Abstract
We deeply sampled the organismal, genetic, and transcriptional diversity in fecal samples collected from a monozygotic (MZ) twin pair and compared the results to 1,095 communities from the gut and other body habitats of related and unrelated individuals. Using a new scheme for noise reduction in pyrosequencing data, we estimated the total diversity of species-level bacterial phylotypes in the 1.2-1.5 million bacterial 16S rRNA reads obtained from each deeply sampled cotwin to be ~800 (35.9%, 49.1% detected in both). A combined 1.1 million read 16S rRNA dataset representing 281 shallowly sequenced fecal samples from 54 twin pairs and their mothers contained an estimated 4,018 species-level phylotypes, with each sample having a unique species assemblage (53.4 ± 0.6% and 50.3 ± 0.5% overlap with the deeply sampled cotwins). Of the 134 phylotypes with a relative abundance of >0.1% in the combined dataset, only 37 appeared in >50% of the samples, with one phylotype in the Lachnospiraceae family present in 99%. Nongut communities had significantly reduced overlap with the deeply sequenced twins’ fecal microbiota (18.3 ± 0.3%, 15.3 ± 0.3%). The MZ cotwins’ fecal DNA was deeply sequenced (3.8-6.3 Gbp/sample) and assembled reads were assigned to 25 genus-level phylogenetic bins. Only 17% of the genes in these bins were shared between the cotwins. Bins exhibited differences in their degree of sequence variation, gene content including the repertoire of carbohydrate active enzymes present within and between twins (e.g., predicted cellulases, dockerins), and transcriptional activities. These results provide an expanded perspective about features that make each of us unique life forms and directions for future characterization of our gut ecosystems.
Keywords: microbial phylogenetic analyses, microbiota, transcriptomics, carbohydrate active enzymes
Human microbiome projects are being initiated throughout the world, with the goal of correlating human physiological phenotypes with the structures and functions of their indigenous microbial communities. Substantial insight into the patterns of variation in the microbiota between body habitats and individuals has been gained using shallow sequencing of 16S rRNA gene amplicons and community DNA. Because of limitations imposed by sequencing costs and throughput, these studies have examined the more abundant species or genes. A timely question is this: What additional insights about the microbial diversity present within a body habit are obtained with deeper sequencing? Moreover, how much of the observed organismal diversity is an artifact of noise introduced during PCR and sequencing of 16S rRNA genes (1–3)? Therefore, in the current study we use a variety of experimental and computational approaches to explore the level of diversity and interpersonal variation in bacterial phylotypes, microbial genes, and their expressed mRNA transcripts within the human gut, home to our largest community of microorganisms.
Results and Discussion
Study Design and Data Collection.
Total community DNA and RNA was initially isolated from two fecal samples, each obtained from 26-year-old, obese, MZ female cotwins (body mass index, 39 and 45 kg/m2). Both cotwins (designated TS28 and TS29) had been vaginally delivered; neither cotwin had any history of intestinal disease, and neither had used antibiotics at least 6 months before providing fecal samples, at which time the cotwins lived 5 km apart (4). A 454 pyrosequencing method was used to obtain 1.2–1.5 million sequencing reads from PCR-amplified V2 regions of bacterial 16S rRNA genes present in each fecal sample (average read length ∼232 nt), and 3.8–6.3 Gbp of single- and paired-end shotgun reads from total fecal community DNA (Table S1). Using a method for rRNA depletion based on a combination of size selection (to remove 5S rRNA and tRNA), and streptavidin bead-based pull-down of biotinylated oligonucleotides hybridized to domains conserved among gut bacterial rRNA genes (5), we enriched for fecal mRNA and then generated 12–16 million sequencing reads representing expressed genes in their microbiomes (Table S2).
Analysis of Bacterial Diversity Present in the Gut Microbiota.
Algorithms for denoising pyrosequencing data: tests using mixtures of bacterial strains.
We analyzed test datasets composed of an unequal mixture of DNA from 90 cloned bacterial 16S rRNA gene sequences (2) or DNA purified from 67 bacterial strains cultured from the human gut and pooled together over a range of relative concentrations (Table S3). These test datasets were used to establish a set of procedures for removing noise from 16S rRNA datasets that arise from PCR and pyrosequencing (SI Text).
Comparison of the fecal microbiota of the deeply sampled MZ co-twins.
