BIOCHEMISTRY Correction for “Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change,” by Ewen Gallagher, Min Gao, Yun-Cai Liu, and Michael Karin, which appeared in issue 6, February 7, 2006, of Proc Natl Acad Sci USA (103:1717–1722; first published January 30, 2006; 10.1073/pnas.0510664103).
The authors note that a portion of Fig. 1 was reprinted from Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, Karin M (2004) Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase itch. Science 306:271–275. Although the reference was included in the original text of the manuscript as ref. 7, the authors neglected to state in the figure legend that the blot used in Fig. 1F is the same blot (probed with a different antibody) shown in Fig. 3E of Gao et al. 2004. The figure and its corrected legend appear below.
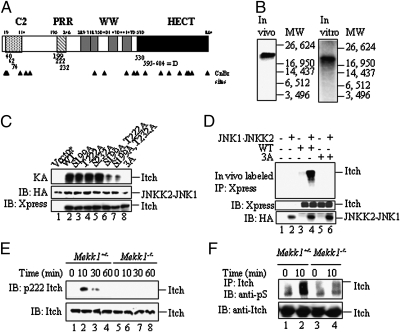
Fig. 1.
Mapping the JNK1 phosphorylation sites of Itch. (A) Schematic representation of Itch and its structural motifs. Locations of possible MAPK phosphorylation sites and the D domain are indicated by the numbers below the bar. CNBr cleavage sites are indicated by arrowheads. (B) Identification of a CNBr cleavage product containing JNK1 phosphorylation sites. (Left) Full-length Itch labeled with 32P in HEK293 cells was immunoprecipitated and digested with CNBr. (Right) H6-ItchΔC2 was incubated with recombinant JNKK2-JNK1 in the presence of [γ-32P]ATP and digested with CNBr. Digests were separated on Tris-Tricine gels along with molecular mass markers. (C) Identification of individual JNK1 phosphorylation sites. (Top) WT Itch and Ala substitution mutants thereof affecting all of the potential MAPK phosphorylation sites within the CNBr fragment identified above were incubated with JNKK2-JNK1 and [γ-32P]ATP . (Middle and Bottom) Itch proteins were also immunoblotted (IB) with anti-Xpress antibody (Bottom) and JNKK2-JNK1 was detected with anti-HA antibody (Middle). (D) In vitro JNKK2-JNK1 phosphorylation sites are also sites for JNK1-mediated phosphorylation in intact cells. WT Itch and the Ala substitution mutants were cotransfected into HEK293 cells along with pSRα-JNKK2-JNK1 or an empty vector. (Top) After 24 h, cells were labeled with [32P]orthophosphate, Itch proteins were immunoprecipitated with anti-Xpress, and the gels were separated and autoradiographed. (Middle and Bottom) Part of each lysate was immunoblotted with anti-Xpress (Middle) or anti-HA (Bottom) antibodies. (E) Itch T222 is phosphorylated in response to T cell receptor ligation. Mekk1ΔKD/ΔKD or Mekk1+/ΔKD thymocytes were stimulated with anti-CD3 and anti-CD28. At the indicated time points, whole-cell lysates were prepared and analyzed by immunoblotting with anti-pT222 Itch (Upper) or anti-Itch (Lower) antibodies. (F) T cell receptor ligation induces Itch Ser phosphorylation. Mekk1ΔKD/ΔKD and Mekk1+/ΔKD thymocytes were stimulated and analyzed as above, except that Itch immunoprecipitates (IP) were immunoblotted with anti-pS (Upper) or anti-Itch (Lower) antibodies. The blot used in this experiment has been used before in ref. 7, Fig. 3E to examine Itch ubiquitination upon T cell receptor ligation. The present results show that Itch Ser phosphorylation correlates with Itch activation.