Abstract
Hundreds of bacterial species produce proteinaceous microcompartments (MCPs) that act as simple organelles by confining the enzymes of metabolic pathways that have toxic or volatile intermediates. A fundamental unanswered question about bacterial MCPs is how enzymes are packaged within the protein shell that forms their outer surface. Here, we report that a short N-terminal peptide is necessary and sufficient for packaging enzymes into the lumen of an MCP involved in B12-dependent 1,2-propanediol utilization (Pdu MCP). Deletion of 10 or 14 amino acids from the N terminus of the propionaldehyde dehydrogenase (PduP) enzyme, which is normally found within the Pdu MCP, substantially impaired packaging, with minimal effects on its enzymatic activity. Fusion of the 18 N-terminal amino acids from PduP to GFP, GST, or maltose-binding protein resulted in their encapsulation within MCPs. Bioinformatic analyses revealed N-terminal extensions in two additional Pdu proteins and three proteins from two unrelated MCPs, suggesting that N-terminal peptides may be used to package proteins into diverse MCPs. The potential uses of MCP assembly principles in nature and in biotechnology are discussed.
Keywords: 1,2-propanediol; B12; carboxysome; Salmonella; protein targeting
Diverse bacteria use proteinaceous intracellular compartments to optimize metabolic reactions having toxic or volatile intermediates (1–3). These compartments, which are polyhedral in shape and about 100–150 nm in cross-section, have been called bacterial microcompartments (BMCs). They are typically built from several thousand polypeptides of 10–20 types and consist of a protein shell that encapsulates metabolic enzymes. There is no evidence for lipid components. Based on sequence analysis, it is estimated that microcompartments (MCPs) are produced by 20–25% of all bacteria and function in seven or more different metabolic processes (1). Different types of MCPs have related protein shells but differ in their encapsulated enzymes (1, 4, 5). The best-studied MCP is the carboxysome, which enhances autotrophic CO2 fixation by confining CO2 in the immediate vicinity of ribulose bisphosphate carboxylase monooxygenase (2, 3, 6). Because of their presence in virtually all cyanobacteria, carboxysomes contribute substantially to global CO2 fixation (7). Other MCPs sequester aldehyde intermediates formed during catabolic processes to prevent toxicity and carbon loss or have unknown functions (1, 3, 8–11). A characteristic of bacterial MCPs is that their shells are mainly formed from a group of related proteins that have one or two BMC domains (Pfam, PF00936). Recent crystallography has shown that BMC-domain polypeptides assemble into extended protein sheets that are perforated by discrete pores thought to mediate transport of enzyme substrates, products, and cofactors between the lumen of the MCP and the cytoplasm of the cell (12–16). Different shell proteins have pores that differ in size and charge, and some may have gated pores tentatively suggesting substrate selectively and regulated movement of metabolites across the shell (12–18). In the literature, MCPs have also been referred to as polyhedral bodies/organelles, enterosomes, and metabolosomes (1, 3, 10).
Salmonella enterica produces an MCP used for B12-dependent 1,2-propanediol (1,2-PD) utilization (Pdu MCP) (19, 20). 1,2-PD is a major product of the anaerobic degradation of the plant sugars rhamnose and fucose and is an important carbon and energy source in natural environments (21, 22). In addition, the capacity to degrade 1,2-PD has been tentatively linked to pathogenesis in Salmonella and Listeria (23–26). The Pdu MCP consists of about 14 different polypeptides, including seven different shell proteins and four enzymes (27) (Fig. 1). The first two steps of 1,2-PD degradation occur in the lumen of the MCP, and the latter steps occur in the cytoplasm of the cell (1). The encapsulated enzymes include coenzyme B12-dependent diol dehydratase and CoA-dependent propionaldehyde dehydrogenase (PduP), which convert 1,2-PD to propionyl-CoA via a propionaldehyde intermediate (8, 27). Propionyl-CoA exits the MCP into the cytoplasm of the cell, where it is converted to 1-propanol and propionate or enters central metabolism via the methylcitrate pathway (19, 27–30). The function of the Pdu MCP is to sequester propionaldehyde formed by the first step of 1,2-PD degradation primarily to protect cells from cytotoxicity and DNA damage (31). It is thought that the shell of the Pdu MCP restricts the outward diffusion of propionaldehyde.