Using these procedures, we determined that most species-level phylotypes were present at low abundance [species defined as organisms sharing ≥97% sequence identity (%ID) in their 16S rRNA genes; Fig. S1]; ∼100,000 16S rRNA sequences were required to observe 60% of the total phylotypes (Fig. 1A). At the 95%ID and 97%ID phylotype cutoffs, rarefaction curves did not completely saturate even when >106 sequences were collected (Fig. 1A), indicating that additional phylotypes remain uncharacterized even at this high level of coverage.
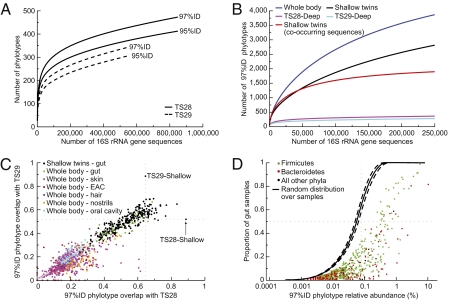
Fig. 1.
Measurements of bacterial diversity in the human fecal microbiota. (A) Rarefaction curves at 97%ID and 95%ID phylotype cutoffs are shown for the deeply sequenced TS28 and TS29 MZ cotwin (“Deep Twins”) datasets. Sequences were classified as chimeric at the 50% probability cutoff. (B) Comparison of diversity within and between gut microbial communities. Curves at 97%ID phylotype cutoff are shown for 250 fecal samples taken from 146 individuals (“Shallow twins”; 1,000 16S rRNA gene sequences were randomly selected from each sample), 250 samples taken from multiple body habitats (“Whole body”; 1,000 randomly selected sequences per sample), and the two deeply sequenced fecal samples (“TS28-Deep” and “TS29-Deep”). Phylotypes found in multiple fecal samples are labeled “co-occurring.” (C) Plot of proportion of 97%ID phylotypes found in TS28 and TS29 across 277 fecal samples (black circles) and 814 samples taken from multiple body habitats in nine individuals [habitat groups are colored green (fecal), purple (skin), red (external auditory canal; EAC), blue (hair), orange (nostrils), and light blue (oral cavity)]. Four EAC and one skin sample did not contain any shared phylotypes with TS28 and TS29. (D) The proportion of the 250 fecal samples containing each 97%ID phylotype plotted as a function of the relative abundance (%) of each phylotype in the combined dataset. Phylotypes are colored according to phylum: Bacteroidetes (red), Firmicutes (green), and other (black). The expected proportion of samples containing each phylotype, assuming a random distribution across samples, is shown (median ± 95% confidence interval).
The total estimated diversity of species-level bacterial phylotypes (97%ID) in the TS28 and TS29 datasets was lower than expected (878 and 768, respectively; Table 1 and Table S4), based on previous studies that did not account for noise. There was notable variation even between these genetically identical cotwins: 35.9% and 49.1% of the species-level phylotypes found in the fecal communities of TS28 and TS29, respectively, were shared between the two samples (39.0% and 52.8% were shared at the 95%ID level).
Table 1.
Number of species-level (97%ID) and 95%ID bacterial phylotypes in the deep and shallow sequenced fecal microbiota of twins, and in the whole body sampling datasets
| Dataset | 16S rRNA seqs | Observed phylotypes (97%ID) | Estimated phylotypes (97%ID Chao)a | Observed phylotypes (95%ID) | Estimated phylotypes (95%ID Chao) |
| TS28-Deep | 848,512 | 473 | 627 | 413 | 538 |
| TS29-Deep | 553,416 | 344 | 558 | 307 | 514 |
| TS28-Shallow | 3,288 | 135 | 375 | 121 | 329 |
| TS29-Shallow | 1,178 | 81 | 127 | 70 | 130 |
| TSAll-Shallow | 250,000 | 2,815 | 4,018 | 1,974 | 2,498 |
| TSAll-Co-occur | 250,000 | 1,898 | 2,043 | 1,221 | 1,283 |
| WholeBody | 250,000 | 3,869 | 4,949 | 2,957 | 3,646 |
aChao’s nonparametric total diversity estimates are given. Phylotypes are grouped based on the degree of sequence identity in the V2 regions of their 16S rRNA genes.
TS28- and TS29-Deep, deeply sequenced cotwin fecal samples; TS28- and TS29-Shallow, shallow sequenced cotwin fecal samples; TSAll-Shallow, 1,000 randomly selected sequences from 250 fecal samples; TSAll-Co-occur, restricted to co-occurring sequences from 250 fecal samples; WholeBody, 250 randomly selected samples from a total of 814 samples obtained from 27 body sites from 9 individuals, 1,000 sequences/sample).
