Abstract
Legionella pneumophila is a Gram-negative opportunistic human pathogen that infects and multiplies in a broad range of phagocytic protozoan and mammalian phagocytes. Based on the observation that small regulatory RNAs (sRNAs) play an important role in controlling virulence-related genes in several pathogenic bacteria, we attempted to identify sRNAs expressed by L. pneumophila. We used computational prediction followed by experimental verification to identify and characterize sRNAs encoded in the L. pneumophila genome. A 50-mer probe microarray was constructed to test the expression of predicted sRNAs in bacteria grown under a variety of conditions. This strategy successfully identified 22 expressed RNAs, out of which 6 were confirmed by northern blot and RACE. One of the identified sRNAs is highly expressed in postexponential phase, and computational prediction of its secondary structure reveals a striking similarity to the structure of 6S RNA, a widely distributed prokaryotic sRNA, known to regulate the activity of σ70-containing RNA polymerase. A 70-mer probe microarray was used to identify genes affected by L. pneumophila 6S RNA in stationary phase. The 6S RNA positively regulates expression of genes encoding type IVB secretion system effectors, stress response genes such as groES and recA, as well as many genes involved in acquisition of nutrients and genes with unknown or hypothetical functions. Deletion of 6S RNA significantly reduced L. pneumophila intracellular multiplication in both protist and mammalian host cells, but had no detectable effect on growth in rich media.
Keywords: Legionella pneumophila genome, microarray, sRNA, sRNA prediction, competition assay
Legionella pneumophila is a Gram-negative opportunistic human pathogen that causes Legionnaires’ disease, a common nosocomial and community-acquired pneumonia. Infection occurs when aerosols of contaminated water are inhaled (1). In humans, alveolar macrophages are the primary type of cell that is infected, whereas in nature, many different species of protists can support L. pneumophila replication. Once internalized, the bacteria prevent phagosome acidification and fusion with lysosomes, and establish a permissive replication niche called the Legionella-containing vacuole (LCV). The LCV acquires membrane material derived from the Golgi and the endoplasmic reticulum (2). This is accomplished primarily by the action of effector proteins delivered to host cells by the Icm/Dot type IVB secretion system (TFBSS), a complex composed of 25 gene products that spans the L. pneumophila cell envelope (3). Currently, there are ∼150 confirmed effector proteins translocated by the Icm/Dot system (4, 5). Various regulatory proteins control the icm and dot genes encoding the TFBSS, as well as several genes encoding effectors. These include a sigma factor, σS; three two-component regulators, CpxR/A, PmrA/B, and LetA/S; and an RNA binding protein, CsrA (reviewed in ref. 6).
Recently, it has become evident that small, noncoding regulatory RNAs (sRNAs) also play important roles in the regulation of genes involved in virulence of several bacterial pathogens (7). sRNAs are short (40–500 nt) RNA molecules that typically do not encode proteins and mainly perform regulatory functions. The vast majority of sRNAs are posttranscriptional regulators that can either inhibit or enhance mRNA translation or affect mRNA stability by base pairing with their target mRNAs (8). In Escherichia coli and other bacterial species, the function of many of the sRNAs depends on their binding to the RNA binding protein Hfq (8). Other sRNAs regulate gene expression by binding to and interfering with regulatory proteins, like the well-known CsrA-CsrB system (9). In L. pneumophila, there are two CsrB homologs, RsmY and RsmZ, and they indirectly control several genes required for intracellular multiplication (6, 10, 11). Another example is the widely distributed 6S RNA, which binds to RNA polymerase (RNAP) holoenzyme σ70 during stationary phase and inhibits its binding to specific promoters, some of which contain an extended -10 element, thus selectively inhibiting transcription (12, 13). In laboratory E. coli strains, deletion of 6S RNA renders cells more resistant to high pH but less able to compete against wild-type bacteria for survival in deep stationary phase (14, 15).
In L. pneumophila, little is known about the existence and roles of sRNAs other than RsmY and RsmZ. Deletion of the hfq gene increases the duration of the lag phase after dilution of stationary-phase culture in fresh rich medium and slightly reduces intracellular growth in human macrophages, suggesting that sRNAs may be involved in regulation of genes related to interactions with the host cell (16). We therefore undertook to identify sRNAs encoded in the L. pneumophila Philadelphia-1 genome. We used computational prediction to identify putative sRNA genes and experimentally determined which ones were expressed by L. pneumophila under different growth and stress conditions. A total of six sRNAs were detected and validated by northern blotting and 5′ RACE, including the widely distributed 6S RNA, encoded by the ssrS gene. An L. pneumophila mutant strain deleted for the ssrS gene exhibited a significant decrease in its ability to grow in both protist and mammalian host cells.
