Abstract
We report a unique mutation in the D-amino acid oxidase gene (R199W DAO) associated with classical adult onset familial amyotrophic lateral sclerosis (FALS) in a three generational FALS kindred, after candidate gene screening in a 14.52 cM region on chromosome 12q22-23 linked to disease. Neuronal cell lines expressing R199W DAO showed decreased viability and increased ubiquitinated aggregates compared with cells expressing the wild-type protein. Similarly, lentiviral-mediated expression of R199W DAO in primary motor neuron cultures caused increased TUNEL labeling. This effect was also observed when motor neurons were cocultured on transduced astrocytes expressing R199W, indicating that the motor neuron cell death induced by this mutation is mediated by both cell autonomous and noncell autonomous processes. DAO controls the level of D-serine, which accumulates in the spinal cord in cases of sporadic ALS and in a mouse model of ALS, indicating that this abnormality may represent a fundamental component of ALS pathogenesis.
Keywords: motor neuron disease, neurodegeneration, linkage analysis, ubiquitinated aggregates, apoptosis
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset neuromuscular disease, characterized by progressive muscle weakness, atrophy, paralysis, and death from respiratory failure. Approximately 5% of ALS cases have an autosomal dominant mode of inheritance, and clinically these cases are indistinguishable from sporadic cases of ALS (SALS). One-fifth of FALS cases are caused by mutations in copper/zinc-dependent superoxide dismutase (SOD1) (1). Recently, mutations have been detected in the gene encoding TAR DNA binding protein 43 (2, 3), a major component of the polyubiquitinated cytoplasmic inclusions characteristic of ALS, and the Fused in sarcoma (FUS) gene, ALS6 (4, 5). Mutations have also been identified in additional genes, causing rare, atypical forms of ALS: vesicle-associated membrane protein/ synaptobrevin-associated membrane protein B gene (VAPB), which is associated both with late-onset spinal muscular atrophy and ALS, ALS8 (6); alsin, associated with juvenile onset recessive ALS, ALS2 (7); and senataxin associated with a slowly progressive form of juvenile onset ALS, ALS4 (8). Additional FALS loci have been reported for classical ALS, ALS3 on 18q identified in a single family (9), ALS with frontotemporal dementia on 9q21-q22 (10), ALS with frontotemporal dementia on chromosome 9p21.3-p13.3 (11, 12), and ALS7 on chromosome 20p (13). Missense mutations have also been reported in both sporadic and familial cases in FIG4, a phosphoinositide phosphatase, which causes a peripheral neuropathy (14) and in angiogenin in cases predominantly of Celtic origin (15). However, at present known genes only account for ≈30% of familial cases of classical ALS and the remaining genes require elucidation.
We carried out a whole genome screen with microsatellite markers in families with confirmed ALS and lacking known FALS mutations and identified a unique FALS locus on chromosome 12 in a single kindred. Candidate genes within the region were screened, and we report here a unique description of a mutation in the D-amino acid oxidase (DAO) gene, located within this locus, which causes classical adult onset familial ALS (FALS). We also provide evidence for the pathogenic effects of this mutation on cell viability, which are associated with the formation of ubiquitinated aggregates. DAO controls the level of D-serine, which accumulates in the spinal cord in sporadic ALS and a mouse model of ALS, indicating that this abnormality may represent a fundamental component of the pathogenesis of both sporadic and familial cases of ALS.
Results
Clinical Description of FALS Patients in a Chromosome 12-Linked Family (MB1).
A diagnosis of ALS was confirmed clinically in affected individuals in this family according to the El Escorial World Federation of Neurology criteria for Diagnosis of ALS with evidence of both upper and lower motor neuron involvement. Bulbar signs were also present. In the index case, the patient presented with weakness in one hand and arm with small muscle wasting. The age at onset was 40 years and the duration of illness was 21 months. A similar rapid disease progression was seen in other affected family members, with an average age at death of 44 years (range 42–55 yr from seven cases). An autopsy was available for individual 3.7, an obligate carrier who died at 73 years from cardiac failure (previously admitted to investigate right-sided weakness and speech difficulties), which showed some loss of motor neurons at the cervical and lumbar level and degeneration of one of the lateral corticospinal tracts visible at all levels. Frontal and temporal lobes were well preserved. TDP-43 staining in motor neurons was clearly localized to nuclei with no TDP-43 positive nuclear or cytoplasmic inclusions (Fig. S1). There was no evidence of cognitive impairment in cases from which DNA was sampled. No mutations were detected in SOD1, VAPB, TAR DNA binding protein, or FUS. This study has been approved by the Riverside Research Ethics Committee (Hammersmith Hospitals NHS Trust) with appropriate informed consent from subjects.
