Abstract
The translocation of large macromolecules through the nuclear pore complex (NPC) of eukaryotic cells is hindered by the phenylalanine-glycine (FG) nucleoporin (Nup) barrier unless molecules are chaperoned by transport receptors. The precise mechanism of facilitated translocation remains unclear due to the challenges of measuring the series of transient interactions between a transport receptor and the FG-Nups. This study developed single-point edge-excitation subdiffraction microscopy to obtain a three-dimensional density map of the transient interactions with a spatiotemporal resolution of 9 nm and 400 μs. Three unique features were observed under real-time trafficking conditions that have escaped detection by conventional electron microscopy: (i) the spatial density of interaction sites between Importin β1 (Imp β1, a major transport receptor) and the FG-Nups gradually increases from both sides of the NPC and is highest in the central pore region; (ii) cargo-free or cargo-bound Imp β1 rarely occupies an axial channel with a diameter of approximately 10–20 nm at its narrowest point through the NPC; and (iii) the pathway of facilitated translocation through the NPC depends more on the interaction sites of the FG-Nups than on the NPC architecture.
Keywords: nucleocytoplasmic transport, single molecule fluorescence, deconvolution, imaging and tracking
Nuclear pore complexes (NPCs) span the nuclear envelope (NE) and enable bidirectional transport between the cytoplasm and nucleus of eukaryotic cells. Proteins destined for nuclear functions, such as nucleic acid polymerases, histones, and splicing and transcription factors, must transit into the nucleus after synthesis on cytoplasmic ribosomes. The major cellular RNAs generated by transcription in the nucleus (mRNAs, tRNAs, and rRNAs) are exported to the cytoplasm. Small molecules (less than ≈20–40 kDa) can transit NPCs without specific recognition (passive diffusion). Large molecules or molecular complexes (up to ≈25–50 MDa) transit through a carrier-mediated, signal-dependent process (facilitated translocation) (1–3).
Electron microscopy has revealed that the pore itself is approximately 40–90 nm in length and about 40–75 nm wide at its narrowest point. Flexible filaments extend out from the pore approximately 50 nm into the cytoplasm, and a filamentous open basket structure extends about 75 nm into the nucleoplasm (3–5). The NPC consists of approximately 30 different nucleoporins (Nups), about one-third of which lack an ordered secondary structure and contain domains rich in phenylalanine-glycine (FG) repeats (3, 6–7). These FG-Nups form a permeable and selective barrier in the NPC that inhibits the efficient translocation of large molecules (> 40 kDa) unless they are chaperoned by transport receptors. Importin β1 (Imp β1), one of the major transport receptors, recognizes a cargo molecule to form a transport complex either directly or indirectly (i.e., via Importin α) (8–10). Imp β1 promotes the movement of the transport complex through the NPC by a series of transient interactions with the FG-Nups. Thousands of FG repeats are distributed within an NPC, and Imp β1 is predicted to have up to 10 binding sites on its surface that interact with the FG repeats (7, 11–12). However, the questions of how these FG repeats spatially distribute along the NPC and how Imp β1 paves the way for facilitated translocation through the FG-Nups barrier under real-time trafficking conditions remain to be answered.
To provide noninvasive imaging of molecules transiting through the NPCs, single-molecule fluorescence microscopy approaches were developed to study the kinetics of nuclear transport. Wide- and narrow-field epifluorescence microscopy have been used to track transport receptors and a model cargo complex through the NPCs at the single-molecule level (13–17). The transport times of Imp β1 (≈7 ms) and its cargo complex (≈9 ms) through the NPCs were successfully visualized. The optimal precision of the spatial localization obtained by the above methods is about 20 nm for immobile molecules and approximately 40–50 nm for moving molecules in the NPCs with a temporal resolution of 2 ms (14, 15). To capture a series of transient interactions between Imp β1 and the FG-Nups in the NPC, higher spatial localization precision and faster temporal resolution are needed.
This study describes a method of single-point edge-excitation subdiffraction (SPEED) microscopy that allows the capture of a series of transient interactions between Imp β1 molecules and the FG-Nups through intact NPCs with a spatiotemporal resolution of 9 nm and 400 μs. With this improved resolution, transport pathways of Imp β1 alone and Imp β1-cargo complexes were imaged. These data were used to obtain 3D spatial density maps of interaction sites between the FG-Nups and the transiting molecules under real-time trafficking conditions.
Results and Discussion
SPEED Microscopy.