Results
Prediction of sRNA-Encoding Genes in Legionella.
Computational prediction of putative sRNA genes in L. pneumophila was based on the identification of Rho-independent terminators in intergenic regions and sequence conservation of the regions preceding them, as was previously described (17, 18) (Methods). This approach led to the identification of 143 putative sRNA genes located in intergenic regions, ending with a predicted Rho-independent terminator and having sequence similarity with at least one other bacterial strain. Although the sequence comparison was carried out against 590 bacterial genome sequences available at the time of the analysis, none of the predicted sRNAs showed sequence conservations with other bacterial species except for other Legionella strains. The majority of predicted sRNAs showed high sequence similarity to all or at least one other L. pneumophila strain. Some predicted sRNAs, located in intergenic regions in the L. pneumophila Philadelphia-1 genome, correspond to small, annotated hypothetical open reading frames (ORFs) in L. pneumophila Paris or Lens. This discrepancy reflects the difficulty of correctly annotating genes encoding low molecular weight proteins (<150 residues) (19). Nonetheless, it is not known whether these small ORFs encode proteins and the corresponding sequences could encode sRNAs and/or polypeptides. Thus, the putative sRNAs located in regions potentially encoding annotated small ORFs in the other L. pneumophila strains were further characterized.
Detection of Transcribed Predicted sRNAs.
We designed a 50-mer oligonucleotide sRNA microarray to monitor transcription of the predicted sRNAs under different growth and stress conditions. We were able to design probes for only 101 of the predicted sRNAs because of cross-hybridization concerns and the possibility of false positives (see Methods for a complete description). RNA samples were prepared from L. pneumophila growing at exponential (E) and postexponential (PE) phases of growth in rich media (AYE) at 37 °C; following exposure to NaCl or hydrogen peroxide; growth at 30 °C; and after heat shock (from 30 °C to 37 °C). These stress conditions were chosen because they are linked to virulence; virulent strains of Legionella are sensitive to salt, oxidative stress is encountered during interaction with host cells, and temperature differences occur during infection of the human host from the inhalation of aerosolized water droplets from the environment. Because the σS factor was shown to be a major regulator of genes that contribute to intracellular growth in L. pneumophila, the level of the predicted sRNAs was also investigated in an rpoS null mutant during E and PE phases of growth. The RNA samples were converted to cDNA and labeled as described in Methods. The labeled cDNA samples were hybridized to the sRNA microarray and the signal intensities were measured as described. Normalized signal intensities for every sRNA under each condition are shown in Fig. 1 (Table S1). Hierarchical clustering of the data revealed two major clusters: (i) the majority of the predicted sRNAs (n = 79) showing weak (<50) and highly variable hybridization signals, and (ii) the remaining (n = 22) predicted sRNAs with much stronger and highly reproducible normalized signals ranging from 51 to 3,889, in at least one condition (black vertical bar in Fig. 1). As a comparison, the average normalized signal intensity of the negative controls was ∼2 and the signal of the positive control, 5S rRNA, was ∼4,600. To minimize false positives, we chose to focus on the most promising sRNAs: the ones found in the second cluster showing a strong and highly reproducible signal. Northern blot experiments were then performed to confirm the expression levels and to estimate the sizes of the 22 detected sRNAs (Table S1). Sixteen were either not detected, were putative 3′ untranslated regions, or were likely to encode putative small ORFs (SI Text and Table S1 and Fig. S5).
Fig. 1.
Detection of predicted sRNAs by microarray. Hierarchical clustering of normalized signal intensities of the predicted sRNAs during (in order, from left): wild-type exponential (E) and postexponential (PE) phases of growth in rich AYE medium at 37 °C; rpoS mutant (rpoS); after hydrogen peroxide stress (H2O2, 0.1 mM, 10 min); after salt stress (NaCl, 100 mM, 10 min); during growth at 30 °C in rich AYE medium; and after heat shock (HS, 30 °C to 37 °C in rich AYE medium). Black bars denote a cluster of putative sRNAs with strong and highly reproducible signal intensity that were studied further. For each condition, the signals from six replicates were analyzed.
Characteristics and Regulation of Six L. pneumophila sRNAs.