However, these values do not account for phylotypes that may be abundant in one sample and rare in another. Overall, shared phylotypes showed a small but positive correlation in relative abundance between samples, and rarely varied by more than two orders of magnitude (R2 = 0.18 for 97%ID and R2 = 0.27 for 95%ID). This observation allowed us to define a normalized overlap between the samples by considering only phylotypes found at a sufficient relative abundance in each sample that they are unlikely to have been missed because of variations in their relative abundance (“Normalized overlap” in SI Text). With this normalization, 68% and 79% of 97%ID phylotypes in TS28 and TS29 were designated as being shared in the other cotwin's microbiota (76.7% and 86.0% at 95%ID).
Comparisons to more shallowly sampled fecal samples obtained from other twin pairs.
To test whether the deep sampling of these cotwins allowed us to capture the bacterial diversity present in fecal samples obtained from other families containing twins, we extended our survey to include 1.1 million bacterial V2 16S rRNA sequencing reads from 281 fecal samples procured from 31 MZ and 23 dizygotic (DZ) twin pairs and their mothers [3,984 ± 232 (mean ± SEM) reads/sample] (4). Like the deeply sampled cotwins, these other twin pairs were born in Missouri, ranged in age from 25 to 32 years, did not have a history of GI pathology, and had not consumed antibiotics before sampling. All 16S rRNA pyrosequencing reads were preprocessed as done above to remove noise and chimeras.
A comparison of the total bacterial diversity found across these fecal samples and the two deeply sequenced samples underscored the much higher level of inter- compared with intrapersonal variation when considering a single body habitat (Fig. 1B). The combined “Shallow” fecal datasets had an estimated 4,018 97%ID phylotypes and 2,498 95%ID phylotypes (2,815 and 1,974 observed, respectively). These values are ∼5-fold higher than in each deeply sequenced fecal microbiota (Table 1). In addition, each sample had a unique collection of 97%ID phylotypes (mean ± SEM, 53.4 ± 0.6% and 50.3 ± 0.5% overlap with TS28 and TS29), whereas the fraction of phylotypes from each sample that were shared with TS28 correlated with the fraction shared with TS29 (R2 = 0.73; Fig. 1C).
Our initial analysis of these fecal samples had indicated that there was no core set of abundant species-level phylotypes found in all individuals (4); this was confirmed after removing PCR and sequencing noise. The proportion of samples containing each phylotype was lower than expected by chance at all levels of relative abundance (Fig. 1D), but within each level of abundance there was a large spread. Only a few phylotypes appeared in the majority of samples: of the 134 species-level phylotypes that had a relative abundance in the combined dataset >0.1%, only 37 appeared in >50% of the samples (28% of the phylotypes, compared with 100% expected by chance). Phylotypes assigned to the Firmicutes phylum were more evenly spread than the Bacteroidetes: 33% with >0.1% relative abundance appeared in 50% of samples, compared with only 12% of the Bacteroidetes phylotypes (Fig. 1D). In addition, one nearly ubiquitous phylotype belonging to the family Lachnospiraceae (phylum Firmicutes) was found in 99% of the samples, representing 5.7% of the sequences in the combined dataset.
Comparisons to bacterial phylotypes present in other human body habitats.
To determine whether phylotypes present in the gut microbiota were detectable in other body habitats, we surveyed V2 16S rRNA sequencing reads obtained from nine unrelated healthy individuals (male and female) who had been sampled at 27 sites, including feces, twice over a 24-h period on two occasions, each occasion separated by 3 months (age range, 30–35 years with the exception of one individual 60 years of age; no recent history of antibiotic use; mean ± SD 1,315 ± 420 reads per sample) (6). All data were subjected to the same denoising procedures described above.
A comparison of the total diversity found across the 27 body habitats to the shallowly sequenced fecal samples and the two deeply sequenced fecal samples demonstrated higher levels of diversity when comparing across multiple body habitats vs. comparisons of the same habitat across multiple individuals (Fig. 1B). The combined 27-body habitats dataset contained an estimated 4,949 species-level phylotypes (97%ID) and 3,646 95%ID phylotypes (3,869 and 2,957 observed, respectively) (Table 1). Although the range of overlapping species-level phylotypes for the fecal samples from the 27-body habitat survey was comparable to the twin fecal cohort (mean± SEM 45.1 ± 1.9% and 41.7 ± 1.4%), the other nongut body habitats showed a significantly reduced overlap (mean ± SEM 18.3 ± 0.3% and 15.3 ± 0.3% with TS28 and TS29; P < 10−17, Student's t test; Fig. 1C). As with the fecal samples from the shallowly sampled twins, the fraction of phylotypes from each sample that were shared with TS28 correlates with the fraction shared with TS29 (R2 = 0.42).