Linkage Analysis.
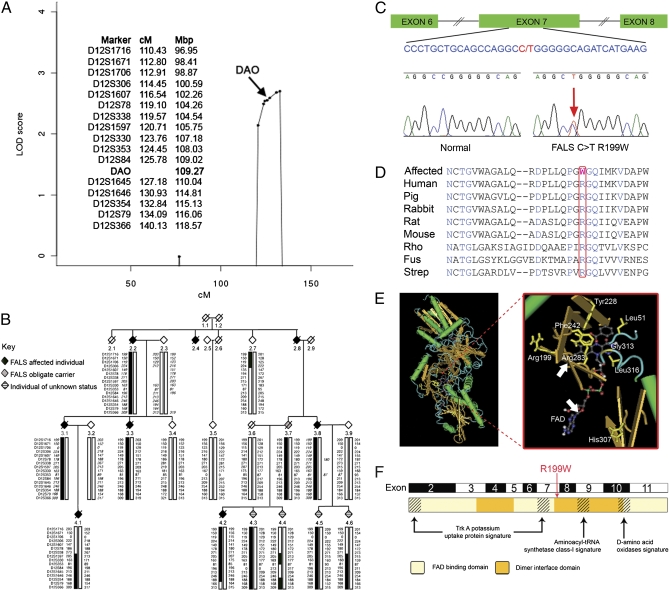
We evaluated a whole genome screen on four FALS kindreds by using microsatellite markers. Multipoint analysis was performed by using Genehunter v2.1 r3 assuming an autosomal dominant model. A maximum logarithm of odds (LOD) score of 2.97 was identified on chromosome 12 (D12S1706 to D12S79), with most of the signal deriving from a three generational FALS kindred (MB1). No LOD scores greater than +0.8 were obtained in chromosomes 1–11 and 13–22 using this family. After fine mapping of chromosome 12, multipoint parametric analysis of family MB1 using MERLIN v1.1.2 revealed significant LOD scores within a region flanked by D12S338 and D12S79 that contains 146 RefSeq genes and 148 Ensembl genes. Maximum LOD scores of 2.70 were obtained at markers D12S1646 and D12S354 (Fig. 1A). NPL analysis highlighted the same region with the highest LOD score of 1.94 (P = 1.4 × 10−3) at marker D12S330.
Fig. 1.
Detection of a R199W mutation in DAO and a protein model. (A) Multipoint analysis of family MB1 (Merlin v1.1.2) showing significant maximum parametric LOD scores between markers D12S338 and D12S79. Marker distances (MAP-O-MAT) obtained from http://compgen.rutgers.edu/mapomat/reference.html and sequence map positions obtained from http://www.ncbi.nlm.nih.gov are given for the region containing significant LOD scores on chromosome 12. (B) Haplotype analysis of the 12q21-24 region in family MB1. Allele data for 16 markers in the 12q21-24 region are shown; inferred genotypes are indicated by italics. Markers are listed on the far left hand side of each generation within the family. The haplotype associated with the disease for the index case 4.2 has been indicated by thick solid “boxing.” Cross-over events are indicated by a change in the boxing. Both males and females are indicated by a diamond symbol. A diagonal line indicates that the individual is deceased. (C) Graphical representation of DAO exons 6–8 showing an expansion of part of exon 7. The R199W point mutation is indicated in red together with the corresponding “normal” and “mutated” sequence electrograph. (D) DAO amino acid conservation of the mutated region among eight species. The position of Arg199 (boxed) is indicated in red and variant amino acids in black. Conserved residues are shown in blue. Rho, Rhodotorula gracilis; Fus, Fusarium Solanii; Strep, Streptomyces Avermitilis. (E) Ribbon representation of porcine DAO homodimer (PDB:1AA8, MMDB:5815) and an expanded view of the DAO active site showing the position of Arg199 and FAD (arrows) in relation to key residues including Tyr228, His307, Leu51, Phe242, Arg283, Gly313, and Leu316. (F) DAO protein domains indicating FAD binding and dimer interaction domains together with other potential functional domains.