The remaining six putative sRNAs were all detected by northern blot and their sizes and hybridization patterns were consistent with bacterial sRNAs. We refer to five of these sRNAs as LprA–LprE (Legionella pneumophila sRNA A–E). The sixth sRNA was found to have a structure similar to the 6S RNA found in many bacterial species and is referred to as 6S RNA. No potential ORFs could be predicted in these sRNA sequences. In all cases, we mapped the 5′ end by RACE using tobacco acid pyrophosphatase to distinguish between 5′ ends of primary transcripts and those originating from processing of longer transcripts (18). The genomic locations and sizes of the sRNAs are listed in Table 1 and the results of structural predictions for LprA–E are shown in Fig. S1. In addition to microarrays, we used northern analysis to examine the expression of the sRNAs we identified under various growth conditions and in two mutant L. pneumophila strains to confirm expression patterns revealed by microarray (Fig. 2 and Fig. S2). In several cases, the differential abundance of these sRNAs observed in these analyses provides some preliminary insights into their potential functions. For example, the abundance of LprA was significantly reduced in a mutant strain lacking RpoS or the transcriptional regulator OxyR and up-regulated by H2O2 during E phase (Fig. 2A), a pattern identical to that seen for the E. coli sRNA OxyS that acts as a key regulator of the oxidative stress response (20). In Legionella, OxyR regulates genes involved in oxidative stress resistance but its activity is not regulated by oxidation, contrary to E. coli OxyR, and its role is therefore unclear (21). LprA seems to be regulated similarly to OxyS, but is larger (265 vs. 109 nt). Interestingly, LprA is encoded between lpg1803 and mutS, which are both predicted to be involved in DNA damage response. Together, these results suggest that LprA may have a role similar to OxyS as an antimutator. LprB expression was also affected by deletion of oxyR, but in contrast to LprA it was up-regulated in an OxyR-minus strain and was not regulated by RpoS (Fig. 2B). We also found structural similarity between LprD and VrrA (Fig. S1D), an sRNA involved in the regulation of ompA translation in Vibrio cholerae (22).
Table 1.
Small RNAs identified in this study
| sRNA | 5′ end* | 3′ end† | Length (nt) | BLAST‡ (% identity) |
| LprA | 2013775 | 2013510 | 265 | C (100), P (98), L (95) |
| LprB | 2022555 | 2022672 | 117 | L (100), C (99) |
| LprC | 978559 | 978676 | 117 | P (100), C (99), L (97) |
| LprD | 3321618 | 3321516 | 103 | C (99), L (98), P (98) |
| LprE | 3339400 | 3339350 | <50 | L (100), P (100), C (96) |
| 6S RNA | 951819 | 951673, 951638 | 147, 182 | P, L, C (100) |
*The 5′ end of small RNA was either identified by 5′ RACE (bold) or estimated based on northern blot observed size. See text for details.
†The 3′ end of small RNA is the last nucleotide of the Rho-independent terminator right before the stretch of U residues. For 6S RNA, the 3′ end was identified by 3′ RACE.
‡BLAST analysis hit only sequences in L. pneumophila strains Paris (P), Lens (L), or Corby (C).
Fig. 2.
Genomic context and expression of five of the detected sRNAs. For each identified sRNA, a northern blot was performed to confirm expression and regulation observed by microarray. E, exponential phase; PE, postexponential phase. Experiments were performed at least three times, and quantification of the northern blots is shown in Fig. S2. 5S rRNA is used as a loading control.
The sixth sRNA we detected by northern analysis is encoded between pntAa and lpg0877 (Fig. 3A) and was the predicted sRNA that produced the highest signal in the microarray analysis (Table S1). It was highly expressed when bacteria reached PE phase, which was confirmed by northern blot (Fig. 3B and Fig. S2F). There were two major products, one of around 180 nt and one of around 150 nt. The start of the primary transcript was determined by 5′ RACE to be at position 951819. Mapping of the 3′ end of the transcript was also performed to investigate the possibility of 3′ processing of the transcript to explain the detection of two major products by northern blotting. Indeed, two 3′ ends were found, one at the end of the predicted Rho-independent terminator and one 35 nt further upstream. Therefore, the upper band and lower band correspond to RNAs of 182 nt and 147 nt, respectively. The shorter fragment is likely generated by 3′ processing because there is no obvious Rho-independent terminator at this position. However, this 3′ trimming does not seem to be mediated by either RNase R or RNase III or to require Hfq, and will require further investigation (Fig. S3C). Secondary structures of both the 182- and 147-nt transcripts were predicted using mFold. The structure of the 147-nt transcript is shown in