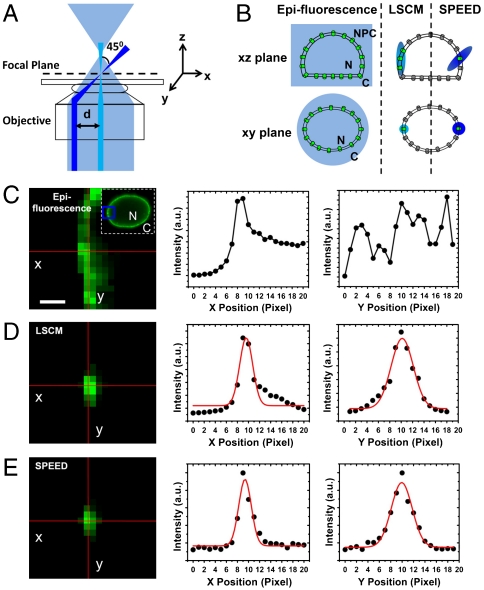
For SPEED microscopy, the method of single-point illumination and detection used was similar to that in the stationary mode of laser scanning confocal microscopy (LSCM) but with several modifications: (i) to obtain spatial information, a CCD camera was used to image the diffraction-limited spot rather than a photomultiplier tube, which is typically used for LSCM; (ii) to reduce the illumination volume in the axial direction, the laser beam was shifted off the center of the objective and focused into an inclined illumination volume in the focal plane (Fig. 1A); and (iii) there was no pinhole in the emission path. Typically, there were few or even no fluorescent objects illuminated outside the focal plane (see below). Thus, pinhole removal did not introduce noticeable background fluorescence but allowed the CCD camera to detect more signal. SPEED microscopy is designed to image only a single fluorescent NPC and to track fluorescent molecules transiting through this NPC.
Fig. 1.
SPEED microscopy. (A) Optics. The simplified optical diagram illustrates the different excitation beam paths of the SPEED (blue), the LSCM (cyan), and the wide-field epifluorescence (light blue) microscopy. The laser beam in the SPEED microscopy was focused into a diffraction-limited spot in the focal plane (dotted line) from the edge of the objective. An angle of 45° was formed between the iPSFs of the SPEED microscopy and the LSCM when the incident laser beam was shifted 237 μm (d) off the center of the objective (Fig. S2). (B) Illumination volumes in the three microscopes. The diagram demonstrates the NPCs inside (green) and outside (gray) the illumination volume in the xy and xz planes. N, nucleus; C, cytoplasm. (C) Multiple GFP-NPCs were excited using wide-field epifluorescence microscopy. The adopted area is enclosed by the blue box in the image of the entire fluorescent nuclear envelope (Inset). (Scale bar, 1 μm.) (D) GFP-NPCs were illuminated by the LSCM. The fluorescent spot was fit by a Gaussian function in both x and y directions. (E) Only a single GFP-NPC was excited in the illumination volume of the SPEED microscopy.
To obtain fluorescent NPCs, green fluorescent proteins (GFP) were conjugated to POM121, one of the Nups that lies at the center of the NPC scaffold (18–20). The centroid of a well-isolated GFP-NPC can be determined from a 2D elliptical Gaussian function (Fig. S1). In normal HeLa cells, the nearest neighbor distance for NPCs is 400–600 nm (21–23). In epifluorescence microscopy, simultaneous excitation of multiple NPCs on the equator of the NE and their greatly overlapped fluorescence prevents a precise localization of each NPC (Fig. 1 B and C). When an incident 488-nm laser beam is focused through the center of the objective as for LSCM, a single GFP-NPC is excited in the lateral dimension of the illumination volume [or the illumination point spread function (iPSF)], but two or more GFP-NPCs are typically excited in the axial dimension due to the lower axial resolution (≈210 nm in the xy plane and ≈540 nm in the z direction based on the Rayleigh criterion, Fig. 1 B and D). To reduce the illumination volume in the z direction, the incident 488-nm laser beam was shifted about 237 μm off the center of the objective to form an inclined iPSF (Fig. 1A and Fig. S2). The inclined iPSF (≈320 nm in the x, y, and z directions) illuminated only a single NPC in three dimensions. The fluorescence of a single GFP-NPC illuminated in this way could be fit well by a 2D elliptical Gaussian function (Fig. 1E). The standard deviation of successive centroid position measurements for 24 NPCs generated precisions of 0.9 ± 0.2 nm in the x direction and 2.5 ± 0.6 nm in the y direction.