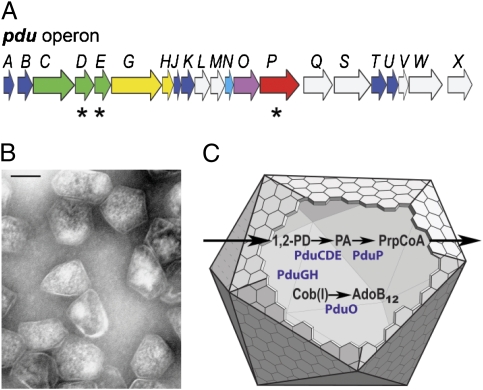
Fig. 1.
Model for the structure and function of the Pdu MCP. (A) Organization of the pdu operon. At least 14 pdu genes (colored) encode proteins that are components of purified Pdu MCPs (27). Asterisks indicate genes that encode polypeptides having potential N-terminal targeting sequences (in the text). Seven genes (blue and cyan) encode shell proteins (27). Homologues of the BMC family of shell proteins are shown in blue. (B) Electron micrograph of purified Pdu MCPs from S. enterica. (Scale bar: 100 nm). (C) Model for B12-dependent 1,2-PD degradation by Salmonella. 1,2-PD is metabolized within the MCP lumen, first to propionaldehyde (PA) and then to propionyl-CoA (PrpCoA). The enzymes that localize to the MCP interior include coenzyme B12-dependent diol dehydratase (PduCDE) and PduP, as well as adenosyltransferase (PduO) and a reactivase (PduGH) that are required for the maintenance of diol dehydratase activity (53–55). The proposed function of the Pdu MCP is to sequester propionaldehyde to minimize its toxicity.
A key to the function of bacterial MCPs is the encapsulation of specific enzymes within a protein shell. Although there has been substantial progress in understanding the structure of bacterial MCPs, particularly carboxysomes, the signals that target proteins to the interior of MCPs are unknown in any system. Prior studies showed that the N-terminal extensions of the PduD and PduE proteins were not required for enzymatic activity, and a possible role in MCP assembly was suggested but not investigated experimentally (32). Here, we present a series of studies that show a short N-terminal peptide of the PduP protein is necessary and sufficient for packaging proteins into the lumen of the Pdu MCP. We also present bioinformatic evidence that suggests N-terminal peptides may have a general role in packaging proteins within diverse MCPs.
Results
A Short N-Terminal Region of PduP Is Required for Efficient Packaging into the Pdu MCP.
A sequence alignment of PduP homologues shows relative agreement over the core sequence regions, with a notable exception at the N termini. Most homologues predicted to be MCP-associated (based on genomic context) have N-terminal extensions of about 20 amino acids that are absent in sequences lacking a predicted MCP association (Fig. 2 and Dataset S1). This suggested to us that the N-terminal region might have a role in packaging PduP into the lumen of the Pdu MCP. To test this, PCR was used to make plasmid constructs for production of PduP lacking 10 or 14 N-terminal amino acids (PduP11–464 and PduP15–464). Full-length PduP and each deletion were produced from vector pLac22 in strains having a deletion of the chromosomal copy of pduP, and the resulting MCPs were purified and analyzed by SDS/PAGE (Fig. 3A). The PduP protein is clearly present in the MCPs purified from the WT strain (lane 1) and the ΔpduP strain producing full-length PduP from pLac22 (lane 3). In contrast, much smaller amounts of PduP11–464 and PduP15–464 were associated with purified MCPs when these proteins were produced from pLac22 in the ΔpduP background (lanes 4 and 5). This finding suggests that deletion of the N-terminal region of PduP impaired its association with the MCP.
Fig. 2.
Multiple sequence alignment of the N-terminal regions of 10 representative PduP homologues from different organisms. Terminal sequence extensions are apparent in four of the five representatives whose genomic context suggests an association with MCP function (BMC-proximal). PduP homologues belong to the conserved NAD(P)+-dependent aldehyde dehydrogenase superfamily (cl11961), whose members oxidize a range of aldehyde substrates in distinct metabolic pathways. The representative homologues were selected from clusters of similar sequences from an alignment of some 100 distinct sequences (the number of sequences in each cluster follows the organism name in parentheses). The National Center for Biotechnology Information gene accession identifications, beginning from the top entry, are 5069459, 16761381, 46907383, 123442957, 188587712, 27366378, 50121254, 110798574, 114563069, and 148378348. The full alignment is provided in Dataset S1. N-terminal methionine residues were omitted to prevent spurious alignment of the divergent termini.