Initially, a limited number of candidate genes in the region were sequenced, including thioredoxin reductase-1, ataxin 2, and D-amino acid oxidase (DAO), which resulted in the identification of a disease-associated mutation in three affected individuals (3.3, 4.1, and 4.2; Fig. 1B) plus one confirmed obligate carrier (3.7) transforming a C to T in codon 199 of DAO, resulting in an amino acid substitution of arginine by tryptophan (R→W) at this position (Fig. 1C). In addition, the mutation was present in three “at risk” individuals (4.3, 4.5, and 4.6) sharing part of the disease haplotype and absent from three unaffected individuals (2.7, 3.4, and 3.5) and spouses (3.9 and 3.6). The ages of the three “at risk” individuals carrying the R199W DAO mutation, 4.3, 4.5, and 4.6, at the time of sampling were 33, 44, and 48 years, respectively, one of whom has been referred to a neurologist to investigate motor symptomatology. We screened for the R199W mutation in a total of 1,002 unrelated individuals (780 controls, 23 sporadic ALS cases, and 199 FALS cases, all of Caucasian origin residing in the United Kingdom, as was the index case and family) to determine its prevalence in the general population, but no further individuals carried this mutation. The R199W mutation in exon 7 was found in 0 of 780 unrelated Caucasian controls and in one index case representing the 12q linked ALS kindred (P < 0.001 using Fisher's exact test). DAO sequencing of a representative affected individual from 196 different FALS families lacking SOD1 mutations (at least 100 individuals per exon) was carried out but no other coding alterations were found. A total of 13 unique and 6 known variants were detected, 17 were intronic/3′ flanking and 2 were exonic (one synonymous SNP and one in the 3′ UTR) (Table S1). Ten of the unique polymorphisms detected were present at low frequencies (<2%).
Other genes and ORFs in the region known to be expressed in spinal cord were also sequenced to investigate the possibility that a second gene mutation in linkage disequilibrium with the DAO mutation was present. The genes sequenced included a serine dehydratase-like enzyme (SDSL) involved in serine metabolism, regulatory factor ×4 (RFX4), a transcription factor involved in transcriptional repression, RNA binding motif protein 19 (RBM19), a nucleolar protein containing six RNA-binding motifs, BRAP, which encodes a protein that binds to the nuclear signal of BRCA1 and other proteins preventing nuclear targeting, glycolipid transfer protein (GLTP) that facilitates glycolipid transfer between membranes, and a vacuolar sorting protein (VPS29) (Table S2), but no further disease-associated mutations were found. Although this sequencing did not yield mutations in genes expressed in spinal cord, other genes present in this chromosomal region cannot be excluded.
DAO (EC.1.4.3.3) is a peroxisomal flavin adenine dinucleotide (FAD)-dependent oxidase present in most organisms and mammalian tissues, which catalyses the oxidative deamination of D-amino acids such as D-alanine, D-serine, and D-proline to their corresponding keto-acids. In the mammalian CNS, DAO is enriched in brainstem and spinal cord. It was only recently recognized that D-amino acids are not only synthesized in the CNS, but also serve essential physiological functions as exemplified by D-serine, which is an activator/coagonist at the N-methyl-D-aspartate (NMDA) glutamate receptor subtype (16). Human DAO is a 39.4-kDa protein consisting of 347 amino acids. Arg199 is highly conserved between mammalian and lower organisms such as yeast, fungi, and bacteria (Fig. 1D). Arg199 lies close to the FAD binding site (Fig. 1E) and between residues Tyr228 and His307, which play a key role in enzyme activity (17). The regions containing FAD binding and dimer interface domains are shown in Fig. 1F. DAO is found in both neuronal and glial cells, the most intense labeling being seen in hindbrain within gigantocellular reticular and facial nuclei, Bergmann glial cells and Purkinje cells of the cerebellum (ref. 18; and confirmed in Fig. S2). In addition, we show here the presence of DAO in gray matter of spinal cord including motor neurons (Fig. 2A).
Fig. 2.