A second laser (633 nm), with an inclined iPSF of approximately 360 nm in the x, y, and z directions, was used to excite single transiting molecules through the illuminated single NPC (Fig. S3). Single-molecule imaging resolution improves with photon counts, which can be enhanced by using a longer detection time or brighter emitters. Using a several-hundred-millisecond detection time or several-hundred-dye-labeled cargo molecules, the spatial localization precision can be improved to several nanometers (24–26). However, a submillisecond temporal resolution and functional fluorescent transiting molecules were needed for our experiments. Thus, neither a longer detection time nor heavily labeled transiting molecules could be used. Instead, a solution was found by squeezing maximum photons from a single lightly labeled molecule within a submillisecond detection time by generating a very high optical density in the illumination volume. At the same time, an on–off mode of laser excitation was adopted to enable a longer photobleaching time of transiting molecules compared to their transport time (Fig. S3). Finally, with 400-μs detection time and an average optical density of 500 kW/cm2 in the illumination area, around 1,100 photons were obtained from a single four-Alexa Flour 647-labeled substrate molecule. The corresponding localization precision was 9 ± 1 nm for the immobile fluorescent molecules and 10 ± 1 nm for the moving molecules (Fig. S4).
Capture of Single-Molecule Trajectories of Imp β1 Through the NPC.
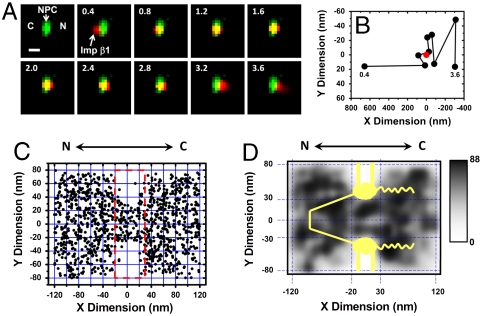
With the above improvements in the localization precision and the temporal resolution, the spatial locations of Imp β1 molecules through single intact NPCs were determined. A single NPC with an orientation perpendicular to the NE was illuminated by SPEED microscopy at the equator of a permeabilized HeLa cell nucleus (Fig. 1 and Fig. S1). Typical transport events of Imp β1 through an illuminated NPC are shown in Fig. 2A and in Movie S1 and S2. Imp β1 molecules were found to transport bidirectionally through the NPC with the same import (4.9 ± 1.9 ms) and export (5.0 ± 2.2 ms) times. These transport times agree with the previously measured dwell time obtained for Imp β1 molecules interacting with the NE without knowing whether the molecules underwent import or export (17). Single-molecule trajectories of Imp β1 were determined and their spatial locations relative to the centroid of the NPC were obtained (Fig. 2B). Inside the NPC, transiting molecules move approximately tenfold slower than in the outside compartments (15). Imp β1 molecules were primarily captured within a region around the centroid of the NPC, as shown in Fig. 2C. In the indicated region, we collected 1,093 spatial locations of Imp β1 undergoing transport through 10 single NPCs of 10 cells. Clearly, the spatial locations of Imp β1 molecules are restricted around the centroid of the NPC, and almost no spot is collected at the location of the NE, as expected. The restricted area has a width of approximately 40–60 nm and a length of approximately 50 nm (Fig. 2 C and D). Such dimensions agree with those of the nuclear central pore scaffold previously determined by electron microscopy (3–5). By application of a Gaussian blur filter function on Fig. 2C, the 2D spatial density of Imp β1 locations was obtained. The density map clearly indicates a nonhomogeneous spatial distribution of Imp β1 inside and outside the nuclear central pore (Fig. 2D).
Fig. 2.
Single-molecule trajectories and 2D spatial locations of Imp β1 in single NPCs. (A) A typical nuclear transport event of Imp β1 molecules from the cytoplasm to the nucleus. First, a single GFP-NPC (green spot) was visualized in the illumination volume. Then, a single fluorescent Imp β1 molecule (red spot) entered the illumination volume, starting in the cytoplasm (C), interacting with the NPC, and entering the nucleus (N). Numbers denote time in milliseconds. (Scale bar, 1 μm.) (B) Single-molecule trajectories of the transport event in A. Based on the centroid (red dot) and the dimensions of the NPC, the Imp β1 molecule was determined to interact with the NPC from 0.8 to 2.8 ms. (C) Superimposed plots of 1,093 spatial localizations of single Imp β1 molecules located primarily within a rectangular area of 240 × 160 nm. N, the nucleoplasmic side of the NPC; C, the cytoplasmic side of the NPC. (D) Two-dimensional spatial density map of Imp β1 locations. The locations of Imp β1 in each 20 × 20 nm area were quantized and filtered by a Gaussian blur function. The highest and lowest densities were 88 locations/μm2 and 0 locations/μm2, as shown by the gray level. A diagram of the NPC architecture (yellow) was superimposed on the density map.