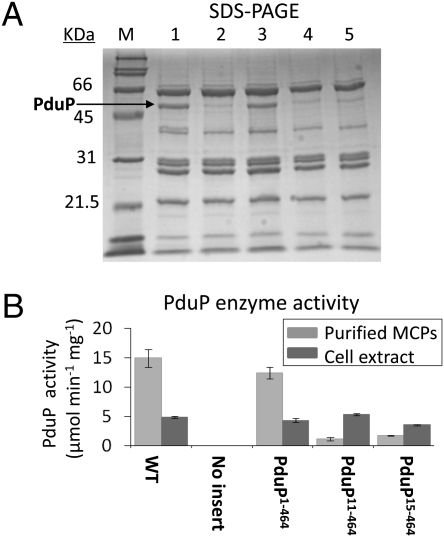
Fig. 3.
Association of PduP with purified Pdu MCPs. (A) SDS/PAGE: (lane 1) WT S. enterica, (lane 2) ΔpduP/pLac22-no insert, (lane 3) ΔpduP/pLac22-pduP1–464 (full-length PduP), (lane 4) ΔpduP/pLac22-pduP11–464, and (lane 5) ΔpduP/pLac22-pduP15–464. Ten micrograms of MCPs was loaded in each well. (B) PduP enzyme activity for the starting cell extracts (dark gray) and purified MCPs (light gray) analyzed in A.
We also measured the PduP enzyme activity in cell extracts and purified MCPs (Fig. 3B). Except for the negative control, cell extracts all had similar activity. This established that PduP11–464 and PduP15–464 are stable and active and eliminated the possibility that they failed to associate with the Pdu MCP because of low production, instability, or misfolding. Enzyme assays also showed that MCPs purified from cells producing PduP11–464 or PduP15–464 had much lower PduP enzyme activity than those purified from cells producing native PduP, supporting the SDS/PAGE analyses that indicated PduP11–464 and PduP15–464 were impaired for MCP association. Given that PduP11–464 and PduP15–464 are stable and active but largely absent from purified MCPs, we infer that a short N-terminal region of PduP is required for efficient packaging of PduP into the Pdu MCP.
A Short Region of the N Terminus of PduP Is Sufficient for Packing Heterologous Proteins into the Lumen of the Pdu MCP.
To determine whether the N-terminal region of PduP was sufficient for packaging and to investigate the minimal sequence length required, we fused 10, 14, 18, or 70 amino acids from the N terminus of PduP to enhanced GFP (eGFP). Each fusion protein (PduP1–10-eGFP, PduP1–14-eGFP, PduP1–18-eGFP, and PduP1–70-eGFP) was produced from pLac22 in a pduP deletion mutant, and Western blots were used to determine whether eGFP was associated with purified MCPs (Fig. 4A). No eGFP was detected in MCPs purified from the WT strain or a pduP deletion mutant expressing native eGFP (lanes 1 and 2). In contrast, each fusion protein was detected in purified MCPs. A faint band was seen for PduP1–10-eGFP, but longer fusion proteins localized more efficiently. When 14 amino acids from the N terminus of PduP were fused to eGFP, the fusion protein was readily detected by Western blotting. These results, in conjunction with the above studies, indicate that a short region of the N terminus of PduP is necessary and sufficient for packing proteins to the lumen of the MCP.
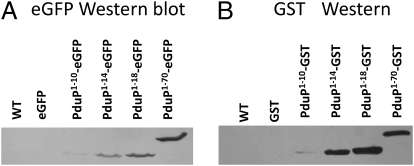
Fig. 4.
Western blots for eGFP, GST, and fusion proteins. (A) Western blot for eGFP. MCPs purified from (left to right) WT S. enterica, ΔpduP/pLac22-eGFP, ΔpduP/pLac22-PduP1–10-eGFP, ΔpduP/pLac22-PduP1–14-eGFP, ΔpduP/pLac22-PduP1–18-eGFP, and ΔpduP/pLac22-PduP1–70-eGFP. (B) Western blot for GST. MCPs purified from (left to right) WT S. enterica, ΔpduP/pLac22-GST, ΔpduP/pLac22-PduP1–10-GST, ΔpduP/pLac22-PduP1–14-GST, ΔpduP/pLac22-PduP1–18-GST, and ΔpduP/pLac22-PduP1–70-GST.