Localization of DAO and functional characterization of WT and R199W DAO. (A) Localization of DAO in mouse lumbar spinal cord. DAO immunoreactivity is widespread in gray matter including motor neurons (arrowheads in the enlarged image). DAPI nuclear counterstain is shown in blue. (Scale bars: Left, 500 μm; Right, 50 μm.) (B) Localization of GFP-tagged wild-type (WT) and R199W DAO in peroxisomes. COS-7 cells were transiently transfected with GFP-tagged DAO and transduced with peroxisomal organelle lights. GFP-tagged WT and R199W DAO (GFP), peroxisomal (Perox) OFP organelle lights and merged (Merge) images with DAPI nuclear staining. (Scale bars: 20 μm.) (C) WT and R199W DAO coimmunoprecipitate. COS-7 cell lysates were cotransfected with untagged and GFP-tagged WT or R199W DAO and immunoprecipitated with anti-GFP antibody. Blotting for DAO identified bands at ≈37 kDa and ≈67 kDa in size, corresponding to untagged and GFP-tagged DAO protein, respectively. (D) Effect of mutation on WT DAO enzyme activity determined using untagged DAO constructs. DAO enzyme activity in COS-7 cells cotransfected with WT and R199W DAO (2 μg of each DNA compared with single transfections using 2 μg of DNA). One-way ANOVA P < 0.0001 with Bonferroni post hoc test indicated. **, P < 0.01. Values are means ± SEM, n = 6. Inset shows a representative experiment using GFP-tagged constructs. Note that higher levels of enzyme activity relative to lysate protein were obtained with the untagged constructs compared with the GFP-tagged constructs because of the greater transfection efficiency.
The effect of the R199W mutation on enzyme activity was examined in post mortem spinal cord available from individual 3.7, an obligate carrier, expressing the R199W mutation, compared with four controls and seven sporadic ALS cases. cDNA amplified from spinal cord mRNA yielded a similar size product present in all spinal cord samples, indicating that DAO mRNA was generated in the individual heterozygous for the R199W mutation (Fig. S3). DAO enzyme activity assayed in control and ALS spinal cord was similar but was markedly reduced in the case expressing the R199W mutation (measured twice by using two different tissue samples) (Table 1), despite normal levels of mRNA expression, which reflects the potent effect of this mutation on the function of the wild-type enzyme. DAO enzyme activity and mRNA levels were much lower in motor cortex than the spinal cord for all cases including individual 3.7. Immunohistochemistry of DAO in multiple regions from this individual confirmed the presence of DAO protein in cell regions both affected (spinal cord) and unaffected (cerebellum and ventral reticular nucleus) by the pathology (Fig. S1).
Table 1.
DAO enzyme activity in spinal cord
| Tissue sample | DAO enzyme activity, units/mg of protein |
| Control spinal cord | 1.501 ± 0.213 × 10−3 (4) |
| Sporadic ALS spinal cord | 1.721 ± 0.295 × 10−3 (7) |
| FALS spinal cord (Individual 3.7) | 0.065 × 10−3 (two separate samples) |
| Rat spinal cord (reference sample) | 1.473 × 10−3 |
DAO enzyme activity was assayed in spinal cord in 4 controls [mean age, 53 yr (range: 20–87 yr) and mean post mortem interval, 27.38 h (range: 14.5–41 h)], seven sporadic ALS cases [mean age, 67 yr (range: 49–89 yr) and mean post mortem interval, 15.29 h (range: 5–30 h)] and individual 3.7 who is heterozygous for the R199W mutation, using a peroxidase coupled method modified from Nagata et al. (26). A rat spinal cord sample was also assayed in parallel. Values are means + SEMs for the number of cases given in parentheses.
GFP-tagged DAO constructs for wild-type and R199W DAO transfected into COS-7 cells yielded sharply delineated punctate staining, indicative of peroxisomal localization (Fig. 2B). The peroxisomal localization was confirmed by transduction with a fluorescent marker containing a peroxisomal signal peptide. Both wild-type and mutant GFP-tagged DAO proteins colocalized with the peroxisomal marker (Fig. 2B). A similar peroxisomal distribution was found for an untagged DAO construct by using a DAO antibody and fluorescently labeled secondary antibody (Fig. S4A). We also evaluated the effect of the mutation on DAO enzyme activity in cell lysates from COS-7 cells transfected with GFP-tagged wild-type DAO, which yielded an activity of 0.29 ± 0.06 mU/100 μg of cell lysate protein (mean ± SD) from three independent experiments. There was no detectable enzyme activity in lysates from untransfected cells, cells transfected with the empty GFP vector, or GFP-tagged R199W DAO, as found in the spinal cord analysis of the autopsy case expressing the R199W mutation.