Conclusions.
Together, these results emphasize the following: (i) despite large interpersonal variations in the composition of the gut microbiota and the absence of a core set of abundantly represented universally shared phylotypes, common phylotypes can be identified through deep sequencing of a small number of individuals; (ii) a surprising amount of phylotypes are shared between distinct body habitats across unrelated individuals (i.e., only five samples did not contain any phylotypes from the deeply sequenced TS28 and TS29 gut microbial communities); and (iii) it seems feasible that future studies that broadly sample humans living in distinct cultural settings will be able to define population-wide gut phylotypes and, as a result, provide a rationale for selecting cultured representatives of these phylotypes for genome sequencing (e.g., start with phylotypes in the top right portion of Fig. 1D).
Deep Shotgun Sequencing of the Fecal Microbiome of the MZ Cotwins: Analyses of Genus-level Phylogenetic Bins.
We turned next to the following questions: Does deep sequencing enable the assembly and binning of “population genomes” from complex microbial communities? How diverse is the gut microbiome in terms of gene content. and how unique are these genes relative to those contained in 122 genomes from cultured human gut isolates? What can we infer about the similarities and differences between MZ cotwins when interrogating their deeply sequenced microbiomes?
Deep shotgun sequencing of total fecal community DNA allowed us to assemble and bin large scaffolds from the TS28 and TS29 microbiomes (Tables S5 and S6 and Phylogenetic binning of microbiome scaffolds in SI Text). A combined assembly of single- and paired-end pyrosequencing reads from TS28 and TS29 yielded 92,104 and 61,460 contigs >500 bp per sample, with 11,780 and 6,392 scaffolds, respectively (scaffolds represent one or more contigs ordered and oriented using paired-end reads). PhyloPythia, a phylogenetic classifier that uses a multiclass Support Vector Machine (SVM) for composition-based characterization of sequence fragments at different taxonomic ranks (7), was trained on 1,775 finished or draft microbial genomes, in addition to 5,548 and 3,391 contigs from TS28 and TS29, respectively, that mapped with high confidence to gut microbial genomes (Table S7). After training, PhyloPythia was used to accurately bin all scaffolds >2 Kbp at the genus- and family-level, resulting in 24–25 bins of scaffolds per fecal sample; these bins contained from 2.0 Kbp to 22.4 Mbp of total sequence (Figs. S2B and S3 and Table S6).
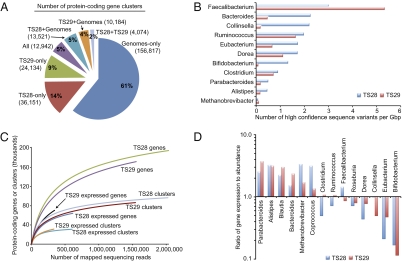
The total number of genes across all microbiome bins from the TS28 and TS29 fecal samples was 88,316 and 64,453, respectively. Clustering of protein sequences from these bins and the 122 gut microbial genomes, revealed 180,550, 257,823, and 334,211 total protein-coding gene clusters at 40%, 60%, and 80% identity cutoffs, respectively (Fig. 2A and Fig. S2A). The largest group of gene clusters at all cutoffs was unique to the reference genomes, whereas 25% of the clusters were found only in the TS28 or TS29 microbiome bins. Overall, 36% of the gut microbiome gene clusters had a representative (60%ID) in the 122 gut microbial genome database, indicating that although sequencing reference genomes from culturable members of the microbiota has already uncovered a substantial proportion of the gene content present in the fecal communities of these cotwins, more reference genome and microbiome sequencing is clearly needed.
Fig. 2.
Diversity of the human fecal microbiome and its metatranscriptome. (A) Distribution of gene clusters across gut microbial genomes and microbiome bins. All protein sequences from 122 gut genomes and the microbiome bins were clustered using cd-hit at 60%ID. (B) Number of sequence variants in each microbiome bin (values normalized by Gbp in bin; all genus-level bins with >100 scaffolds are shown). (C) Rarefaction analysis of the number of genes, gene clusters, expressed genes, and expressed gene clusters in the fecal microbial communities of TS28 and TS29 as a function of sequencing depth. The total number of protein-coding genes in the set of 122 gut genomes and the microbiome bins is 525,329, representing 257,823 gene clusters. (D) Ratio of gene expression to gene abundance (relative abundance of cDNA sequences divided by relative abundance of DNA sequences) mapped to a subset of the bacterial taxa in the fecal microbiome. Taxa with >1,000 mapped cDNA and DNA sequencing reads in both samples are shown.
