Abstract
We identified a p53 target gene, phosphate-activated mitochondrial glutaminase (GLS2), a key enzyme in conversion of glutamine to glutamate, and thereby a regulator of glutathione (GSH) synthesis and energy production. GLS2 expression is induced in response to DNA damage or oxidative stress in a p53-dependent manner, and p53 associates with the GLS2 promoter. Elevated GLS2 facilitates glutamine metabolism and lowers intracellular reactive oxygen species (ROS) levels, resulting in an overall decrease in DNA oxidation as determined by measurement of 8-OH-dG content in both normal and stressed cells. Further, siRNA down-regulation of either GLS2 or p53 compromises the GSH-dependent antioxidant system and increases intracellular ROS levels. High ROS levels following GLS2 knockdown also coincide with stimulation of p53-induced cell death. We propose that GLS2 control of intracellular ROS levels and the apoptotic response facilitates the ability of p53 to protect cells from accumulation of genomic damage and allows cells to survive after mild and repairable genotoxic stress. Indeed, overexpression of GLS2 reduces the growth of tumor cells and colony formation. Further, compared with normal tissue, GLS2 expression is reduced in liver tumors. Thus, our results provide evidence for a unique metabolic role for p53, linking glutamine metabolism, energy, and ROS homeostasis, which may contribute to p53 tumor suppressor function.
Keywords: glutathione antioxidant, glutaminolysis, tumor suppression, apoptosis
Recent evidence implicates p53 in regulation of cell metabolism, energy production, autophagy, and levels of reactive oxygen species (ROS) (1–3). In cancer cells, glucose uptake is much higher than in most normal tissues, and glycolysis persists even under aerobic conditions, a process known as the Warburg effect (4, 5). p53 plays several roles in regulating this process. For example, p53 decreases the glycolytic rate through inhibiting expression of glucose transporters (6) and phosphoglycerate mutase (7) while increasing the expression of TIGAR that reduces fructose-2,6-bisphosphate levels (8). Nevertheless, some studies report that p53 can promote at least some steps in glycolysis (9, 10). In addition, p53 has the ability to help maintain mitochondria (11, 12) and drive oxidative phosphorylation through the transcriptional activation of subunit I of cytochrome c oxidase (13), increased expression of cytochrome c oxidase (SCO2) (14), and induction of the ribonucleotide reductase subunit p52R2 (15).
Glutamine metabolism is another target for alteration in cancer development. Both glutamine uptake and the rate of glutaminolysis (i.e., catabolism of glutamine to generate ATP and lactate in the mitochondria) are known to increase in tumors (1, 16). In glutamine metabolism, mitochondrial glutaminase (GLS) is central in the conversion of glutamine to glutamate. Glutamate participates in regulation of mitochondrial bioenergetics in many normal and cancer cells via the tricarboxylic acid cycle for ATP production as well as antioxidant defense through GSH synthesis. The two different phosphate activated GLS isoforms GLS1 (kidney-type) and GLS2 (liver-type) in mammals are encoded by separate genes on different chromosomes (17–19). Previous data have suggested that GLS1 up-regulation is associated with increased rates of proliferation, whereas GLS2 prevalence seems to correspond with resting or quiescent cell states (17). Here we found that GLS2 is a p53-inducible gene that functions to regulate the energy supply and to protect against oxidative stress.
Results
GLS2 Is a p53 Target Gene.