DAO has been shown to exist as a homodimer in man, being the major form of both the apoenzyme and holoenzyme. To determine whether heterodimers were formed between wild-type and R199W DAO and whether this interaction affected enzyme activity, GFP-tagged wild-type DAO and untagged R199W DAO were cotransfected in COS-7 cells. Cell lysates from these cotransfections were immunoprecipitated with anti-GFP antibody and blotted with anti-DAO antibody. As expected, GFP immunoprecipitation pulled down the 67-kDa GFP-tagged protein, whether wild-type or R199W, but in addition the 37-kDa untagged protein was also pulled down (Fig. 2C). As the wild-type and mutant DAO coimmunoprecipitated, we investigated whether wild-type protein enzyme activity was affected by coexpression with R199W DAO. COS-7 cells were cotransfected with untagged wild-type and R199W DAO, and enzyme activity was measured in the resulting lysates. Cotransfection caused a significant attenuation in enzyme activity compared with wild-type DAO, despite the presence of similar levels of DAO protein in the double transfection compared with the combined single transfections (Fig. S4B), indicating that DAO was not exclusively present as homodimers but that heterodimers were also formed, which contributed to the reduced enzyme activity, suggesting a potential dominant negative effect of the mutation on the wild-type protein (Fig. 2D).
To determine whether R199W DAO affected cell survival, we assayed TUNEL labeling in primary rat motor neuron cultures transduced with lentiviral vectors expressing wild-type and R199W DAO (Fig. S5). Motor neurons expressing R199W DAO showed a 5-fold increase in the proportion of TUNEL-positive cells compared with untransduced motor neurons. Motor neurons expressing either wild-type DAO or the control marker gene, deltaLNGFR (∆LNGFR) showed a 2- to 2.5-fold increase in TUNEL-positive cells, which was not significantly different from untransduced cells (Fig. 3A). However, the number of TUNEL-positive cells expressing R199W was significantly increased compared with cells expressing wild-type DAO and control cells expressing the vector (Fig. 3A and Fig. S5). Because DAO is known to be abundantly expressed in astrocytes as well as motor neurons, primary astrocytes were transduced with lentiviral vectors expressing wild-type DAO, R199W DAO, or ∆LNGFR and primary motor neurons were layered above. Fewer TUNEL-positive cells were detected in cocultures compared with motor neuron cultures, (2.56% and 3.81%, respectively, in uninfected cells). However, as with motor neuron cultures, a significant increase in the number of TUNEL-positive cells was detected in cocultures with astrocytes infected with R199W compared with wild-type DAO (Fig. 3A). The reason for the lack of difference between the vector and the mutant in the coculture experiment despite obtaining a significant difference in infected motor neurons alone, where the effect of mutation is significant over both wild-type DAO and ∆LNGFR, is unknown. One possible reason might be that expressing ∆LNGFR molecules on the surface of astrocytes might interfere with motor neuron/astrocyte interactions.
Fig. 3.
Effect of R199W DAO on cell viability. (A) Effect of R199W DAO in lentiviral vector transduced motor neurons and cocultures of untransduced motor neurons with lentiviral vector transduced astrocytes. Motor neurons or astrocytes were transduced with the following lentiviruses at DIV1: WT DAO, R199W DAO or a negative control, ΔLNGFR and TUNEL-positive cells quantified at DIV3 (fold change over TUNEL-positive cells in untrans-duced cells). One-way ANOVA for motor neurons alone (n = 4) and coculture of untransduced motor neurons plated on transduced astrocytes (n = 3) was P = 0.043 and P = 0.0241, respectively, with Bonferroni post hoc test value indicated. *, P < 0.05. Values are means ± SEM. (B) Effect of R199W DAO on ubiquitin aggregates. NSC-34 cells expressed GFP-tagged WT or R199W DAO 72 h after transfection. Ubiquitin (UBQ) staining with aggregates in GFP-positive cells are indicated by arrows and merged (Merge) images with DAPI nuclear staining. (Scale bars: 20 μm.) (C) Effect of R199W DAO on number of ubiquitin aggregates and NSC-34 cell morphology. Cells containing ubiquitin aggregates (≥0.5 μm in diameter) were quantified after treatment with DMSO vehicle control (n = 4) (Vehicle) and 1 μg/mL tunicamycin (n = 3) (Tunicamycin), giving one-way ANOVA values of P = 0.0057 and P = 0.044, respectively. Cells with atypical morphology (rounded, shrunken, and with fewer processes) were quantified (Morphology). One-way ANOVA P = 0.0128 (n = 4) with Bonferroni post hoc test indicated. *, P < 0.05; **, P < 0.01. Values are means ± SEM.