A similar set of studies was done by fusing N-terminal sections of PduP to GST and maltose binding protein (MBP). Either, 10, 14, 18, or 70 amino acids from the N terminus of PduP were fused to GST. Native GST failed to associate with the MCP, but each fusion protein did so (Fig. 4B). In the case of MBP, one fusion protein was constructed having 18 amino acids from the N terminus of PduP. This fusion protein was clearly present by Western blotting, whereas WT MBP was not detected (Fig. S1). We also note that PduP1–18-MBP, PduP1–70-eGFP, and PduP1–70-GST were easily detected as MCP components by SDS/PAGE, followed by Coomassie staining showing that relatively large amounts were present (Fig. S2). However, the lower molecular mass fusion proteins were obscured in PAGE gels by comigrating MCP proteins, necessitating detection by Western blot. The fact that GFP, GST, and MBP all associated with the MCP when fused to a short region of PduP is evidence against a nonspecific interaction peculiar to a particular fusion protein and favors the interpretation that fusion proteins specifically interact with the MCP via the sequences derived from PduP.
Fusion Proteins Compete with PduP for MCP Association.
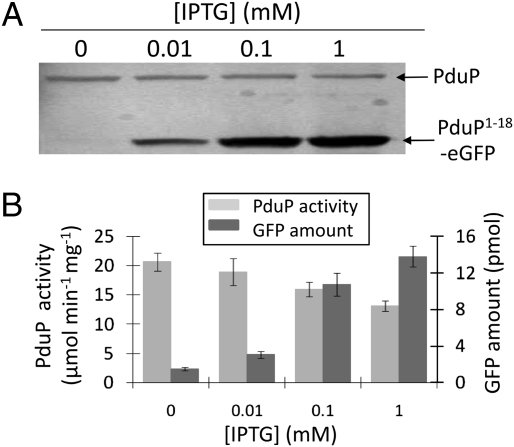
To test the specificity of the interaction between the PduP1–18-GFP and the MCP, a competition experiment was performed. PduP1–18-GFP was produced via an isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoter in a WT background that also produced PduP. MCPs were purified, and Western blotting was used to detect the PduP1–18-GFP and PduP (Fig. 5A). With increasing concentrations of IPTG, the amount of PduP1–18-GFP present in purified MCPs increased and the amount of PduP decreased, indicating that this fusion protein competed with PduP for MCP association. The PduP and PduP1–18 eGFP present in purified MCPs were also quantified by enzyme assay and relative fluorescence (Fig. 5B). The MCP-associated PduP activity decreased with increasing concentrations of IPTG and vice versa for PduP1–18-eGFP fluorescence. A similar experiment was performed by producing PduP1–18-MBP in place of PduP1–18-GFP. Western blots showed that PduP1–18-MBP also competed with PduP for MCP association (Fig. S3). Thus, competition studies indicate that PduP1–18-eGFP and PduP1–18-MBP associate with the Pdu MCP by binding to the same site/region that PduP would otherwise occupy.
Fig. 5.
Competition between fusion proteins and PduP for MCP binding. (A) Increasing IPTG concentrations (0, 0.01, 0.1, and 1 mM) were used to increase production of PduP1–18-GFP by S. enterica/pLac22-PduP1–18-eGFP. MCPs were purified, and Western blots were used to detect GFP and PduP. (B) Amount of PduP and eGFP associated with MCPs purified from S. enterica/pLac22-PduP1–18-eGFP grown with different IPTG concentrations based on PduP enzyme activity and relative fluorescence.
Immunoprecipitation Indicates That the Fusion Proteins Are Encapsulated Within the Pdu MCP.
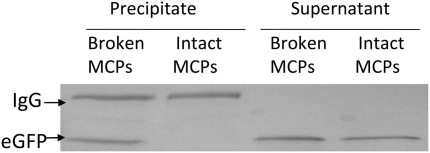
To test whether the fusion proteins were encapsulated within the shell, MCPs were purified from a strain producing PduP1–18-eGFP and a portion was broken by dialysis and sonication. Intact and broken MCPs were incubated with anti-GFP antibody. Protein A-immobilized beads were used to precipitate the anti-GFP antibody and associated proteins. Western blots showed that PduP1–18-eGFP coprecipitated with protein A-immobilized beads incubated with anti-GFP antibody and broken MCPs but not with protein A beads incubated with anti-GFP antibody and intact MCPs (Fig. 6). These findings support the idea that PduP1–18-eGFP is encapsulated within the shell of the Pdu MCP, blocking its interaction with the anti-GFP antibody, and oppose the idea that PduP1–18-eGFP is associated with the outer surface of the MCP.