Fig. 3C because the only difference between the two structures was a 3′ tail comprising the Rho-independent terminator for the 182-nt transcript. The predicted structure was highly similar to the published consensus structure of the widely distributed 6S RNA that binds to RNAP (23, 24) (see SI Text for details). To confirm that this sRNA is 6S RNA, coimmunoprecipitation experiments were then performed, as previously described (25). For unknown reasons, only the 147-nt fragment was detectable after the coimmunoprecipitation reaction was carried out. Because the reaction was performed on ice and an RNase inhibitor was used, this may be due to processing by the same mechanism involved in vivo. This sRNA coimmunoprecipitated with RNAP-specific antibody (α-RpoB) but not with α-FliC antibody or protein A Sepharose beads alone (Fig. 3D). In contrast, 5S rRNA did not coimmunoprecipitate with the α-RpoB antibody. Because the signal of the coimmunoprecipitated 6S RNA on the northern blot was weak, we confirmed this result by RT-PCR. This weak signal is unlikely the result of low coimmunoprecipitation efficiency because other RNAP subunits were also coimmunoprecipitated with the α-RpoB antibody, as visualized by SDS/PAGE. Together, the structure prediction and coimmunoprecipitation studies strongly suggest that this sRNA is the L. pneumophila 6S RNA. Therefore, its coding sequence was named ssrS in compliance with published nomenclature recommendations (23). The microarray data suggested that L. pneumophila 6S RNA is induced by H2O2 and NaCl stresses in E phase, but this was not confirmed by northern blot (Table S1 and Fig. S3 A and B). Possible regulation by RpoS, OxyR, or LetS was investigated by northern blot analysis (Fig. S3 A and B), but none seemed to have a substantial effect on 6S RNA levels.
Fig. 3.
The sixth detected sRNA is the L. pneumophila 6S RNA. Genomic context of 6S RNA gene ssrS is shown in A. Northern blot was used to monitor expression of 6S RNA during growth in rich AYE medium over 24 h (B). The growth phase represented by each time point can be visualized on the growth-curve graph shown above the northern blot. This experiment was performed three times, and quantification of the northern blots is shown in Fig. S2. (C) Structure prediction of L. pneumophila 6S RNA reveals a structure similar to E. coli 6S RNA. (D) L. pneumophila 6S RNA coimmunoprecipitates with the core RNA polymerase subunit RpoB. 6S RNA and the negative control 5S rRNA were visualized by northern blot and the result was confirmed by RT-PCR. Lysates were obtained from PE-phase bacteria; the ssrS deletion strain was used as a control for specificity. Immunoprecipitaion with protein A Sepharose beads alone (Beads) and α-FliC antibody were used as negative controls. This experiment was performed four times with similar results.
6S RNA Regulates Genes Related to Host Interactions and Stress.
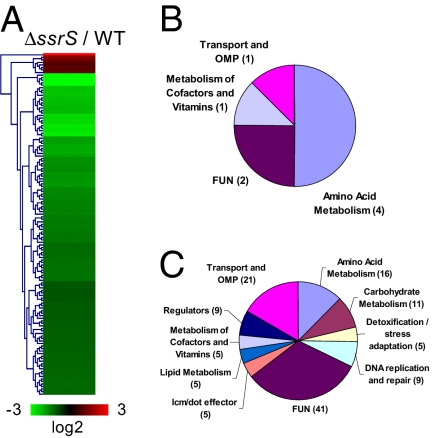
To investigate the effects of 6S RNA on global Legionella gene expression, a precise deletion of the ssrS gene was constructed by allelic replacement with a kanamycin marker (Fig. S3D). The steady-state level of gene transcripts during the PE phase of growth in the ssrS strain was compared with the wild-type strain by using a 70-mer oligonucleotide microarray that represents all annotated ORFs (6). The expression levels of 135 genes out of 3,005 (5%) were affected (±2-fold, P < 0.005) by the absence of 6S RNA (Fig. 4A and Table S2). Following functional grouping of the affected genes, it appears that four out of the eight negatively affected genes (showing increased expression in the mutant) are involved in amino acid metabolism (Fig. 4B). The majority of the positively affected genes (showing reduced expression in the mutant) clustered in three main functional groups: genes of unknown function, transport, and amino acid metabolism. Smaller functional groups included genes involved in detoxification and stress adaptation and in DNA repair and replication, as well as regulators and Icm/Dot effectors (Fig. 4C). Quantitative PCR (qPCR) was used to validate the expression pattern of four genes in the mutant and in a strain carrying a wild-type copy of ssrS under the control of its own promoter (including 300 bp upstream) on a plasmid (Fig. S3D). Regulation of groES, sdhC, and two TFBSS effectors, vipA and legC5, by 6S RNA was confirmed by qPCR because their expression was decreased in the mutant strain and was similar to the wild type in the complemented strain (Fig. S3E). Taken together, these results suggest that 6S RNA regulates a number of cellular functions related to intracellular multiplication. However, consistent with the lack of a major effect on icm/dot gene expression, TFBSS effector translocation activity was not impaired in the ssrS mutant (Fig. S4A).