The present results clearly indicate that the disease-associated mutation R199W reduces cell viability. To explore these mechanisms further we analyzed the effect of the mutation on protein aggregation, ubiquitination, response to ER stress, and cell morphology by using the motor neuron cell line NSC-34. At 72 h after transfection, cells expressing R199W DAO showed a variety of abnormal features including atrophied or shrunken cells and loss of projections affecting the pattern of DAO staining (Fig. S6), compared with cells expressing wild-type DAO. A prominent feature of R199W cells was the presence of ubiquitinated protein aggregates (0.5 μm or greater in diameter) (Fig. 3B) and significant increases were detected between wild-type and R199W DAO expressing cells when aggregate-containing cells were quantified (Fig. 3C). After application of tunicamycin, an ER stress inducer, the significant increase in the number of ubiquitin positive aggregates was maintained in cells expressing R199W DAO compared with controls (Fig. 3C). The number of atypical shrunken atrophic cells lacking projections (under control conditions) was also increased in R199W DAO compared with wild-type and control cells (Fig. 3C).
Discussion
The kindred described in this paper predominantly exhibits a severe phenotype with death occurring in the early 40s in 75% of cases. Incomplete penetrance was also evident in an obligate carrier, but neuropathological assessment of this case at autopsy demonstrated motor neuron loss and corticospinal tract degeneration at multiple levels. We identified a mutation in DAO in the chromosomal region segregating with disease and screened for mutations in other genes in this region that are expressed in spinal cord but found no variants associated with disease. However, we cannot exclude the possibility that another mutation may be in linkage disequilibrium with the R199W DAO mutation. Although a causal role for the R199W mutation in DAO in ALS remains to be established unequivocally, increasing evidence supports the involvement of D-serine in ALS. We have demonstrated here that this disease-associated mutation in DAO, R199W, not only causes almost total loss of enzyme activity but also impairs cell viability, promotes the formation of ubiquitin aggregates, and increases apoptosis in neuronal cells. Increased apoptosis was also observed when motor neurons were cocultured on transduced astrocytes expressing R199W, indicating that motor neuron cell death induced by this mutation can be mediated by both cell autonomous and noncell autonomous processes. We have also found evidence of the interaction of the mutant protein with the wild-type protein. It is not possible at present to determine whether the neurotoxic effect of R199W DAO results from the accumulation of the aberrant protein or whether the impaired enzyme activity contributes to pathogenesis. The major consequence of this loss of enzyme activity is the buildup of D-amino acids, of particular interest being D-serine, a coagonist at the glycine site of the NMDA glutamate receptor enhancing glutamate transmission. Animals lacking DAO activity (a spontaneous mutant G181R DAO, ddY/DAO−) have elevated levels of D-serine in the CNS and show enhanced hippocampal LTP, improved performance in spatial learning tests (19), enhanced NMDA-receptor-mediated excitatory postsynaptic currents in spinal cord dorsal horn neurons (20), and reduced locomotor activity (21).
In ALS spinal cord and in the G93A SOD1 mouse model of ALS, levels of D-serine and serine racemase (SR), which converts L-serine into D-serine (22), are markedly elevated (23). D-serine is detected early in disease in G93A SOD1 mice, accumulating around vacuolated motor neurons. Subsequently, D-serine levels increase substantially, initially in astrocytes and towards the end stage in activated microglia, where SR is mainly localized. The initial trigger for induction of SR in the mouse model may be the accumulation of mutant SOD1 protein and/or local inflammatory conditions, which are known to induce SR through the c-Jun-N-terminal kinase pathway (24). Glutamate action at AMPA receptors also induces SR and D-serine release from astrocytes (25). Interestingly, enzymes involved in D/L serine metabolism (e.g., phosphoserine phosphatase and 3-phosphoglycerate dehydrogenase) are among the earliest transcriptional changes detected in motor neurons in the G37R SOD1 mouse (26).