Fig. 6.
Western blot for eGFP following immunoprecipitation of eGFP from broken and intact MCPs. MCPs were purified from ΔpduP/plac22-PduP1–18-eGFP, and a portion was broken by dialysis and sonication. Rabbit anti-GFP was incubated with broken and intact MCPs and then precipitated with protein A-immobilized beads. The proteins that coprecipitated with the beads and the supernatant fractions were analyzed by Western blot for eGFP. IgG was also detected by the secondary antibody used for the Western blot.
Fluorescence and EM Further Support Localization of PduP1–18-eGFP and PduP1–18-GST to the Pdu MCP.
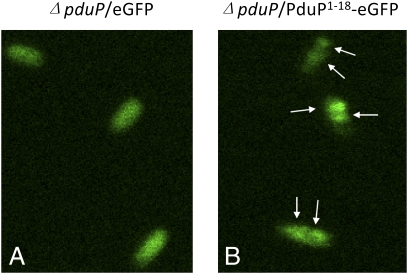
We examined subcellular localization of the PduP1–18-eGFP and PduP1–18-GST by fluorescence and EM. In cells producing eGFP without the packaging sequence, green fluorescence was distributed relatively evenly in 96% of the cells and only 4% of cells showed localized brighter green fluorescence (Fig. 7). In cells producing PduP1–18-eGFP, there was localized brighter green fluorescence in 80% of cells and distributed fluorescence in 20% of cells, consistent with the association of PduP1–18-eGFP with MCPs in living cells. The fluorescence was further supported by immunogold labeling, which showed that PduP1–18-eGFP and PduP1–18-GST localized to the MCP, whereas eGFP and GST did not (Fig. S4). Cumulatively, the series of studies presented here demonstrates that a short N-terminal peptide is necessary and sufficient for association of proteins with the Pdu MCP and likely mediates packaging within the interior of the MCP.
Fig. 7.
Localization of fusion proteins by fluorescence microscopy of cells producing eGFP (A) and PduP1–18eGFP (B). Arrows point to areas of localized brighter green fluorescence.
Effects of PduP Targeting on 1,2-PD Degradation.
We also examined the effects of PduP localization on growth of S. enterica on 1,2-PD. To do this, we compared the growth of WT with that of strain BE1375, which carries a chromosomal deletion of the pduP gene and produces PduP lacking 18 N-terminal amino acids from a plasmid pLac22. This strain has near-normal PduP enzymatic activity, but the majority of that activity is found in the cell cytoplasm rather than in the Pdu MCP. BE1375 grew more slowly than WT S. enterica (a doubling time of 9.1 h compared with 8.1 h) and accumulated propionaldehyde to a level about 2-fold higher than WT (6.5 mM compared with 3 mM) during growth on 1,2-PD. As an additional control, we examined growth on 1,2-PD and propionaldehyde production by strain BE270, which has a chromosomal deletion of pduP and produces WT PduP enzyme from plasmid pLac22. For BE270, PduP localized to the MCP (Fig. 3) and growth rate and propionaldehyde production were indistinguishable from those of WT S. enterica. These results are consistent with prior studies indicating that the function of the Pdu MCP is to optimize growth by minimizing propionaldehyde toxicity (8, 27, 31).
Bioinformatic Analysis Indicates That N-Terminal Sequences Are Part of a General Mechanism for Packaging Proteins into Diverse Bacterial MCPs.
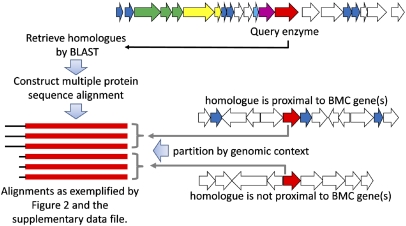
To expand on the finding that a short N-terminal sequence targets PduP to the Pdu MCP, we searched the National Center for Biotechnology Information genomic database for other proteins that might be targeted to MCPs by a similar mechanism. We asked whether protein families could be identified in which extended N-terminal sequences are present only in homologues that are associated with MCP function (Fig. 8). A protein was treated as functionally associated with an MCP if its encoding gene was within 10 genes upstream or downstream of a gene for a BMC protein; proteins from this conserved family comprise the shells of diverse MCPs. Multiple sequence alignments showed that in addition to PduP, five other putative MCP enzymes had N-terminal extensions compared with homologues unassociated with MCPs: diol dehydratase small and medium subunits (PduD and PduE) from the Pdu MCP (33), the ethanolamine ammonia lyase small subunit (EutC) and ethanol dehydrogenase (EutG) from the Escherichia coli K-12 Eut MCP (34–36), and the pyruvate formate lyase (Pfl2) thought to be part of a yet uncharacterized Vibrio furnissii MCP (37). Collectively, these results raise the intriguing possibility that N-terminal sequences are part of a general mechanism for packaging proteins into diverse bacterial MCPs, although other mechanisms may also be important because N-terminal extensions appear to be absent from some MCP-associated enzymes such as PduC (Dataset S1).