3D Spatial Locations of Imp β1 in the NPC.
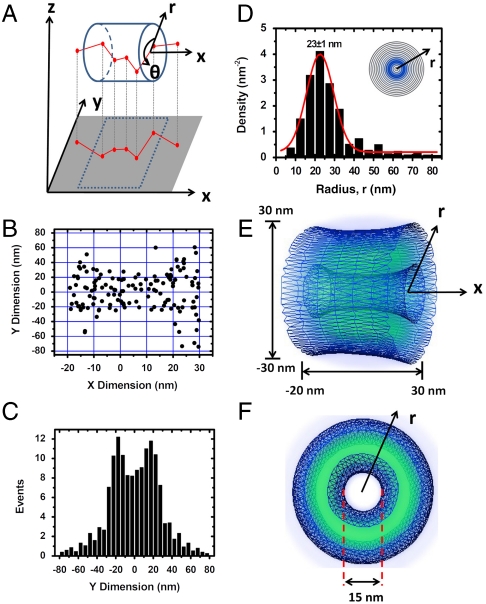
The obtained 2D spatial distribution of Imp β1 is a projection of its actual 3D spatial locations in the xy plane (Fig. 3A). These 3D spatial distributions can be described in a cylindrical coordinate system (r,θ,x). Coordinate (r,θ) represents each location at the cross-section of an NPC, and x refers to the position along the NPC axis (Fig. 3A). Regarding the determination of both the centroid and NPC orientation, the coordinates (r,x) of the Imp β1 3D spatial locations in the NPC can be defined, but not coordinate θ due to the lack of a reference point. If Imp β1 locations collected from multiple NPCs are superimposed based on overlapping centroids and aligned NPC orientations, the spatial distributions of Imp β1 in both the x and r dimensions could be enhanced; however, the distribution in the θ dimension would be almost a constant at each (r,x) coordinate. Thus, the cylindrical coordinate system can be simplified as (r,x). A deconvolution process was subsequently conducted to convert the Cartesian coordinates (x,y) to the simplified cylindrical coordinates (r,x) (Fig. S5). Histogram evaluation of Imp β1 locations in the nuclear central pore region suggests a nonuniform but symmetrical distribution in the y dimension (Fig. 3 B and C). By deconvoluting the histogram in the y dimension, the spatial density of the Imp β1 locations in the r dimension was obtained, and the Gaussian fitting of the spatial density revealed a maximal density at 23 ± 1 nm from the axis of the NPC (Fig. 3D). The corresponding 3D spatial density distribution of Imp β1 in the nuclear central pore was then extracted (Fig. 3E). In the central pore region, these results showed that Imp β1 rarely occupies the approximately 15-nm-diameter axial channel and instead primarily locates at the periphery of the channel (Fig. 3F).
Fig. 3.
A 2D to 3D deconvolution process. (A) A diagram to show that the obtained 2D spatial locations of Imp β1 are a projection effect of the actual 3D spatial locations of Imp β1 in the xy plane. (B) Two-dimensional spatial locations of Imp β1 in the central pore region enclosed in the red box in Fig. 2C. (C) Histogram of Imp β1 locations in the central pore region in the y dimension. (Bin size, 5 nm.) (D) The obtained histogram of spatial densities along the radius (r) at the cross-section of NPC in the central pore. The radii were used to plot concentric rings. The darker the ring, the higher the density. (Bin size, 5 nm.) (E and F) The obtained 3D spatial densities of Imp β1 (blue shaded region and isolated surface lines, brighter green color indicates higher density) in the central pore.