D-serine plays a key role in ALS pathogenesis, indicating the contribution of astrocytes and microglia to disease progression, the effects of which would be potentiated through the R199W DAO mutation. It is also likely that protein aggregation in motor neurons (seen in this study) would occur before glial activation and play a significant role in the initiation of disease in familial cases. The role of different cell types in disease progression is also clearly seen in the SOD1 mouse model of ALS where onset and progression of disease are determined by motor neurons and microglia, respectively (27). Future studies should focus establish the role of R199W DAO in the pathogenesis of ALS by using in vivo models in order to determine the potential of DAO as a molecular target in ALS and other age-related neurodegenerative disorders.
Materials and Methods
Linkage Analysis.
For all individuals used in the linkage study, genomic DNA was extracted from whole blood or buffy coat by standard techniques. The whole genome screen was performed with a total of 378 microsatellite markers spaced ≈10 cM apart. Multipoint parametric linkage analysis of genotyping data were performed by using Genehunter version 2.1 r3. Subjects were classified as affected (2), unaffected (1), and unknown (0). The analysis was carried out by using an affected only model assuming that FALS was inherited in an autosomal dominant manner. The frequency of the mutant allele was taken as 0.0001. Simple counting estimates were used to determine marker allele frequencies from at least 52 individuals. A total of 33 microsatellite markers spanning 104.37 cM were used to fine map the linked region on chromosome 12. Simulation calculations by using SIMLINK version 4.12 were used to demonstrate the statistical power of the family to detect genetic linkage. Multipoint parametric and nonparametric linkage (NPL) analysis of the fine-mapped region was conducted by using MERLIN (see SI Text for additional details). NPL linkage analysis used all of the affected individuals including the obligate carrier (3.3, 4.1, 4.2, and 3.7), unaffected individuals >63 years (2.7, 3.4, and 3.5), unaffected spouses (3.6 and 3.9), and 6 at risk. A reduced pedigree was used for the parametric linkage analysis with one less at risk individual.
Mutation Analyses.
See SI Text for additional details.
DAO Enzyme Activity of Human Spinal Cord, Human Motor Cortex, and Rat Spinal Cord.
Tissue was homogenized in 0.02 M sodium pyrophosphate pH 8.3 containing 1.4 × 10−5 M FAD, centrifuged for 1 min at 4 °C and supernatant was removed for DAO enzyme assay determination by using a peroxidase coupled system modified from Nagata et al. (28). One enzyme unit is defined as the amount of enzyme required to oxidize one micromole of D-serine per min at 37 °C and pH 8.3 (see SI Text for additional details).
DAO Constructs and Lentiviral Vectors.
Cell Culture of COS-7 and NSC-34 Cells.
See SI Text for additional details.
Tissue Immunohistochemistry.
See SI Text for additional details.
Motor Neuron and Motor Neuron-Astrocyte Cocultures.
Immunostaining and TUNEL assay of Primary Cultures.
See SI Text for additional details.
Western Blotting and Immunoprecipitation.
See SI Text for additional details.
DAO enzyme activity in cell lysates.
Enzyme activity was determined by using transfected COS-7 cell lysates, and the protocol was developed by Gabler et al. (32) by using D-proline as substrate (see SI Text for additional details).
Statistical Analysis.
Cell culture data (mean ± SEM) was tested for normality before analysis. Statistical differences between groups were established by using one-way ANOVA followed by the Bonferroni post hoc test. In all cases differences were considered significant if P < 0.05.
Supplementary Material
Acknowledgments
We thank Juliet Greenwood, Ian Gardiner, Richard Orrell, Derek Anane, and Ruth Jones for their contribution to our FALS research. We also thank Drs. Luigi Naldini and Mario Amendola (San Raffaele Telethon Institute, Milan) for providing the bidirectional lentiviral vector plasmids, Dr. Susan Greensmith (Institute of Neurology, London) and Prof. Mimoun Azzouz (Sheffield University, Sheffield, UK) for help and protocols for the primary motor neuron cultures, Prof. Silvia Arber (Basel University, Basel) for providing the Hb9 antibody, Prof. Neil Cashman for providing the NSC-34 cell line and Drs. Roncaroli and Gentleman (Imperial College London) for TDP-43 staining. We are grateful to the Smith's Charity, the American ALS Association, the Haywood Foundation, the Motor Neurone Disease Association, the Garfield Weston Foundation, and Land Securities Group for funding our research into FALS and to the patients and their relatives involved in this research for their willing cooperation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914128107/DCSupplemental.