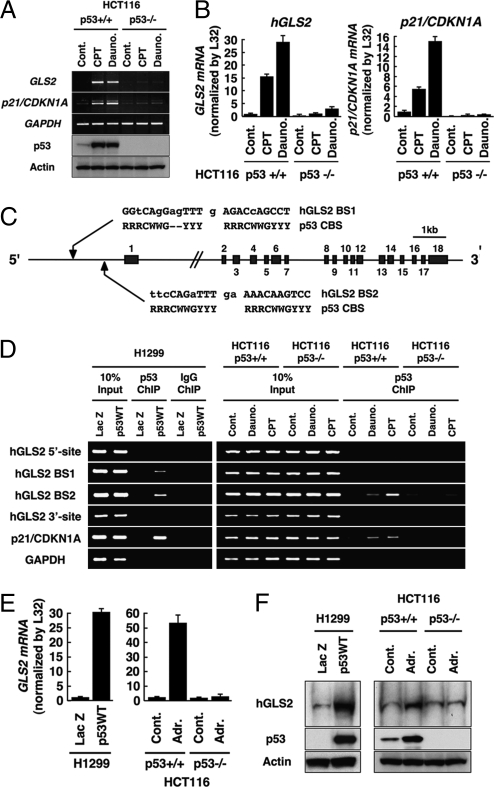
We used a cDNA microarray system to screen for p53-inducible genes in unstressed cells (Fig. S1A). Previously described and undescribed p53 targets were identified (Fig. S1B). Among potential new targets, phosphate activated GLS (GLS2; accession no. NM_013267) was significantly induced (approximately 7.5-fold) under these conditions (Fig. S1B). Consistently, GLS2 mRNA was induced in HCT116 (p53+/+) but not (p53−/−) cells treated with camptothecin or daunorubicin, and to approximately the same extent as was p21/CDKN1A (Fig. 1A and 1B). p53 did not induce GLS1 mRNA expression in these cells (Fig. S2). Mouse Gls2 was induced by daunorubicin in mouse embryonic fibroblasts (MEFs) expressing p53 (p53+/+) but not in MEFs lacking p53 (p53−/−; Fig. S3).
Fig. 1.
Identification of phosphate-activated GLS (GLS2) as a p53-inducible gene. (A) HCT116 (p53+/+) or (p53−/−) cells were treated with camptothecin (CPT; 300 nM) or daunorubicin (Dauno; 200 nM). RT-PCR analysis for GLS2, p21/CDKN1A, and GAPDH expression (top three panels) and immunoblotting to detect p53 (DO1) and actin (Sigma; bottom two panels) were performed. (B) HCT116 cells were treated with indicated agents as in A. Total RNA was subjected to real-time RT-PCR analysis. Expression levels of GLS2 (Left) and p21/CDKN1A (Right) RNAs were determined by the comparative threshold cycle method and then normalized by L32 expression. (C) Genomic structure of human GLS2 with its exon/intron organization and two potential p53 binding sites upstream of the first exon (GLS2 BS1 and GLS2 BS2) compared with the canonical p53 binding site. R, purine; Y, pyrimidine; W, adenine or thymine. (D) H1299 cells infected with adenoviruses expressing either LacZ or p53 (p53WT) for 24 h (Left) and HCT116 (p53+/+) or (p53−/−) cells either not treated (Control) or treated with daunorubicin (200 nM) or CPT (300 nM) for 24 h (Right) were processed for ChIP assays using anti-p53 (DO1/1801) or control IgG (Santa Cruz Biotechnology), followed by the amplification of p53 binding sites as indicated. (E) H1299 cells (Left) were infected with adenoviruses expressing either LacZ or p53 (p53WT) or HCT116 cells (p53+/+; Right) or (p53−/−) cells were treated with doxorubicin (Adr., 0.3 μM) for 24 h. Expression level of GLS2 mRNA was determined as in B. (F) Cells were treated as in E and subjected to mitochondrial fractionation. Immunoblotting was performed to detect GLS2 protein in the mitochondrial fraction and p53 and actin from whole extracts.
The human GLS2 gene, located on chromosome 12q13, contains 18 coding exons and two possible p53 binding sites, approximately 1.4 kb (GLS2 BS1; −1,437/1,415) and 0.5 kb (GLS2 BS2; −584/−575) upstream of the first exon (Fig. 1C). Adenovirally transduced p53 (Adp53) bound to both BS1 and BS2 whereas endogenous p53 associated with only BS2 (Fig. 1D). Neither source of p53 was associated with two negative sites. These results indicate that, once activated, p53 induces expression of GLS2 mRNA by directly associating with a response element (BS2) in the GLS2 promoter.