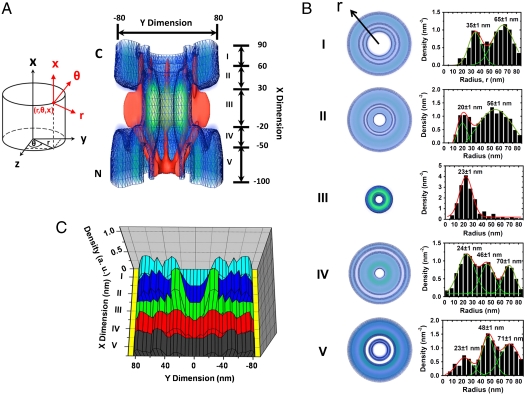
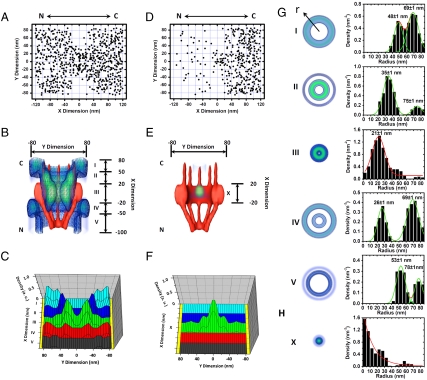
Such a deconvolution procedure was expanded to the 2D spatial locations of Imp β1 outside the central pore region, and a complete 3D spatial density map of Imp β1 locations within the NPC was extracted as shown in Fig. 4 and Movie S3. Based on the dimensions of the NPC physical structure (3–5), a 3D schematic of the NPC was plotted and superimposed onto the 3D spatial density map of Imp β1 (Fig. 4A). Five regions with distinct spatial location groups for Imp β1 along the NPC axis were found. Outside the central pore, the axial channel that is seldom occupied by Imp β1 still remains, and more Imp β1 location groups appear on the cytoplasmic and nucleoplasmic sides of the NPC. On the cytoplasmic side, one more group with a highest density at 56 ± 1 nm appears in range II, and two new groups emerge at 35 ± 1 nm and 65 ± 1 nm in range I (Fig. 4B). On the nucleoplasmic side, besides the location group at 23 nm, two more sets at 46 ± 1 nm and 70 ± 1 nm, respectively, were found in ranges IV and V (Fig. 4B). Outside the above regions (> 90 nm or < -100 nm), the distribution of the Imp β1 locations becomes random, and no further location groups were found (Fig. S6). By normalizing the spatial densities of Imp β1 locations in the above five regions, it was found that the density gradually increases from both sides and becomes highest in the central pore region (Fig. 4C).
Fig. 4.
A 3D spatial density map of interaction sites between Imp β1 and the FG-Nups. (A) Cutaway view of the 3D spatial density map of Imp β1 (blue shaded region and isolated surface lines, brighter green color indicates higher density) superimposed on the NPC architecture (red). Five regions with distinct spatial location groups for Imp β1 were marked from I to V with relative distances from the centroid of the NPC. Numbers denote the distance in nanometers. The Cartesian and cylindrical coordinate systems are shown. C, the cytoplasmic side of NPC; N, the nucleoplasmic side of NPC. (B) Histograms of spatial densities along the radii (r) at the cross-section of NPC in the range I–V. Major peaks were obtained by Gaussian fittings (green and red lines). (Bin size, 5 nm.) (C) Normalized densities of the interaction sites between Imp β1 and the FG-Nups in range I–V.
Imp β1 has multiple binding pockets by which it can interact with various FG-Nups (27–32), and numerous models have proposed that these interactions could promote the diffusion of Imp β1-cargo complexes through the NPC (33–37). However, the spatial distribution of these interactions within the NPC under real-time trafficking conditions remains unknown. As shown in Fig. 4, Imp β1 forms spatial location groups in regions I–V and becomes randomly distributed outside this range. The results in Fig. 5 indicated that cargo molecules alone possess a random distribution within the NPC and assemble into spatial location groups only when they form complexes with Imp β1. Therefore, the formation of spatial location groups may be driven by the interactions between the FG-Nups and Imp β1. The spatial location groups of Imp β1 in regions I–V comprise the actual pathway of Imp β1 through the NPC. This pathway may depend more on the spatial distribution of the interaction sites between Imp β1 and the FG-Nups than on the NPC architecture (Fig. 4A). A higher spatial density of Imp β1 locations in the central pore region may suggest that more or longer effective interactions between Imp β1 and the FG-Nups occur in the central pore than on the cytoplasmic and nucleoplasmic sides of the NPC (Fig. 4C).
Fig. 5.