A total of 25 genus- and family-level bins were identified in the TS28 fecal microbiome dataset, and 24 in the TS29 dataset; 22 of these bins were found in both samples (bins unique to one sample only contained nine of the 16,554 total scaffolds). There were strong correlations between the two fecal microbiomes with respect to the number of scaffolds, their aggregate length, and the number of genes found in each bin (R2 = 0.94, 0.74, and 0.69, respectively; Table S6). As expected from our bacterial 16S rRNA analyses, the genus-level bins with the largest number of scaffolds were the Ruminococcus, Bacteroides, Clostridium, and Eubacterium (members of the Bacteroidetes and Firmicutes phyla). However, substantial assemblies were also obtained from Methanobrevibacter [M. smithii is reported to be the dominant archaeon in the human gut microbiome; (8)] and from Bifidobacterium (the former is missed with primers for amplification of bacterial 16S rRNA genes, whereas the current version of V2-directed bacterial primers miss members of the latter taxa; Fig. S4). When sequencing reads from each sample were mapped to the microbiome bins from that sample to identify high-confidence sequence variants in each bin, we found that the Faecalibacterium had the highest relative level of variation, whereas the Methanobrevibacter had the lowest (Fig. 2B).
Taken together, these results suggest that “population genomes” can be constructed and reliably binned even from diverse microbial communities given enough sequencing depth, although rare members of the community will be missed (e.g., the TM7 phylum). The bins provided the basis for a more in-depth analysis, annotation, and transcriptional profiling than a standard gene-centric (i.e., sequencing read–based) approach, revealing 36,151 and 24,134 gene clusters unique to TS28 and TS29, respectively, and not represented in any of the 122 reference gut genomes (Fig. 2A). Comparisons of the abundance of shared clusters between TS28 and TS29 revealed a stronger average correlation than the shared species-level phylotypes (mean R2 = 0.37 vs. R2 = 0.18). Rarefaction analysis disclosed that the number of genes and gene clusters in the gut microbiomes continues to increase even after 2 million mapped reads (Fig. 2C), with an estimated plateau of 242,023 and 234,661 genes, corresponding to 115,216 and 112,522 gene clusters in the TS28 and TS29 fecal microbiomes, respectively (Table S8).
The Diversity of Carbohydrate Active Enzymes in the Human Gut Microbiome and Evidence of Genes with Predicted Cellulolytic Activity.
The human genome lacks the large repertoire of glycoside hydrolases and polysaccharide lyases required to cleave the many glycosidic linkages present in complex dietary polysaccharides (9). Because processing of these polysaccharides is a major function of the distal gut microbiota (10), we annotated the predicted proteins from each genus- and family-level microbiome bin using procedures described in the Carbohydrate-Active EnZyme database [CAZy (9)] (Table S9 and S10 and Fig. S5). In total, we observed 143 CAZy families representing 5,145 genes in the gut microbiomes of these cotwins.
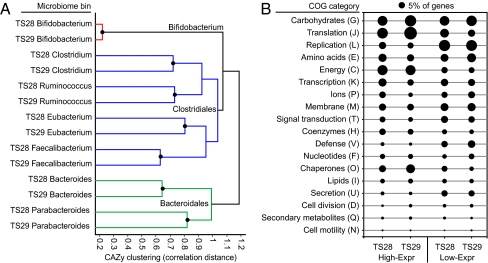
In general, the relative abundance of genes assigned to each CAZy family was consistent across genus-level bins from both individuals (Fig. 3A and Table S10). However, one notable exception was found: the Faecalibacterium bin from TS28 contained 42 genes predicted to encode dockerins, which are small proteins involved in the assembly of extracellular cellulosomes (11). None of these genes were identified in the Faecalibacterium bin from her cotwin's fecal microbiome, nor in the genome of F.prausnitzii isolate M21/2. However, 30 dockerins were identified across the Ruminococcus and Eubacterium bins of the two samples (Table S9). In agreement with the predicted formation of cellulosomes, the Faecalibacterium dockerins from TS28 were found with a number of genes predicted to encode cellulases (GH5,GH9,GH44,GH48), beta-mannanases (GH26), xyloglucanases (GH74), and polysaccharide lyases (PL), none of which were observed in the Faecalibacterium bin from TS29 (χ2 test, P < 10−4). Finally, a cohesin-encoding gene (the cognate molecule for dockerins) was identified in the Faecalibacterium bin from TS28, further supporting the existence of human gut cellulosomes.