References
- 1.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 4.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 5.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimura AL, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadano S, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 8.Chen YZ, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hand CK, et al. A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am J Hum Genet. 2002;70:251–256. doi: 10.1086/337945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosler BA, et al. Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA. 2000;284:1664–1669. doi: 10.1001/jama.284.13.1664. [DOI] [PubMed] [Google Scholar]
- 11.Morita M, et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66:839–844. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- 12.Vance C, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 13.Sapp PC, et al. Identification of two novel loci for dominantly inherited familial amyotrophic lateral sclerosis. Am J Hum Genet. 2003;73:397–403. doi: 10.1086/377158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow CY, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenway MJ, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 16.Snyder SH, Kim PM. D-amino acids as putative neurotransmitters: Focus on D-serine. Neurochem Res. 2000;25:553–560. doi: 10.1023/a:1007586314648. [DOI] [PubMed] [Google Scholar]
- 17.Todone F, et al. Active site plasticity in D-amino acid oxidase: A crystallographic analysis. Biochemistry. 1997;36:5853–5860. doi: 10.1021/bi9630570. [DOI] [PubMed] [Google Scholar]
- 18.Moreno S, Nardacci R, Cimini A, Cerù MP. Immunocytochemical localization of D-amino acid oxidase in rat brain. J Neurocytol. 1999;28:169–185. doi: 10.1023/a:1007064504007. [DOI] [PubMed] [Google Scholar]
- 19.Maekawa M, Watanabe M, Yamaguchi S, Konno R, Hori Y. Spatial learning and long-term potentiation of mutant mice lacking D-amino-acid oxidase. Neurosci Res. 2005;53:34–38. doi: 10.1016/j.neures.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Wake K, et al. Exaggerated responses to chronic nociceptive stimuli and enhancement of N-methyl-D-aspartate receptor-mediated synaptic transmission in mutant mice lacking D-amino-acid oxidase. Neurosci Lett. 2001;297:25–28. doi: 10.1016/s0304-3940(00)01658-x. [DOI] [PubMed] [Google Scholar]
- 21.Almond SL, et al. Behavioral and biochemical characterization of a mutant mouse strain lacking D-amino acid oxidase activity and its implications for schizophrenia. Mol Cell Neurosci. 2006;32:324–334. doi: 10.1016/j.mcn.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Wolosker H, Blackshaw S, Snyder SH. Serine racemase: A glial enzyme synthesizing D-serine to regulate glutamate-N-methyl-D-aspartate neurotransmission. Proc Natl Acad Sci USA. 1999;96:13409–13414. doi: 10.1073/pnas.96.23.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasabe J, et al. D-serine is a key determinant of glutamate toxicity in amyotrophic lateral sclerosis. EMBO J. 2007;26:4149–4159. doi: 10.1038/sj.emboj.7601840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S, Barger SW. Induction of serine racemase by inflammatory stimuli is dependent on AP-1. Ann N Y Acad Sci. 2004;1035:133–146. doi: 10.1196/annals.1332.009. [DOI] [PubMed] [Google Scholar]
- 25.Kim PM, et al. Serine racemase: Activation by glutamate neurotransmission via glutamate receptor interacting protein and mediation of neuronal migration. Proc Natl Acad Sci USA. 2005;102:2105–2110. doi: 10.1073/pnas.0409723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobsiger CS, Boillée S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc Natl Acad Sci USA. 2007;104:7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boillée S, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 28.Nagata Y, Shimojo T, Akino T. Two spectrophotometric assays for D-amino acid oxidase: For the study of distribution patterns. Int J Biochem. 1988;20:1235–1238. doi: 10.1016/0020-711x(88)90225-x. [DOI] [PubMed] [Google Scholar]
- 29.Amendola M, Venneri MA, Biffi A, Vigna E, Naldini L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat Biotechnol. 2005;23:108–116. doi: 10.1038/nbt1049. [DOI] [PubMed] [Google Scholar]
- 30.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 31.Gingras M, Gagnon V, Minotti S, Durham HD, Berthod F. Optimized protocols for isolation of primary motor neurons, astrocytes and microglia from embryonic mouse spinal cord. J Neurosci Methods. 2007;163:111–118. doi: 10.1016/j.jneumeth.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 32.Gabler M, Hensel M, Fischer L. Detection and substrate selectivity of new microbial D-amino acid oxidases. Enzyme Microb Technol. 2000;27:605–611. doi: 10.1016/s0141-0229(00)00262-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.