Fig. 4.
Identification of genes regulated by 6S RNA. A 70-mer microarray representing all annotated ORFs was used to identify genes differentially regulated in a ΔssrS strain compared with the wild-type strain during PE phase of growth (A). Three biological replicates were used. Genes negatively (B) or positively (C) affected by the presence of 6S RNA were classified by their known or putative function, according to the L. pneumophila genome annotation. Shown is the fraction of affected genes in the different categories. FUN, genes of unknown function; OMP, outer membrane protein.
6S RNA Is Required for Optimal Intracellular Multiplication.
Expression of 6S RNA during infection of human cultured macrophage-like THP-1 cells was monitored by qPCR and induced 4-fold at late time points during intracellular growth relative to exponential growth in AYE broth, in which 6S RNA was expressed at its lowest level (Fig. S4 C and D). Deletion of the ssrS gene or an additional plasmid copy of the ssrS gene had no effect on Legionella growth in AYE rich medium (Fig. S4B). Wild-type and mutant strains were used to infect THP-1 cells and the protozoan host Acanthamoeba castellanii. The ssrS mutant strain showed a 10-fold reduction in intracellular growth after 72 h compared with the wild-type strain in both host cells (Fig. S4 E and F). Introduction of a wild-type copy (pssrS) in trans fully complemented this phenotype. Deletion or introduction of additional copies of the ssrS gene had no detectable effect on L. pneumophila cytotoxicity toward THP-1 cells as monitored by the MTT [1-(4,5-dimethylthiazol-2-yl)-3,5-diphenylformazan] assay (Fig. S4G).
Because the differences between the abilities of the wild-type and ssrS deletion strains to replicate in host cells were relatively small, we used competition assays to more precisely study the intracellular growth phenotype of the ssrS deletion, as described previously (26). In E. coli, the phenotype associated with loss of 6S RNA was discovered by using a competition assay for growth in bacteriological media (15). To control for the effect of the antibiotic resistance markers, we designed the assay using a set of four strains (ssrS+-CmR, ssrS+-KnR, ΔssrS-CmR, and ΔssrS-KnR). The wild-type strain (KS79) used to construct the ssrS deletion strain is a marker-less comR deletion mutant that renders the bacteria hypercompetent for DNA uptake. We therefore used an isogenic comR deletion mutant that contains either a kanamycin or chloramphenicol cassette in place of comR. We also constructed a ΔssrS deletion strain with a chloramphenicol cassette by allelic exchange of the kanamycin cassette from the original ΔssrS deletion strain. Host cells were infected at a multiplicity of infection of 0.1 with mixtures of these strains as stated in the legend of Fig. 5, and the colony-forming unit of each strain was determined by spreading samples on plates containing kanamycin or chloramphenicol (15). As can be seen in Fig. 5 A and B (black and blue lines), coculture of the two wild-type strains (ssrS+-CmR and ssrS+-KnR) or the two mutant strains (ΔssrS-CmR and ΔssrS-KnR) resulted in equal representation of each differentially marked strain [log competitive index (CI) ≈ 0] in either A. castellanii or THP-1 cells. This shows that the CmR or KmR markers had no significant effect on the ability of the strains to replicate intracellularly in either type of host cell. As expected, the wild-type strain (ssrS+-CmR) greatly outcompeted the dotA mutant (Log CI < −7) in both hosts (Fig. 5 A and B, purple line). Coculture of the ssrS+ strains with the ΔssrS strains resulted in decreasing CI over time (Log CI < −4 at 96 h) in both hosts, regardless of the marker used (Fig. 5 A and B, green and red lines). Because we used two sets of strains in which the CmR or KmR marker were reversed, it is unlikely that the observed effect was due to their presence. Because the intracellular growth experiments included several rounds of replication, we also asked whether the differences in the abilities of the ssrS+ strains and ΔssrS to replicate in host cells were due to a general decreased ability of the ΔssrS strain to grow or to transition between multiple growth cycles in rich media. We grew mixtures of the strains in rich AYE medium (Fig. 5C) and exposed the mixed populations to three cycles of 1:100 dilution with fresh AYE following stationary phase (Fig. 5D). Coculture of the differentially marked ssrS+ strains or the differentially marked ΔssrS strains and coculture of ssrS+ with ΔssrS strains resulted in equal representation of each strain (Log CI ≈ 0) in all cases (Fig. 5 C and D). Therefore, we conclude that 6S RNA performs a function that is necessary for optimal competitive growth of L. pneumophila in both mammalian and protist hosts but is dispensable for competitive growth in rich media.