Three-dimensional pathways for the import cargo complex and cargo alone. (A) Superimposed plots of 938 spatial localizations of single import cargo complexes located primarily within a rectangular area of 240 × 160 nm. N, the nucleoplasmic side of the NPC; C, the cytoplasmic side of the NPC. (B) Cutaway view of the 3D spatial density map of the import cargo complex (blue shaded region and isolated surface lines, brighter green color indicates higher density) superimposed on the NPC architecture (red). (C) Normalized densities of the interaction sites between the import cargo complexes and the FG-Nups in the range I–V. (D) Superimposed plots of 417 spatial localizations of single cargo molecules located within a rectangular area of 240 × 160 nm. (E) A 3D view of the inhibition barrier for cargo molecules (blue and green clouds) within the NPC architecture (red). (F) Normalized spatial densities of cargo locations. (G) Histograms of spatial densities for import complexes along the radii (r) at the cross-section of NPC in range I–V. (Bin size, 5 nm.) (H) Histogram of the spatial densities for cargo alone at the cross-section of NPC in range X. The histogram can be roughly fitted by an exponential decay function (red line). (Bin size, 5 nm.)
Clearly, Imp β1 rarely enters the NPC axial channel (Fig. 4A). Extensive simulations have further confirmed this observation (Figs. S7 and S8). Previous in vitro experiments have demonstrated that the FG-Nup153 (a Nup located at the nuclear basket) can be collapsed by Imp β1 binding and then released by the addition of RanGTP (38). Thus, the formation of the unoccupied axial channel could be caused by an insufficient presence of FG-Nups in the channel or by a collapse of FG-Nups initiated through the interaction with Imp β1. However, in permeabilized cells, it is unclear whether the collapse–release effect on the FG-Nups also occurs. To address this question, the 3D pathway of Imp β1 was measured in the presence of RanGTP. Eight hundred and fifteen spatial locations of Imp β1 from eight NPCs of eight cells were collected, and a 3D pathway was computed, as shown in Fig. S9. The obvious change is that the spatial density of Imp β1 on the nucleoplasmic side becomes less than that on the cytoplasmic side of the NPC, which agrees with the understanding that RanGTP facilitates Imp β1 dissociation from the nuclear basket (29, 39). However, the dimensions of the unoccupied axial channel and the spatial location of Imp β1 with the addition of RanGTP remained nearly the same as those of Imp β1 alone. The results suggested that the unoccupied central channel observed in the presence of Imp β1 may not have been generated by the Imp β1-mediated collapse of FG-Nups, although the possibility that the conformational change of the FG-Nups caused by Imp β1 and RanGTP was too slight or brief to be observed under the current experimental conditions could not be completely excluded.
3D Spatial Locations of Imp β1-Facilitated Translocation Through the NPC.
Large molecules are repelled from the NPC and require transport receptors to mediate their passage through the NPC. Whether the pathway of Imp β1-facilitated translocation through the NPC follows that of cargo-free Imp β1 is also of great interest. To map the routes taken by the Imp β1-cargo complex, single-molecule trajectories of a model import complex (Imp β1/Imp α/NLS-2xGFP) were tracked. NLS-2xGFP consists of a nuclear localization signal (PPKKKRKV) and two copies of green fluorescence protein. This model substrate for the classical import pathway has been well characterized (13–15). Following the same experimental procedure as employed for cargo-free Imp β1, 938 spatial locations of Imp β1-assisted four-Alexa Flour 647-labeled NLS-2xGFP through the NPC from nine NPCs of nine cells were collected, and a 3D pathway for the transiting cargo complexes was computed (Fig. 5 A and B, and Movie S4). The results indicated that the 3D pathway of import cargo complexes also consists of five effective interaction regions through the NPC. In region III, a single group of interaction sites with a peak at 21 ± 1 nm was found for the import cargo complexes (Fig. 5G). Clearly, the cargo-Imp β1 complex occupied more space of the central channel than did Imp β1 alone (Fig. 5B). In contrast, import cargo complexes in the other regions left a much larger unoccupied axial channel and had several different location groups compared to those of Imp β1 alone (Fig. 5G). As for the normalized spatial densities of interaction sites, the spatial density of the import cargo complex in the central pore was also the highest. The spatial densities on the nucleoplasmic side were found to be less than those on the cytoplasmic side of the NPC. As when cargo-free Imp β1 was treated with RanGTP, RanGTP added to the import system could also enhance the dissociation of the import complex from the nuclear basket (Fig. 5C) (29, 39).