An anti-GLS2 antibody recognized a single polypeptide species of approximately 65 kDa, consistent with the previously reported size of GLS2 (Fig. S4) (17). As GLS2 was shown to be a mitochondrial enzyme (18), we isolated mitochondria, and immunoblotting showed that GLS2 protein induced by p53 was in that fraction (Fig. 1F). Both ectopically expressed FLAG-tagged GLS2 in H1299 cells and endogenous GLS2 in daunorubicin-treated human aorta endothelial cells (HAECs) displayed a cytoplasmic particulate staining pattern, indicative of mitochondrial localization (Fig. S5 A and B).
Normal cells (HAEC and TIG-7) as well as tumor cell lines expressing WT p53 (HCT116, MCF-7, and U2OS) were subjected to treatments that cause oxidative genotoxic damage, and GLS2 induction was assessed by real-time PCR analysis (Fig S5 C–F). GLS2 was confirmed to be a p53 target gene in both nontumor and tumor cells, although the extent of its up-regulation differed depending on the cell type or the levels and nature of the stress.
p53 and GLS2 Modulate Intracellular ROS Levels and the GSH/GSSG Ratio in Cells.
p53 has been reported to have opposing roles in the regulation of ROS through transactivation of antioxidant and prooxidant genes, each of which contribute to the tumor suppressor function of p53 (19, 20). As previously reported (19), basal ROS levels were suppressed (approximately by half) in exponentially cultured HCT116 (p53+/+) cells or in cells treated with low concentrations of doxorubicin (0.03–0.3 μM) compared with HCT116 (p53−/−) cells (Fig. 2A). In contrast, at higher doses of doxorubicin (>1 μM), p53+/+ cells showed a relative increase in ROS, indicating that p53 differentially regulates ROS levels depending on the severity of the stress signal. Accordingly, whereas GLS2 and p21/CDKN1A were induced at lower concentrations of doxorubicin, a prooxidant gene such as PUMA was induced only at relatively high concentrations of this drug (Fig. 2B). Thus, WT p53-expressing cells are more tolerant of low doses of oxidative stress compared with p53-negative cells. During acute stress conditions, however, p53 facilitates increased intracellular ROS levels and apoptosis.
Fig. 2.
Modulation of GLS2 or p53 expression affects intracellular ROS levels. (A) HCT116 cells that either express p53 (p53+/+ in green) or lack p53 (p53−/− in red) were treated with the indicated doses of doxorubicin for 24 h and then subjected to DCF staining followed by FACS analysis. (B) HCT116 p53+/+ cells were treated with indicated dose of doxorubicin as in A. Expression levels of GLS2, p21/CDKN1A, and PUMA mRNAs were determined as in Fig. 1B. (C) U2OS cells were transfected with luciferase RNAi (Luci RNAi; gray), p53 RNAi (blue), or GLS2 RNAi (red) for 24 h and then cells were either not treated (stress −; Left) or treated with doxorubicin (100 nM; Right) for 24 h. DCF staining was followed by FACS analysis. (D) U2OS cells were transfected with indicated RNAi and then treated with doxorubicin as in C. Expression levels of GLS2 mRNA were determined as in Fig. 1B. (E) HAEC cells were transfected as in C and then either not treated (stress −) or treated with daunorubicin (100 nM) or H2O2 (0.1 mM) for 24 h. DCF staining was followed by FACS analysis. (F) Cells were transfected with indicated DNA for 48 h. After the selection in the presence of 600 μg/mL G418 for 5 d, cells were split, cultured for 24 h, and then not treated (stress −) or treated with doxorubicin or CPT for another 24 h. DCF staining was followed by FACS analysis.