Fig. 5.
The ssrS mutant strain is unable to compete with the wild type during intracellular multiplication. THP-1 cells (A) or A. castellanii cells (B) were infected with a 1:1 mixture of differentially marked ssrS+ and ΔssrS strains, as well as a mixture of icm+/dot+ and dotA mutant as control (see text for details). The mixtures were also grown in rich AYE medium at 37 °C (C) and exposed to cycles of dilution (1:100) into fresh AYE medium after growth to PE phase (D). Mixtures of bacterial strains are as follows: black, ssrS+-Cmr/ssrS+-Knr; blue, ΔssrS-Cmr/ΔssrS-Knr; red, ssrS+-Cmr/ΔssrS-Knr; green, ssrS+-Knr/ΔssrS-Cmr; purple, ssrS+-Cmr/ΔdotA-Knr. Values represent the average ± SD of three experiments; *P < 0.0001.
Discussion
The main goal of this study was to identify and characterize small RNAs expressed by the intracellular pathogen L. pneumophila. To this end, we conducted an in silico search for sRNA-encoding genes in intergenic regions and then used custom-made microarrays produced from oligonucleotides spotted on glass slides for experimentally testing the expression of 101 predicted L. pneumophila sRNAs (Fig. 1). This flexible and relatively inexpensive approach allowed us to detect 22 intergenic transcripts as well as gain important insights into their condition-dependent regulation. Transcripts corresponding to the remaining 79 predicted loci displayed weak or hypervariable signals under all of the experimental conditions tested and were not studied further. These may only be expressed at levels detectable by microarray under specific growth conditions that were not included in our study. In addition, some of the predicted sRNA-encoding genes are likely false-positive hits, such as those that may correspond to conserved untranslated regions of mRNAs.
We were able to confirm six sRNAs (LprA–E and 6S RNA) expressed by L. pneumophila (Figs. 2 and 3 and Table 1). None of them was found in other genera by BLAST analysis, even when using permissive search parameters. This is not surprising, because sRNA sequence similarity has been only observed between closely related species (27). Of note, the Rfam website (http://rfam.sanger.ac.uk) contains a large database of 1,013 predicted 6S RNA sequences, based on the algorithm of Barrick et al. (23), which constructed covariance models based on manual multiple alignments of 6S RNA sequences. The predicted sequence for Legionella 6S RNA is located at position 951829–951668, which is in agreement with our finding (Table 1).
Based on expression pattern, LprA could be involved in oxidative stress response or act as an antimutator like OxyS. The functions of LprB–E are currently unknown and will require more work. Another sRNA we identified coimmunoprecipitated with core RNAP and its predicted structure highly resembles the structure of E. coli 6S RNA, strongly suggesting it corresponds to the L. pneumophila 6S RNA (Fig. 3). In E. coli, 6S RNA is encoded in a polycistronic operon upstream of the ygfA ORF and is generated by cleavage of the transcript. In contrast, L. pneumophila 6S RNA is not part of an operon and possesses its own promoter and a putative Rho-independent terminator at its 3′ end, as also found in certain groups of bacteria (23). We also note that L. pneumophila 6S RNA is processed at its 3′ end by an as yet unknown mechanism. It is currently unknown whether the 182-nt fragment is also able to bind to RNAP because it was not detected after the coimmunoprecipitation reaction. Processing of 6S RNA has been observed in other bacteria, but whether this processing has functional implications remains unclear (23).
