptides that were phosphorylated in vitro by a kinase 5-fold over the median value for all peptides were analyzed, using the whole peptide library for background correction (SI Materials and Methods). Results of this analysis were used as input into the weblogo application (weblogo.berkeley.edu) for computing motifs using sequence entropies (26, 27).
Model of PknB Structure in Complex with an Ideal Peptide Substrate.
M. tuberculosis PknB kinase domain (1o6y) (28) and phosphorylase kinase (Phk)-peptide substrate complex (2phk) (29) crystal structures were aligned using PyMOL (DeLano Scientific) to add missing PknB–peptide contact residues in the activation loop. Activation loop residues from Phk and its substrate were changed to the corresponding PknB and peptide substrate residues so that this model should represent the conformation of the active form of PknB. The Maestro molecular modeling package was used to minimize protein and substrate side chain conformations, and the protein/substrate H-bond constraints with PknB atoms were frozen. Possible contacts between PknB active site residues and the substrate peptide were predicted using a 4 Å radius around peptide residues.
Supplemental Data.
Detailed methods descriptions, supplemental figures, mass spectra of phosphopeptides, lists of peptides, and in vitro phosphorylation data are available as supplemental files.
Supplementary Material
Acknowledgments
We thank Yin Yin Lin and Nurhan Ozlu for help with mass spectrometry; Wiebke Timm and Flavio Monigatti for help with data analysis and management; Mauricio Anaya for initial production of recombinant kinases; and Mark Fleming and Christopher Locher for helpful discussions. This work was funded by research grants from Vertex Pharmaceuticals Incorporated, the National Institutes of Health, and the Potts Memorial Foundation (to R.N.H.), and from the US Department of Energy Genomic Sciences Program and the Bill and Melinda Gates Foundation (to G.M.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The entire dataset of the chromatography tandem mass spectrometry results is in an Excel file that is part of the supplemental material. Raw spectral data files are available at http://www.researchcomputing.org/Husson/Mtb_Phosphoproteome_spectra.zip
This article contains supporting information online at www.pnas.org/cgi/content/full/0913482107/DCSupplemental.
References
- 1.Deutscher J, Saier MH., Jr Ser/Thr/Tyr protein phosphorylation in bacteria—for long time neglected, now well established. J Mol Microbiol Biotechnol. 2005;9:125–131. doi: 10.1159/000089641. [DOI] [PubMed] [Google Scholar]
- 2.Pérez J, Castañeda-García A, Jenke-Kodama H, Müller R, Muñoz-Dorado J. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc Natl Acad Sci USA. 2008;105:15950–15955. doi: 10.1073/pnas.0806851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehenkel A, et al. Mycobacterial Ser/Thr protein kinases and phosphatases: Physiological roles and therapeutic potential. Biochim Biophys Acta. 2008;1784:193–202. doi: 10.1016/j.bbapap.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Kang CM, et al. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: Substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154:725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 6.Dasgupta A, Datta P, Kundu M, Basu J. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology. 2006;152:493–504. doi: 10.1099/mic.0.28630-0. [DOI] [PubMed] [Google Scholar]
- 7.Cowley S, et al. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol. 2004;52:1691–1702. doi: 10.1111/j.1365-2958.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- 8.O'Hare HM, et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70:1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 9.Nott TJ, et al. An intramolecular switch regulates phosphoindependent FHA domain interactions in Mycobacterium tuberculosis. Sci Signal. 2009;2:ra12. doi: 10.1126/scisignal.2000212. [DOI] [PubMed] [Google Scholar]
- 10.Grundner C, Gay LM, Alber T. Mycobacterium tuberculosis serine/threonine kinases PknB, PknD, PknE, and PknF phosphorylate multiple FHA domains. Protein Sci. 2005;14:1918–1921. doi: 10.1110/ps.051413405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 12.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 13.Soufi B, et al. The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics. 2008;8:3486–3493. doi: 10.1002/pmic.200800069. [DOI] [PubMed] [Google Scholar]
- 14.Macek B, et al. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics. 2007;6:697–707. doi: 10.1074/mcp.M600464-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Macek B, et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Ravichandran A, Sugiyama N, Tomita M, Swarup S, Ishihama Y. Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics. 2009;9:2764–2775. doi: 10.1002/pmic.200800655. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 18.Camus JC, Pryor MJ, Médigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 19.Boitel B, et al. PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol Microbiol. 2003;49:1493–1508. doi: 10.1046/j.1365-2958.2003.03657.x. [DOI] [PubMed] [Google Scholar]
- 20.Durán R, et al. Conserved autophosphorylation pattern in activation loops and juxtamembrane regions of Mycobacterium tuberculosis Ser/Thr protein kinases. Biochem Biophys Res Commun. 2005;333:858–867. doi: 10.1016/j.bbrc.2005.05.173. [DOI] [PubMed] [Google Scholar]
- 21.Molle V, et al. Characterization of the phosphorylation sites of Mycobacterium tuberculosis serine/threonine protein kinases, PknA, PknD, PknE, and PknH by mass spectrometry. Proteomics. 2006;6:3754–3766. doi: 10.1002/pmic.200500900. [DOI] [PubMed] [Google Scholar]
- 22.Bendt AK, et al. Towards a phosphoproteome map of Corynebacterium glutamicum. Proteomics. 2003;3:1637–1646. doi: 10.1002/pmic.200300494. [DOI] [PubMed] [Google Scholar]
- 23.Villarino A, et al. Proteomic identification of M. tuberculosis protein kinase substrates: PknB recruits GarA, a FHA domain-containing protein, through activation loop-mediated interactions. J Mol Biol. 2005;350:953–963. doi: 10.1016/j.jmb.2005.05.049. [DOI] [PubMed] [Google Scholar]
- 24.Liang X, Van Doren SR. Mechanistic insights into phosphoprotein-binding FHA domains. Acc Chem Res. 2008;41:991–999. doi: 10.1021/ar700148u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durocher D, et al. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 26.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz-Lombardía M, Pompeo F, Boitel B, Alzari PM. Crystal structure of the catalytic domain of the PknB serine/threonine kinase from Mycobacterium tuberculosis. J Biol Chem. 2003;278:13094–13100. doi: 10.1074/jbc.M300660200. [DOI] [PubMed] [Google Scholar]
- 29.Lowe ED, et al. The crystal structure of a phosphorylase kinase peptide substrate complex: Kinase substrate recognition. EMBO J. 1997;16:6646–6658. doi: 10.1093/emboj/16.22.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.