Fig. 8.
Approach for the bioinformatic detection of potential terminal targeting sequences in MCP proteins. For a given MCP protein, homologous sequences were retrieved and classified according to genomic context, with genomic proximity to a BMC shell protein (blue) taken to indicate a likely association with MCP function. For some proteins involved in MCP function (like the one illustrated in the hypothetical example), the BMC-proximal homologues encode amino-terminal sequence extensions of ≈20–35 residues, which are postulated to serve as MCP targeting sequences.
Discussion
The use of short N-terminal sequences for the formation of large protein assemblies is somewhat atypical, because multiprotein complexes often form via substantial subunit interfaces (38, 39). However, various short peptides are known to mediate the formation of protein assemblies. For example, PDZ domain proteins typically bind short carboxyl-terminal peptides of membrane proteins mediating the formation of higher order complexes (40). In a nanocompartment in Thermotoga, a short carboxyl-terminal peptide mediates encasement of the DyP protein within a shell assembled from 60 encapsulin subunits (41). In this report, we present evidence that short N-terminal sequences direct proteins to the lumen of bacterial MCPs. Thus, various evolutionarily distinct systems use short peptides for the formation of protein assemblies. A common feature of these systems is the anchoring of proteins to a biological surface. Hence, N- and C-terminal peptides may have a wider role than generally recognized in arraying proteins on surfaces such as lipid membranes, viral capsids, bacterial and archaeal S-layers, and bacterial MCPs. The use of N-terminal sequences to target certain proteins to the interior of MCPs also parallels systems used to target proteins to organelles in higher organisms as well as the periplasmic space of bacteria (42). Because systems for localizing proteins to various subcellular compartments and organelles have distinct targeting sequences, it is likely that they arose independently and converged to analogous functions during evolution. The discovery of a growing number of such systems demonstrates that N-terminal sequence extensions have provided a facile route for the evolution of macromolecular assemblies.
The mechanism by which N-terminal sequences direct packaging of PduP into the MCP is uncertain. Presumably, the N-terminal extensions of PduP bind to the interior surface of a particular shell protein. This could help to explain the diversity of shell proteins used to construct the Pdu MCP; each lumen protein might have a partner shell protein that directs its packaging (19, 27). Such a system would offer an advantage by allowing the MCP lumen enzymes to be arrayed on the inner surface of the shell with an architecture that properly orients sequential catalytic sites as well as the pores proposed to act as metabolite conduits through the shell.
With regard to biotechnology, it has been suggested that bacterial MCPs might find application as specialized reaction chambers for the production of industrial chemicals (13, 43–47). A major challenge in engineering pathways for the production of biofuels, green chemicals, and pharmaceuticals is balancing enzyme activities to limit the accumulation of pathway intermediates, particularly those that may be toxic to host cells. A promising solution to this problem is anchoring enzymes to synthetic scaffolds to promote substrate channeling as is found in some natural systems such as polyketide synthases and cellulosomes (43, 45). Bacterial MCPs might also be useful for enhancing engineered metabolic pathways by providing reaction chambers for channeling substrates, increasing enzyme efficiency, and limiting the toxicity of pathway intermediates. Understanding the molecular basis of enzyme encapsulation into MCPs is an important step toward this possibility.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions.
The bacterial strains used in this study are derivatives of S. enterica serovar Typhimurium LT2 (Table S1). The rich medium used was LB/Lennox medium (Difco) (48). The minimal media used were no-carbon-E with 1 mM MgSO4 and 0.3 mM each valine, isoleucine, leucine, and threonine (49, 50). Growth of S. enterica on 1,2-PD and measurement of propionaldehyde were performed as described (31).