We identified a number of genes whose expression is positively affected by 6S RNA, including genes involved in amino acid metabolism, stress adaptation, and a subset of Icm/Dot effectors (Fig. 4). The effects of 6S RNA on these genes could be indirect, because several regulatory genes were also found to be positively regulated. The fact that many more genes are positively affected by the presence of 6S RNA is somewhat contradictory to the current view of 6S RNA function, which is to inhibit the activity of σ70-containing RNAP (Eσ70) at σ70-dependent promoters. A recent study (13) found that E. coli promoters negatively regulated by the presence of 6S RNA contain a weak -35 element and an extended -10 element. It is currently unknown whether this is also the case in other bacterial species. Our data suggest that the 6S RNA of L. pneumophila does not act as a main repressor of genes during stationary phase, because relatively few genes are negatively affected by 6S RNA. In E. coli, genes known to be positively regulated by 6S RNA are σS-dependent (15). The exact mechanism is currently the subject of debate and is presumably a result of the sequestration of Eσ70 that allows the remaining core polymerase to bind to other σ factors (12, 28). Comparison of genes regulated by σS (6) and genes regulated by 6S RNA in Legionella reveals 17 genes (out of 749 genes affected by rpoS deletion or ssrS deletion) that are significantly affected in both mutants (Fig. S3F). However, only 4 were positively regulated by σS and 6S RNA and 1 was negatively regulated by both. The remaining 12 were affected in an opposite manner in each mutant (Fig. S3F). Although σS is required for the increased expression of 310 genes in PE phase (6), the roles of σ70 or other σ factors and their competition for core RNAP are not clear. Our results suggest that in PE phase, 6S RNA influences transcription by RNAP holoenzymes other than EσS because the majority of genes positively affected by 6S RNA are not σS-dependent. In E. coli, the vast majority of 6S RNA is bound to RNAP (25) whereas in Legionella only a fraction seems to coimmunoprecipitate with it (Fig. 3D), perhaps due to weaker binding. Taken together, our results suggest that the mechanism underlying regulation of gene expression by 6S RNA in Legionella could be different from in E. coli, adding a new level of complexity to the current view of 6S RNA mainly being a negative regulator. Understanding the mechanisms by which 6S RNA affects genes in L. pneumophila will require further studies.
The lack of 6S RNA resulted in a decreased ability to replicate in phylogenetically diverse protist and mammalian hosts (Fig. S4 E and F) and to compete against the wild type for growth inside cells (Fig. 5). This effect could be the result of decreased survival or replication of the ssrS mutant. These assays cannot distinguish between those two possibilities. However, because many genes involved in nutrient acquisition were positively affected by 6S RNA, we suggest that the mutant is in a nutritionally disadvantaged state inside cells, which results in a slower growth rate. Further experiments will be required to fully understand the mechanism involved. Because there is such a strong selection for Legionella pneumophila to survive and replicate in protists in the environment, even the modest effect of 6S RNA on intracellular replication indicates that it plays a critical role for Legionella’s ability to survive or replicate in the environment and act as a pathogen.
Methods
Prediction of Candidate sRNAs.
The predictive algorithm first identified the intergenic regions in the genome of the L. pneumophila Philadelphia-1 genome, and then searched them for Rho-independent terminators. To this end, we used the search module for the Rho-independent terminator developed by us previously based on their sequence and structure properties (18), as well as two other search strategies, RNAMotif (29) and TransTerm (30), implemented in the sRNAPredict2 algorithm (17). The sequence upstream of the terminator was then checked for conservation against the whole-genome sequences of 590 bacterial strains available at the time of the analysis, including the other three sequenced strains of L. pneumophila: Paris, Lens, and Corby. Therefore, if Rho-independent terminators have conserved upstream sequences in at least one other bacterial strain, this sequence is considered as a putative sRNA gene. Conservation was checked using BLAST version WUBLAST 2.0. An alignment was considered conserved if the e-value was smaller than 10−5 and the percent identity was above 70%. The maximum space allowed between the 3′ end of the conserved region and the 5′ end of a predicted Rho-independent terminator was 20 nt.
sRNA Microarray Design, Hybridization, and Data Analysis.
A microarray was designed for the detection of the predicted sRNAs by using OligoWiz version 2.1.3 (31, 32). The prokaryotic setting was used to design one 50-mer probe located, when possible, at the 3′ end of the predicted sRNA gene. For many genes, the score of the only possible probe was not perfect (0.7 < score < 0.9), because the target sequence was in many cases very small (<300 nt), resulting in low but significant cross-hybridization potential. For 42 predicted sRNAs, the score of the only possible probe was very low (<0.7), and to minimize false positives we decided not to print probes for those predicted sRNA genes. Probes for 10 negative controls, representing 10 genes of the L. pneumophila Paris plasmid, were also designed. Probes (Illumina) were dissolved in 50% DMSO to a final concentration of 30 μM and printed in triplicate on UltraGAPS-coated glass slides (Corning) using a PerkinElmer SpotArray microarray printer. Fifteen micrograms of total RNA was labeled during cDNA synthesis using SuperScript II reverse transcriptase (Invitrogen) and amino-allyl dUTP (Sigma). Bacterial genomic DNA was used as the reference channel on each slide to allow comparison of each time point and different samples (33). Five micrograms of genomic DNA (gDNA) was labeled with amino-allyl dUTP by using Klenow fragments and random primers (Invitrogen) at 37 °C for 18 h. DNA was subsequently coupled to the succinimidyl ester fluorescent dye Alexa Fluor 546 (for cDNA) or Alexa Fluor 647 (for gDNA) (Invitrogen) following the manufacturer's protocols. Hybridization and data acquisition were performed as previously described (6). Very low density arrays, like the sRNA microarray used here, cannot be normalized with common procedures such as total intensity or LOWESS. Local background was removed from spot signal intensity and the noise signal was estimated by recording the average signal intensity of 10 negative controls printed on the chip. Normalization of signal intensity was carried out by calculating the fold increase over the noise signal value. Correlation of replicates using this normalization procedure was ≥0.95.