Fig. 3.
Clustering of fecal microbiome bins and the annotation of differentially expressed genes. (A) UPGMA clustering was performed on the relative abundance of CAZy families across each microbiome bin. Number of genes assigned to each CAZy family was normalized to the total number of genes in each sequence bin (all bins with >30 CAZy family assignments in both samples are shown). Black circles represent clustered nodes after z-score normalization across all bins (inconsistency threshold = 0.75, “cluster” function in Matlab v7.7.0). (B) Percentage of genes with high relative expression (High-Expr) or low relative expression (Low-Expr) assigned to each COG category. Percentages are represented by the area of each circle (black circle labeled 5% provides reference).
To assess the distribution of genes predicted to encode dockerins across microbiomes from other twins, we compared 18 fecal microbiome datasets (mean ± SEM 535,232 ± 23,294 sequencing reads per sample; 118.7 ± 8.7 Mb/sample) obtained from six MZ twin-pairs and their mothers (4) to the protein-coding gene sequences from the microbiome bins obtained from the deeply sampled MZ twins. This analysis revealed that the identified dockerin-encoding genes are widely distributed across gut microbiomes but vary in abundance: all 18 microbiomes contained reads with significant sequence similarity to these genes (mean number of genes 12.4, range 1–55 genes; and mean number of sequencing reads 54.3, range 2–222 reads). However, only sequences from TS28 contained reads matching the identified cohesin-encoding gene.
Together, these results expand the known diversity of CAZymes in the human gut microbiome and reveal a suite of genes with predicted cellulolytic activity. The fact that the latter genes were highly enriched in the Faecalibacterium bins found in the microbiome of TS28 and not in her genetically identical cotwin highlights another level of genetic variation between humans. Future research will be necessary to characterize the enzymatic activity of these systems, the breadth of their organismal distribution, the host and environmental parameters (including diet) that determine their abundance in a given human gut microbiome, and their contributions to host nutrient/energy harvest.
The Metatranscriptome Viewed from the Perspective of Phylogenetic Bins.
To characterize gene expression in the gut microbiome, we analyzed cDNA and DNA datasets obtained from sequencing total community cDNA and DNA prepared from the two fecal samples of TS28 and TS29. All sequencing reads were mapped against the database of 122 gut microbial genomes and the microbiome bins (Metatranscriptome analysis in SI Text). The results revealed marked differences in gene abundance and expression (Figs. S6 and S7). In all cases, technical replicates of each microbiome and metatranscriptome (n = 3–4) clustered together; this clustering was robust to subsampling by COG functional categories (Fig. S6). Microbiome profiles showed the highest average correlation between individuals (R2 = 0.37), relative to metatranscriptomes (R2 = 0.12) and the relative abundance of species-level phylotypes (R2 = 0.18). As with the microbiome, rarefaction analysis of the metatranscriptome revealed that the number of expressed genes and gene clusters continues to increase even after 500,000 mapped reads (Fig. 2C), with an estimated plateau of 85,099 and 173,309 genes, corresponding to 35,781 and 58,339 gene clusters in TS28 and TS29, respectively (Table S8).
We subsequently calculated the ratio of the relative abundance of cDNA sequences in each microbiome bin to the relative abundance of DNA sequences in that bin, for each fecal community (12). Even at the genus-level, there were detectable differences in relative gene expression: six bins showed higher relative expression than gene abundance, whereas the Bifidobacterium had the lowest level of relative expression in both microbiomes (Fig. 2D).
We then compared cDNA and DNA profiles at the level of individual genes to determine the relative expression of each gene compared with its abundance (12). Genes were defined as “High Relative Expression” (High-Expr) or Low-Expr based on the ratio of cDNA to DNA relative abundance. A 10-fold difference was chosen as the threshold cutoff based on all pairwise comparisons of technical replicate datasets obtained from cDNA or DNA sequencing of each sample (Fig. S8A, n = 3–4 replicates per sample per method).