We next examined the effect of p53 or GLS2 silencing on ROS levels with or without cellular stress in both carcinoma and normal cells (Fig. 2 C–E). GLS2 siRNA reduced GLS2 levels by approximately 90% without affecting p53 protein levels or p53 target gene induction (Fig. S6). Down-regulation of p53 or GLS2 increased ROS levels significantly in both unstressed and doxorubicin-treated U2OS cells (Fig. 2 C and D). Similarly, p53 or GLS2 silencing in normal primary HAEC cells led to increased ROS in both untreated and DNA damaged cells (Fig. 2E) whereas expression of FLAG-tagged GLS2 led to a significant reduction of both basal ROS levels and ROS levels in DNA-damaged H1299 cells (Fig. 2F). Thus, p53 and GLS2 are directly involved in ROS regulation and antioxidant defense. The antioxidant or prooxidant outcomes of p53 activation are likely to depend on the differential regulation of p53 targets, including GLS2.
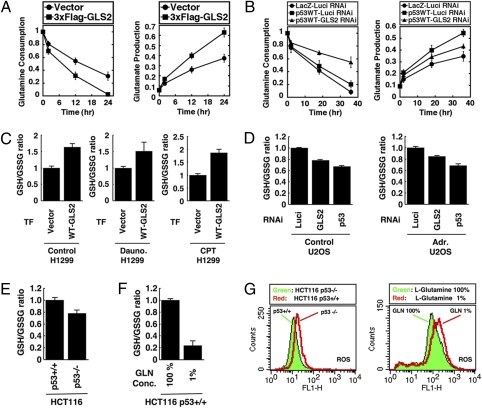
Glutamine is catabolized by GLS2 to glutamate, one of the precursor amino acids in the biosynthesis of GSH, and GSH/oxidized glutathione (GSSG) is the major redox couple that determines the antioxidative capacity of cells (21). Glutamine preserves total GSH levels after oxidative damage, making it a component of the cellular antioxidant defense (22). GLS2 plays a critical role by up-regulating GSH levels upon oxidative stress (23, 24). Overexpression of 3×FLAG–GLS2 increased glutamine consumption and facilitated the production of glutamate detected in the culture medium of H1299 cells, thus confirming its enzymatic function to catabolize glutamine to glutamate (Fig. 3A). Conversely, down-regulation of GLS2 decreased the glutamine consumption rate in H1299 cells expressing Adp53 at low levels in which apoptosis was not induced (Fig. 3B Left). Consistently, the induction of p53 led to increased glutamate production, whereas silencing of GLS2 attenuated the glutamate production rate significantly in H1299 cells (Fig. 3B Right), supporting the role of GLS2 in p53-dependent intracellular glutamine metabolism.
Fig. 3.
GLS2 controls glutamine metabolism and GSH antioxidant capacity to decrease intracellular ROS levels. (A) Cells were transfected with indicated plasmids for 24 h and then switched to fresh medium. Glutamine consumption is represented as a ratio to initial concentration and graphs show the mean of six measurements from two independent experiments, with error bars representing SD. (B) Cells were transfected with luciferase RNAi (closed circles and squares) or GLS2 RNAi (closed triangles) for 12 h and then cells were infected with adenoviruses expressing either LacZ or WT p53 (p53WT) at a multiplicity of infection of 2 for another 24 h. Cultures were switched to fresh medium and glutamine consumption and glutamate production was calculated as in A. (C) Cells were transfected with indicated plasmids for 48 h. After the selection in the presence of 600 μg/mL G418 for 5 d, cells were split and cultured for 24 h, and then not treated (control) or treated with daunorubicin or CPT for another 24 h. Cells were collected and subjected to GSH and GSSG assay as described in Materials and Methods. Graphs show the mean of two independent experiments, with error bars representing SD. (D) Cells were transfected with indicated RNAi for 24 h and then either not treated (control; Left) or treated with doxorubicin (100 nM; Right) for another 24 h. Cells were collected and subjected to GSH and GSSG assay as in C. (E) HCT116 cells that either express p53 (p53+/+) or lack p53 (p53−/−) were cultured exponentially and then subjected to GSH and GSSG assay as in C. (F) HCT116 (p53+/+) cells were cultured exponentially and then switched to normal medium containing 100% (584 mg/L) L-glutamine (GLN), or glutamine depletion medium containing 1% (5.8 mg/L) GLN, and then cultured for 36 h. Assay of GSH and GSSG was performed as in C. (G) HCT116 cells that either express p53 (p53+/+) or lack p53 (p53−/−) were cultured exponentially (Left) or HCT116 p53+/+ cells were switched to normal medium (GLN; 100%) or glutamine depletion medium (GLN; 1%) for 36 h as in F. DCF staining was followed by FACS analysis.