The fundamental similarities between the pathways of the Imp β1-cargo complex and Imp β1 alone suggest that the interactions of cargo-bound Imp β1 with the FG-Nups through the NPC are similar to those of cargo-free Imp β1. The significant differences between the two pathways may be caused by a complicated combination of several factors: (i) RanGTP can induce a conformational change of Imp β1 or of the FG-Nups and thus affect the binding affinities between Imp β1 and the FG-Nups (27, 31, 32, 39, 40); (ii) the cargo and Imp α bound to Imp β1 in the cargo complex could affect the binding affinities of Imp β1 for the FG-Nups (27, 28, 30); and (iii) the location and orientation of the labeled cargo in the Imp β1-cargo complex could also result in a slightly different route from that of the Imp β1 of the complex.
To seek the location of the inhibition barrier in the NPC for large cargo molecules, a control experiment designed to measure the spatial location of NLS-2xGFP within the NPC without addition of Imp β1 and other transport cofactors was conducted. Based on 417 spatial locations of cargo molecules collected from eight NPCs of eight cells, the majority of captured spatial locations were on the cytoplasmic side of the NPC; approximately 10% of them were found on the nucleoplasmic side. The results are consistent with previous measurements showing that NLS-2xGFP alone has a very low transport rate through the NPC (13). However, no location groups for cargo molecules, such as those observed for Imp β1, were found on either side of the NPC, although the spatial locations of the cargo molecules were recorded in these regions (Fig. 5 D–F). The data strongly suggest that, without forming a complex with Imp β1/Imp α, NLS-2xGFP itself cannot make effective interactions with the FG-Nups, and, therefore, its movements are merely passive diffusion. Along the pathway, the cargo molecules are gradually barricaded on the cytoplasmic side and further inhibited in the central pore region (Fig. 5 E, F, and H; Movie S5).
Conclusions
SPEED microscopy allows 3D spatial density maps of transient interactions to be obtained for facilitated translocation through the NPC deep within permeabilized cells with a spatiotemporal resolution of 9 nm and 400 μs. This study demonstrated that the actual pathway for facilitated translocation might depend more on the interaction sites of the FG-Nups than on the architecture of the NPC. Furthermore, both cargo-free and cargo-bound Imp β1 were found to seldom occupy an axial channel through the NPC, and passive diffusion of the cargo alone (57 kDa) was inhibited in this channel. The channel could be the primary pathway for the passive diffusion of smaller molecules (≤ 20–40 kDa), as suggested by the spaghetti oil model and the reduction of dimensionality model (34, 36). It is unlikely that such a channel could be formed in the hydrogel meshwork barrier, as proposed by the selective phase model (35, 41). The spatial densities of interactions between the FG-Nups and cargo-free or cargo-bound Imp β1 were found to gradually increase from both sides to the center of the NPC. Complementary to the proposal by the virtual gating model that a single entropic barrier exists in the nuclear pore (37), these data may suggest an entropic barrier with a symmetrical gradient in the NPC (Fig. S10). The symmetrical gradient barrier includes a higher entropic barrier in the central pore and several lower barriers on either side. This gradient entropic barrier allows much easier diffusion of Imp β1-cargo complex from the cytoplasmic or the nucleoplasmic side to the center of the NPC while the NPC still provides selectivity.
The methodologies described herein can also be used to measure spatial density maps for more transport receptors and their cargo complexes in the NPCs. With a complete map of the 3D dynamic transport pathways for different-sized molecules and the spatial distribution of FG-Nups, our understanding of nuclear transport mechanism will be fundamentally advanced. This method could also be applied to the measurement of fast kinetics in other cellular cavities.
Materials and Methods
Further details of materials and methods are included in SI Text
A HeLa cell line stably expressing the GFP-conjugate of POM121 was adopted, and freshly split cells were grown overnight on coverslips in DMEM supplemented with 10% FCS. For microscopy, flow cells were constructed with a top coverslip and two lines of silicone grease as spacers. Cells were washed with transport buffer (20 mM Hepes, 110 mM KOAc, 5 mM NaOAc, 2 mM MgOAc, 1 mM EGTA, pH 7.3), permeabilized for 2 min with 40 μg/mL digitonin in transport buffer, and washed again with transport buffer supplemented with 1.5% polyvinylpyrrolidone (PVP; 360 kDa). PVP was included in all transport buffer solutions after digitonin treatment to prevent osmotic swelling of the nuclei.