These comparisons revealed 6,961 genes with high or low relative expression in the fecal microbiome of TS28 (4,816 High-Expr and 2,145 Low-Expr) and 7,893 genes in TS29 (5,476 High-Expr and 2,417 Low-Expr; Tables S11 and S12). As expected, many of these genes came from bins with an overall higher relative expression (Fig. 2D), including Parabacteroides, Alistipes, Methanobrevibacter, and Bacteroides, or bins with a lower relative expression (the Bifidobacterium bin contained 962 Low-Expr genes in sample TS29 and 112 in TS28). However, some notable exceptions were found; the Bacteroides had 1,416 High-Expr genes in the TS28 microbiome, despite having overall similar levels of cDNA and DNA assignments across the entire bin (ratio 1.5).
The distribution of genes assigned to COG functional categories was then calculated using each set of High- or Low-Expr genes (Fig. 3B), as well as the set of genes that were observed only with cDNA or DNA sequencing (Fig. S9A). A disproportionate number of High-Expr genes encoded hypothetical proteins without predicted functions [33.9% (TS28) and 31.2% (TS29) of the High-Expr genes, comprising 77.9% (TS28) and 75.1% (TS29) of the total hypothetical genes with either a high or low relative expression]. High-Expr genes from both microbiomes were more frequently assigned to COG categories for translation (J), energy metabolism (C), and chaperones (O) (Fig. 3B and Tables S11 and S12), whereas Low-Expr genes were more frequently assigned to COG categories for secretory systems (U), replication, recombination, and repair (L), and membrane proteins (M) (Fig. 3B). In addition, many of these High-Expr genes have predicted functions related to fermentation and carbohydrate metabolism: e.g., ABC-type transport systems for carbohydrate import and metabolism plus genes involved in methanogenesis and acetogenesis (key pathways in the clearance of the hydrogen end-product of fermentation, and thus important determinants of fermentation efficiency).
To better characterize specific pathways represented by genes with high or low relative expression, we annotated each gene in the 122 gut microbial genomes and the microbiome bins using the KEGG annotation scheme (v52) (13). The relative abundance of KEGG pathways was tallied across genes defined as High- or Low-Expr in TS28 and TS29 or found to be unique to the cDNA or DNA datasets, and used for UPGMA clustering. Both microbiomes showed consistent trends, including high relative expression of genes assigned to pathways for essential cell processes, e.g., “RNA polymerase,” “Ribosome,” “Pyruvate metabolism,” and “Glycolysis” (Fig. S8B). We extended these analyses to five additional samples from two sets of MZ cotwins and one unrelated individual (Samples labeled “TSDA” in Fig. S9 and Additional microbiomes and meta-transcriptomes in SI Text) and found similar results, including the higher relative expression of genes assigned to COG categories for transcription, energy metabolism, defense mechanisms, and chaperones (Fig. S9A), in addition to KEGG pathways involved in carbohydrate metabolism (e.g., fructose/mannose metabolism), nucleotide metabolism, and vitamin metabolism/biosynthesis (e.g., folate biosynthesis) (Fig. S9B).
Prospectus.
Our results indicate that a majority of species-level phylotypes are shared between these deeply sampled MZ cotwins, despite large variations in the abundance of each phylotype. The genetic and transcriptional diversity of the human gut microbiome is remarkable. Much of this diversity has not been previously identified through sequencing cultured human gut isolates; 64% of the gene clusters present in our microbiome bins had no representative in a set of 122 human gut microbial genomes, and only 17% were shared between the two cotwins. This diversity, even between genetically identical individuals, provides an expanded view of our multicellularity and interpersonal genetic variation. Features of the genus-level bins within the gut microbiome were distinctive in many ways, ranging from differences in gene content and transcriptional activity, to the extent of sequence variation within each population. Identifying the factors that determine such between-taxon differences will provide an important step toward understanding the functions (niches) of these organisms in the human gut microbial community, with the ultimate goal of linking the presence of specific organisms to gene content and activity. Our results and the accompanying datasets also provide a framework for future studies of human and environmental microbiomes. As noted above, 16S rRNA gene sequence datasets can be used to prioritize genomes for isolation and sequencing, starting with the most abundant phylotypes found across the most individuals, and working toward the rare members of the gut microbiota. The reduced level of organismal diversity in a single individual implies that it may be soon be possible to identify all strains present in a single gut (fecal) microbiota. The fraction of shared phylotypes between MZ cotwins, between unrelated individuals, and between body habitats provides an important context for designing studies of the assembly, dynamic operations, and host effects of “model” human gut microbiota/microbiomes, composed of sequenced cultured gut isolates, in gnotobiotic mice. Finally, the application of transcriptional profiling to the study of human body habitat-associated microbial communities will enable correlations to be made between genes expressed by our microbiomes and our physiologic and metabolic phenotypes.