Further, overexpression of 3×FLAG–GLS2 increased GSH/GSSG ratios in both unstressed and daunorubicin or camptothecin-treated H1299 cells (Fig. 3C). In contrast, down-regulation of GLS2 or p53 suppressed the GSH/GSSG ratio in unstressed and doxorubicin-treated U2OS cells (Fig. 3D). Last, consistent with previous results, HCT116 (p53+/+) cells had a higher GSH/GSSG ratio and more intracellular ROS than HCT116 (p53−/−) cells (Fig. 3 E and G Left). Depletion of glutamine in the medium decreased the GSH/GSSG ratio strongly and up-regulated intracellular ROS levels in HCT116 (p53+/+) cells (Fig. 3 F and G Right). Together, these data indicate that GLS2 regulates glutamine metabolism to control ROS through the GSH-dependent antioxidant system.
GLS2 Protects Cells from DNA Oxidation and ROS-Sensitive Apoptosis.
High levels of ROS lead to formation of 8-hydroxy-2′-deoxyguanosine (8-OH-dG), the main source of oxidation-associated mutagenesis (25). Consistent with the intracellular ROS levels seen in HCT116 (p53−/−) cells in unstressed conditions (Fig. 3G Left), these p53-negative cells displayed approximately twofold higher 8-OH-dG levels compared with HCT116 (p53+/+) cells (Fig. 4 A and B). Following daunorubicin treatment, which significantly up-regulated ROS levels in these cells (Fig S7A), 8-OH-dG levels in HCT116 (p53−/−) cells increased to a greater extent than in HCT116 (p53+/+) cells (Fig. 4 A and B). Down-regulation of p53 or GLS2 increased 8-OH-dG to levels comparable to those observed in the HCT116 (p53−/−) cells in both untreated and daunorubicin-treated cells (Fig. 4C) or U2OS cells that contain WT p53 (Fig. 4D). Furthermore, overexpression of GLS2 in H1299 cells suppressed 8-OH-dG levels (Fig S7B). Thus, p53-mediated induction of GLS2 contributes to the antioxidant function of p53 by lowering intracellular ROS levels and thereby preventing DNA oxidation.
Fig. 4.
p53 and GLS2 regulate DNA oxidation and ROS-mediated apoptosis. (A) HCT116 cells were untreated (control) or treated with daunorubicin (Dauno.; 100 nM) for 24 h and then fixed and stained using anti–8-OH-dG antibody and anti-p53 polyclonal antibody (FL) and visualized using Alexa Fluor–488– and -594–conjugated secondary antibodies. Nuclei were counterstained with DAPI and images were taken using a Keyence microscope. (B) HCT116 cells were treated as in A. Intensity of 8-OH-dG (Left) and p53 (Right) staining was quantified using Keyence software. The average of six random visual fields from two independent experiments is shown, with error bars representing SD. (C) HCT116 cells expressing WT p53 (p53+/+) were transfected with indicated RNAi for 24 h and then cells were either not treated (untreated) or treated with daunorubicin (100 nM) for another 24 h. DNA oxidation was detected as in A and quantified as in B. (D) U2OS cells were transfected with luciferase RNAi or GLS2 RNAi and then treated with daunorubicin (100 nM) as in C. DNA oxidation was detected and quantified as in C. (E) HCT116 (p53+/+) or (p53−/−) cells were transfected with luciferase RNAi (Luci) or GLS2 RNAi for 24 h and then were either not treated (control) or treated with daunorubicin (300 nM) for another 36 h. The amount of sub–G0/G1 cells was calculated using the Cell Quest program for FACS. Average of three independent experiments is shown, with error bars indicating SD. (F) Model for regulation of intracellular ROS levels by GLS2. Upon oxidative stress or DNA damage, p53 is stabilized and activated to induce several targets including antioxidant and prooxidant genes. One such target, GLS2, catalyzes the hydrolysis of glutamine to produce glutamate and NH4+ and functions as an antioxidant protein. In response to severe cellular stress or irreparable damage p53 transactivates prooxidant genes (PUMA, PIG3, Proline Oxidase), resulting in the elevation of intracellular ROS, and apoptosis. The balance between anti- and prooxidant genes and the differential regulation of p53 targets can determine the choice of cellular outcomes.