For the efficient nuclear import of 1 nM NLS-2xGFP, the buffer contained the import cofactors 1 mM GTP, 0.5 μM importin α, 0.5 μM Imp β1, 2 μM Ran, and 1 μM NTF2. More details can be found in previously published methods (13–15). Labeled Imp β1 at 1 nM (without importin α and NTF2) was used to conduct single-molecule measurements of 3D pathways for cargo-free Imp β1 through the NPCs in the absence or presence of RanGTP (2 μM Ran and 1 mM GTP).
Supplementary Material
Acknowledgments.
We thank S.M. Musser for the plasmids of all proteins; B. Burke for the GFP-POM121 HeLa cells; P. Lu for the 488-nm and 633-nm lasers at the initial stage of the project; S.M. Musser, P. Lu, G. Bullerjahn, and P. Morris for critical comments on the manuscript. This project was supported by the Research Capacity Enhancement Grant (Bowling Green State University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0908269107/-/DCSupplemental.
References
- 1.Weis K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 2.Fried H, Kutay U. Nucleocytoplasmic transport: Taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fahrenkrog B, Aebi U. The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 4.Beck M, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 5.Rout M-P, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cronshaw J-M, et al. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denning D-P, et al. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart M. Molecular mechanism of the nuclear import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 9.Palmeri D, Malim M. Importin beta can mediate the nuclear import of an Arginine-Rich nuclear localization signal in the absence of Importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagstaff K, Jans D. Importins and beyond: Non-conventional nuclear transport mechanisms. Traffic. 2009;10:1188–1198. doi: 10.1111/j.1600-0854.2009.00937.x. [DOI] [PubMed] [Google Scholar]
- 11.Isgro T-A, Schulten K. Binding dynamics of isolated nucleoporin repeat regions to importin-β. Structure. 2005;13:1869–1879. doi: 10.1016/j.str.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Bednenko J, Cingolani G, Gerace L. Importin ß contains a COOH-terminal nucleoporin binding region important for nuclear transport. J Cell Biol. 2003;162:391–401. doi: 10.1083/jcb.200303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Gelles J, Musser S-M. Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA. 2004;101:12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W, Musser S-M. Visualizing single molecules interacting with nuclear pore complexes by narrow-field epifluorescence microscopy. Methods. 2006;39:316–328. doi: 10.1016/j.ymeth.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W, Musser S-M. Nuclear import time and transport efficiency depend on importin beta concentration. J Cell Biol. 2006;174:951–961. doi: 10.1083/jcb.200605053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubitscheck U, et al. Nuclear transport of single molecules: Dwell times at the nuclear pore complex. J Cell Biol. 2005;168:233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dange T, et al. Autonomy and robustness of translocation through the nuclear pore complex: A single-molecule study. J Cell Biol. 2008;183:77–86. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallberg E, Wozniak R-W, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122(3):513–521. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 20.Funakoshi T, et al. Two distinct human POM121 genes: Requirement for the formation of nuclear pore complexes. FEBS Lett. 2007;581:4910–4916. doi: 10.1016/j.febslet.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 21.Daigle N, et al. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldherr C-M, Kallenbach E, Schultz N. Movement of a karyophilic protein through the nuclear pores of oocytes. J Cell Biol. 1984;99:2216–2222. doi: 10.1083/jcb.99.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubitscheck U, et al. Single nuclear pores visualized by confocal microscopy and image processing. Biophys J. 1996;70:2067–2077. doi: 10.1016/S0006-3495(96)79811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 25.Yildiz A, Tomishige M, Vale R-D, Selvin P-R. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 26.Yildiz A, et al. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 27.Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 28.Conti E, Müller C-W, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16(2):237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Liu S-M, Stewart M. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-β homologue, Kap95p. J Mol Biol. 2005;349:515–525. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Bayliss R, et al. GLFG and FxFG nucleoporins bind to overlapping sites on Importin β. J Biol Chem. 2002;277:50597–50606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- 31.Otsuka S, et al. Individual binding pockets of importin-beta for FG-nucleoporins have different binding properties and different sensitivities to RanGTP. Proc Natl Acad Sci USA. 2008;105:16101–16106. doi: 10.1073/pnas.0802647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel S-S, Belmont B-J, Sante J-M, Rexach M-F. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 33.Rout M-P. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters R. Translocation through the nuclear pore complex: Selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 35.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macara I-G. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rout M-P, Aitchison J-D, Magnasco M-O, Chait B-T. Virtual gating and nuclear transport: The hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Lim R-Y, et al. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- 39.Lee S-J, Matsuura Y, Liu S-M, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 40.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295(5564):2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 41.Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.