Protein and Molecular Methods.
PAGE was performed using Bio-Rad Ready gels and Bio-Rad Mini-Protean II electrophoresis cells according to the manufacturer's instructions. Following gel electrophoresis, Coomassie Brilliant Blue R-250 was used to stain proteins. The protein concentration of solutions was determined by using Bio-Rad protein assay reagent. The Pdu MCP was purified as described (27). PduP enzyme assays were carried out as previously described (51). Standard methods for DNA manipulation and Western blots were used (52). Some additional experimental details are presented in SI Text.
Immunoprecipitation.
Purified MCPs were dialyzed with 50 mM Tris (pH 8.0), 50 mM KCl, and 5 mM MgCl2 overnight at 4 °C. The dialyzed MCPs were then broken by sonication. Protein A-Sepharose 4B Fast Flow beads (Sigma) were used to precipitate rabbit IgG. Beads were washed with cold PBS buffer three times, diluted 10-fold with PBS, mixed with anti-GFP, and shaken gently for 1 h at 4 °C. Broken or intact MCPs were added and gently shaken for 1 h at 4 °C. Mixtures were centrifuged for 2 min at 2,000 × g to collect the beads, and the supernatant was saved for analysis later. The beads were mixed with PBS buffer and placed on ice for 10 min, and the supernatant was removed. This was repeated three times. Next, the beads were mixed with Hepes (pH 7.0) centrifuged for 2 min at 2,000 × g, and the supernatant was removed. This was also repeated three times, and the washed beads were used for analysis. The supernatant and precipitate fractions were analyzed by Western blots for eGFP.
GFP Quantification.
GFP was quantified by means of the GFP Quantification Kit (Biovision) following the manufacturer's instructions with purified eGFP as the standard. Fluorescence was monitored using a BioTek Synergy microplate reader and 48-well flat-bottom plates (Falcon) (excitation/emmision = 488/507 nm).
Fluorescence Microscopy.
For fluorescence microscopy, cells were grown as described above for MCP purification. The cells were mounted on agarose-covered microscope slides and examined with a Leica SP5× confocal microscope system with an inverted microscope front end at a resolution of 1,024 × 1,024 pixels. The green fluorescent images were created using 470- to 490-nm excitation and 515 long-pass detection.
Supplementary Material
Acknowledgments
We thank Dr. Jianqiang Shao at the University of Iowa for help with EM. We also thank Dr. Margie Carter at Iowa State University for help with fluorescence microscopy. We thank Neil King and Dr. Richard Llewellyn at the University of California, Los Angeles for suggestions on the bioinformatic analysis. This project was funded by National Science Foundation Grant MCB0616008 (to T.A.B.) and by National Science Foundation Grant MCB0843065 and National Institutes of Health Grant AI081146 (to T.O.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913199107/DCSupplemental.
References
- 1.Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: Their properties and paradoxes. BioEssays. 2008;30:1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): Functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 3.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 4.Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- 5.Cannon GC, et al. Microcompartments in prokaryotes: Carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: Polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 7.Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: Molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- 8.Havemann GD, Sampson EM, Bobik TA. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2. J Bacteriol. 2002;184:1253–1261. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penrod JT, Roth JR. Conserving a volatile metabolite: A role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol. 2005;187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stojiljkovic I, Bäumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: Nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein MG, et al. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–333. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka S, et al. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 14.Kerfeld CA, et al. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 15.Sagermann M, Ohtaki A, Nikolakakis K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci USA. 2009;106:8883–8887. doi: 10.1073/pnas.0902324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heldt D, et al. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem J. 2009;423:199–207. doi: 10.1042/BJ20090780. [DOI] [PubMed] [Google Scholar]
- 17.Crowley CS, Sawaya MR, Bobik TA, Yeates TO. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure. 2008;16:1324–1332. doi: 10.1016/j.str.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka S, Sawaya MR, Phillips M, Yeates TO. Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci. 2009;18:108–120. doi: 10.1002/pro.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeter RM. Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium. J Gen Microbiol. 1990;136:887–896. doi: 10.1099/00221287-136-5-887. [DOI] [PubMed] [Google Scholar]
- 21.Lin ECC. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Niedhardt FD, editor. Washington, DC: American Society for Microbiology; 1987. pp. 244–284. [Google Scholar]