Intracellular ROS levels can affect the sensitivity of cells to p53-dependent apoptosis (26). Although GLS2 down-regulation slightly increased apoptosis in p53-deficent cells, reduced GLS2 expression led to a larger increase in apoptosis in both unstressed and daunorubicin-treated (p53+/+) cells (Fig. 4E). Accordingly, the antioxidant compound N-acetylcysteine lowered GLS2 siRNA-enhanced apoptosis in U2OS cells (Fig. S8). Based on these data, we propose that GLS2 functions to reduce cellular sensitivity to ROS-associated apoptosis (Fig. 4F).
A Potential Tumor-Suppressor Role for GLS2.
Overexpression of FLAG-tagged GLS2 in H1299 cells led to a significant reduction of growth (Fig. 5A) as well as colony formation ability (Fig. 5B). Importantly, expression of GLS2 mRNA was significantly decreased in most of 12 specimens from hepatocellular carcinomas and metastatic liver tumors from colon cancers compared with liver tissues with chronic hepatitis (n = 6) or adjacent normal liver tissues (n = 6; Fig. 5C). Along with the fact that the expression of GLS2 is reduced in many brain tumors such as glioblastoma and anaplastic astrocytomas (27), these data suggest that GLS2 plays a role in tumor suppression.
Fig. 5.
GLS2 inhibits tumor cell growth and colony formation and GLS2 expression is decreased in liver tumors. (A) Cells were transfected as indicated for 48 h. Cells were then split and subjected to the cell growth analysis (Left) or colony formation assay visualized by crystal violet staining (Right). (B) Loss or reduction of GLS2 mRNA expression in human liver tumors. N, normal liver; L, tumor adjacent tissues with chronic hepatitis; HCC, hepatocellular carcinoma; MC, liver-metastatic tumor of colorectal carcinomas. The expression of GLS2 mRNA were determined by real-time PCR and normalized by actin expression.
Discussion
Although GLS2 was originally thought to be present only in adult liver tissue (28), emerging evidence has revealed that GLS2 expression also occurs in extrahepatic tissues, such as brain, pancreas, and breast cancer cells, as well as many other cell types (29). GLS2 localizes to the inner mitochondrial membrane to catalyze the hydrolysis of the γ-amino group of GLN forming glutamate and ammonia (27). This ammonia may be used to form carbamoyl phosphate or may diffuse from the mitochondria and the cell. Glutamate can be further deaminated to form α-ketoglutarate and thus enter the citric acid cycle for energy metabolism. Glutamate also preserves total GSH levels after oxidative stress (22, 30). Our data indicate that p53-inducible GLS2 regulates intracellular glutamine metabolism and ROS levels and promotes antioxidant defense through controlling the GSH/GSSG ratio, although we do not exclude the additional possibility that regeneration of GSH from GSSG is increased by GLS2 expression.
The modulation of intracellular ROS levels in cells is important in controlling the development and maintenance of tumors. A number of p53-induced antioxidant genes have been previously reported, including sestrins (SESN1 and SESN2) (31), aldehyde dehydrogenase 4 (ALDH4) (32), and TIGAR (14). Sestrins (SENS1 and SENS2) are essential for regulation of overoxidized peroxiredoxins (31). TIGAR blocks glycolysis, leading to elevated NADPH generation that results in increased GSH levels, thus promoting consumption of ROS (8). ALDH4 is a mitochondrial-matrix NAD+-dependent enzyme converting L-glutamic-γ-semialdehyde to glutamate via the proline degradation pathway (32). Here we have identified another metabolic role for p53 in the control of glutamine metabolism through GLS2. It is interesting that the activities of TIGAR, ALDH4, and GLS2 proteins converge onto a common mechanism in their regulation of intracellular ROS levels.