Materials and Methods
Sequencing of 16S rRNA Gene Amplicons.
Fecal samples were stored at −80°C before processing. DNA was extracted by bead beating followed by phenol-chloroform extraction as described previously (4). The V2 region was targeted for amplification by PCR (with primers 8F-338R) and multiplex GS FLX pyrosequencing (4). In addition, six control pools were constructed with equimolar or variable concentrations of purified genomic DNA from 67 cultured reference human gut–derived strains; the V2 regions of 16S rRNA genes present in these pools were then amplified and sequenced.
Assembly of the Human Gut Microbiome.
Shotgun sequencing runs were performed on libraries prepared from total fecal community DNA using the 454 GS FLX Titanium single- and paired-end protocols. For all analyses involving unassembled reads, sequencing reads with degenerate bases (“Ns”) were removed along with all replicate sequences using the following parameters: 0.9 (90%ID), length difference requirement = 0, and 3 beginning bases checked (14). Each deeply sequenced dataset (TS28 and TS29) was assembled separately using the 454 GS de novo assembler software (Newbler v2.0.00.22), and all scaffolds were used for subsequent analysis. High-confidence sequence variants were identified using the 454 GS Reference Mapper software (v2.0.00.20).
Metatranscriptome Analysis.
Microbial RNA sequencing (RNA-Seq) was performed as described previously (5). Briefly, total RNA was extracted from each fecal sample. The sample was subjected to rigorous DNase digestion to remove residual gDNA, depleted for rRNA and tRNA, converted to cDNA, and sequenced using the Illumina GAII platform. A total of 36 nucleotide reads produced from the each run were trimmed at their beginning and ends to remove bases with a quality score <20. Adapter sequences and sequencing reads with a length <20 nucleotides were subsequently eliminated from further analysis. All trimmed reads were mapped with SSAHA2 (15) to phylogenetic bins constructed from microbiome scaffolds and to 122 sequenced human gut-associated microbial genomes (SSAHA2 parameters: -best 1 -score 20 -solexa). Gene clusters were defined by grouping all protein sequences from the database using the program cd-hit [parameter -c 0.6 -n 4 (16)]. Gene and gene cluster counts were normalized based on the total number of mapped sequencing reads. Genes from the database with significant homology (BLASTN e-value <10−30) to noncoding transcripts from the 122 gut microbial genomes were excluded from subsequent analysis. Ties representing sequences matching multiple reference genes with the same score were split evenly, whereas ties matching multiple gene clusters were weighted according to the frequency of unique (nontie) matches to each cluster.
Details concerning (i) phylogenetic binning of microbiome scaffolds, (ii) analysis of gene, bin, and transcript abundance, (iii) development and validation of methods for 16S rRNA gene sequence analysis, and (iv) additional cohorts of humans analyzed are given in SI Text.
Supplementary Material
Acknowledgments
We thank S. Wagoner, J. Manchester, J. Hoisington-López, A. Heath, S. Marion, and D. Riches for their technical and other assistance during various phases of this work, as well as A. Goodman and A. Reyes for many helpful suggestions. This work was supported in part by National Institutes of Health Grants DK78669 and DK70977 (to J.I.G.), the Crohn's and Colitis Foundation of America, and Groupe Danone.
Footnotes
The authors declare no conflict of interest.
Data deposition: Datasets from shotgun sequencing projects have been deposited at DDBJ/EMBL/GenBank under accession number 43253. 454 and Illumina sequencing reads have been deposited in the NCBI Short Read Archive.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002355107/DCSupplemental.
References
- 1.Lahr DJ, Katz LA. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques. 2009;47:857–866. doi: 10.2144/000113219. [DOI] [PubMed] [Google Scholar]
- 2.Quince C, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 3.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1 doi: 10.1126/scitranslmed.3000322. 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 8.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 11.Bayer EA, Lamed R, White BA, Flint HJ. From cellulosomes to cellulosomics. Chem Rec. 2008;8:364–377. doi: 10.1002/tcr.20160. [DOI] [PubMed] [Google Scholar]
- 12.Frias-Lopez J, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 15.Ning Z, Cox AJ, Mullikin JC. SSAHA: A fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.