- 22.Obradors N, Badía J, Baldomà L, Aguilar J. Anaerobic metabolism of the L-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J Bacteriol. 1988;170:2159–2162. doi: 10.1128/jb.170.5.2159-2162.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchrieser C, Rusniok C, Kunst F, Cossart P, Glaser P Listeria Consortium. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: Clues for evolution and pathogenicity. FEMS Immunol Med Microbiol. 2003;35:207–213. doi: 10.1016/S0928-8244(02)00448-0. [DOI] [PubMed] [Google Scholar]
- 24.Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heithoff DM, et al. Coordinate intracellular expression of Salmonella genes induced during infection. J Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph B, et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havemann GD, Bobik TA. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2003;185:5086–5095. doi: 10.1128/JB.185.17.5086-5095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horswill AR, Escalante-Semerena JC. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J Bacteriol. 1999;181:5615–5623. doi: 10.1128/jb.181.18.5615-5623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios S, Starai VJ, Escalante-Semerena JC. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J Bacteriol. 2003;185:2802–2810. doi: 10.1128/JB.185.9.2802-2810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, et al. PduL is an evolutionarily distinct phosphotransacylase involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar typhimurium LT2. J Bacteriol. 2007;189:1589–1596. doi: 10.1128/JB.01151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobimatsu T, Kawata M, Toraya T. The N-terminal regions of beta and gamma subunits lower the solubility of adenosylcobalamin-dependent diol dehydratase. Biosci Biotechnol Biochem. 2005;69:455–462. doi: 10.1271/bbb.69.455. [DOI] [PubMed] [Google Scholar]
- 33.Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. Propanediol utilization genes (pdu) of Salmonella typhimurium: Three genes for the propanediol dehydratase. J Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faust LR, Connor JA, Roof DM, Hoch JA, Babior BM. Cloning, sequencing, and expression of the genes encoding the adenosylcobalamin-dependent ethanolamine ammonia-lyase of Salmonella typhimurium. J Biol Chem. 1990;265:12462–12466. [PubMed] [Google Scholar]
- 35.Roof DM, Roth JR. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1989;171:3316–3323. doi: 10.1128/jb.171.6.3316-3323.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kofoid E, Rappleye C, Stojiljkovic I, Roth JR. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wackett LP, Frias JA, Seffernick JL, Sukovich DJ, Cameron SM. Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1. Appl Environ Microbiol. 2007;73:7192–7198. doi: 10.1128/AEM.01785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grueninger D, et al. Designed protein-protein association. Science. 2008;319:206–209. doi: 10.1126/science.1150421. [DOI] [PubMed] [Google Scholar]
- 39.Janin J, Miller S, Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 40.Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 41.Sutter M, et al. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 42.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 43.Dueber JE, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 44.Menon BB, Dou Z, Heinhorst S, Shively JM, Cannon GC. Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species. PLoS One. 2008;3:e3570. doi: 10.1371/journal.pone.0003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papapostolou D, Howorka S. Engineering and exploiting protein assemblies in synthetic biology. Mol Biosyst. 2009;5:723–732. doi: 10.1039/b902440a. [DOI] [PubMed] [Google Scholar]
- 46.Parsons JB, et al. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem. 2008;283:14366–14375. doi: 10.1074/jbc.M709214200. [DOI] [PubMed] [Google Scholar]
- 47.Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Self-assembly in the carboxysome: A viral capsid-like protein shell in bacterial cells. Biochem Soc Trans. 2007;35:508–511. doi: 10.1042/BST0350508. [DOI] [PubMed] [Google Scholar]
- 48.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
- 49.Berkowitz D, Hushon JM, Whitfield HJ, Jr, Roth J, Ames BN. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: Partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 51.Leal NA, Havemann GD, Bobik TA. PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol. 2003;180:353–361. doi: 10.1007/s00203-003-0601-0. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 53.Johnson CLVJ, et al. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J Bacteriol. 2001;183:1577–1584. doi: 10.1128/JB.183.5.1577-1584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mori K, Tobimatsu T, Toraya T. A protein factor is essential for in situ reactivation of glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. Biosci Biotechnol Biochem. 1997;61:1729–1733. doi: 10.1271/bbb.61.1729. [DOI] [PubMed] [Google Scholar]
- 55.Mori K, Toraya T. Mechanism of reactivation of coenzyme B12-dependent diol dehydratase by a molecular chaperone-like reactivating factor. Biochemistry. 1999;38:13170–13178. doi: 10.1021/bi9911738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.