Additional Methods.
Bacterial strains (Table S3), primers (Table S4), growth conditions, RNA extraction and purification, northern blot analysis, global gene expression profiling by microarray, qPCR, RACE, structure prediction, coimmunoprecipitation, and infection of host cells were all performed as described in SI Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant AI064481 (to H.A.S.). H.A.S. was supported during the beginning of the project as a Visiting Professor of the Lady Davis Foundation at The Hebrew University. S.P.F. is a recipient of a postdoctoral fellowship from the National Sciences and Engineering Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE19451).
This article contains supporting information online at www.pnas.org/cgi/content/full/0911764107/DCSupplemental.
References
- 1.Fraser DW, et al. Legionnaires’ disease: Description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 2.Shin S, Roy CR. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 2008;10:1209–1220. doi: 10.1111/j.1462-5822.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 3.Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burstein D, et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franco IS, Shuman HA, Charpentier X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 2009;11:1435–1443. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 6.Hovel-Miner G, et al. σS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J Bacteriol. 2009;191:2461–2473. doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 10.Sahr T, et al. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06677.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasis M, Segal G. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol. 2009;72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 12.Wassarman KM. 6S RNA: A regulator of transcription. Mol Microbiol. 2007;65:1425–1431. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- 13.Cavanagh AT, Klocko AD, Liu X, Wassarman KM. Promoter specificity for 6S RNA regulation of transcription is determined by core promoter sequences and competition for region 4.2 of σ70. Mol Microbiol. 2008;67:1242–1256. doi: 10.1111/j.1365-2958.2008.06117.x. [DOI] [PubMed] [Google Scholar]
- 14.Trotochaud AE, Wassarman KM. 6S RNA regulation of pspF transcription leads to altered cell survival at high pH. J Bacteriol. 2006;188:3936–3943. doi: 10.1128/JB.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trotochaud AE, Wassarman KM. 6S RNA function enhances long-term cell survival. J Bacteriol. 2004;186:4978–4985. doi: 10.1128/JB.186.15.4978-4985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNealy TL, Forsbach-Birk V, Shi C, Marre R. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J Bacteriol. 2005;187:1527–1532. doi: 10.1128/JB.187.4.1527-1532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Argaman L, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 19.Rudd KE, Humphery-Smith I, Wasinger VC, Bairoch A. Low molecular weight proteins: A challenge for post-genomic research. Electrophoresis. 1998;19:536–544. doi: 10.1002/elps.1150190413. [DOI] [PubMed] [Google Scholar]
- 20.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 21.LeBlanc JJ, Brassinga AK, Ewann F, Davidson RJ, Hoffman PS. An ortholog of OxyR in Legionella pneumophila is expressed postexponentially and negatively regulates the alkyl hydroperoxide reductase (ahpC2D) operon. J Bacteriol. 2008;190:3444–3455. doi: 10.1128/JB.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song T, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol. 2005;12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- 25.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 26.Beuzón CR, Holden DW. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 2001;3:1345–1352. doi: 10.1016/s1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 27.Pichon C, Felden B. Small RNA gene identification and mRNA target predictions in bacteria. Bioinformatics. 2008;24:2807–2813. doi: 10.1093/bioinformatics/btn560. [DOI] [PubMed] [Google Scholar]
- 28.Gildehaus N, Neusser T, Wurm R, Wagner R. Studies on the function of the riboregulator 6S RNA from E. coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA-directed de novo transcripts. Nucleic Acids Res. 2007;35:1885–1896. doi: 10.1093/nar/gkm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macke TJ, et al. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. Prediction of transcription terminators in bacterial genomes. J Mol Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen HB, Wernersson R, Knudsen S. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 2003;31:3491–3496. doi: 10.1093/nar/gkg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wernersson R, Nielsen HB. OligoWiz 2.0—Integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res. 2005;33:W611–W615. doi: 10.1093/nar/gki399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talaat AM, et al. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 2002;30:e104. doi: 10.1093/nar/gnf103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.