Recently, several studies have shown that p53 has a role in the regulation of both glycolysis and oxidative phosphorylation. p53 slows glycolysis by inhibiting the expression of the glucose transporters GLUT1, GLUT4, and GLUT3 and decreasing the levels of phosphoglycerate mutase (PGM) while increasing the expression of TIGAR. However, these findings are confounded by other studies showing apparently opposing activities: for example, the presence of p53-responsive elements in the promoters of PGM and hexokinase II suggests that p53 can promote at least some steps in glycolysis (3). Another study has shown that p53 induces expression of SCO2 (synthesis of cytochrome c oxidase 2) that participates in the assembly of cytochrome c oxidase (COX) in mitochondria, implicating p53 in the regulation of oxygen consumption and mitochondrial respiration (14). Similar to SCO2, GLS2 expression may cause subsequent metabolic changes in mitochondrial respiration as its glutamate product can eventually be further deaminated to form α-ketoglutarate and thus enter the TCA cycle. Indeed, the overexpression of GLS2 increased ATP production in H1299 cells whereas GLS2 silencing inhibited ATP levels in U2OS cells (Fig. S9). These findings connecting p53 to the regulation of energy production are rather complicated, and it is very likely that the roles of p53 in responding to and effecting alterations in metabolism will have consequences beyond cancer, influencing other aspects of normal life and disease. Future investigations should provide more information as to how p53 is able to coordinate the actions of multiple intracellular metabolic networks to exert its tumor suppressive function.
During oncogenesis, as cells accumulate defects in the p53 pathway, multiple intracellular metabolic safety mechanisms are bypassed. Energy supply and consumption systems, including glutamine metabolism and glycolysis, proceed at full capacity and the normal restraints on tumor growth are lost. It is thus possible that GLS2 expression might be under positive selection in tumors as a result of GLS2’s control of energy metabolism. Indeed, it was shown that GLS activity is positively correlated with malignancy in tumors (23). Recently, it was shown that c-Myc up-regulates GLS1 through its ability to repress miR-23a and miR-23b (33). These data seem to be at odds with our findings that p53 may be supplying the TCA cycle through its related target, GLS2. As both enzymatic forms of GLS have distinct kinetic and molecular characteristics (34), we speculate that the differential regulation of GLS1 and GLS2 may reflect their possibly distinct functions or requirements in different tissues or cell states (33).
Finally, we have demonstrated that GLS2 reduces cellular sensitivity to ROS-associated apoptosis possibly through GSH-dependent antioxidant defense processes. Yet it was shown that increased ROS levels lead to stabilized and activated p53 (35). As we cannot exclude the possibility that GLS2 expression affects certain p53 target gene(s) or some specific function of p53, which leads to the modulation of ROS-associated apoptosis, further investigation is required to clarify the relationship among ROS levels, glutamine metabolism, and p53-dependent apoptotic response. At present, the duality of p53 function as an inducer of antioxidant genes, including GLS2 and TIGAR, while also activating genes that enhance oxidative stress, remains to be elucidated. Perplexingly, both antioxidant and prooxidant outcomes of p53 transcription are proposed to contribute to tumor suppression. Nevertheless, our results that GLS2 inhibits tumor cell growth and is underexpressed in liver tumors implicate GLS2 as a contributor to p53-mediated tumor suppression and should provide impetus for further studies on this topic.
Materials and Methods
Cell Lines, Cell Culture, Western Blot Analysis, and Antibodies.
For detailed description of cell lines and antibodies, immunoblotting, real-time RT-PCR, FACS, and colony formation assays, please refer to SI Materials and Methods.
Chromatin Immunoprecipitation Assay.
H1299 cells infected with a recombinant adenovirus expressing p53 (Ad-p53; 30 MoI) and HCT116 cells (p53+/+ and p53−/−) were treated with daunorubicin (220 nM) for 24 h. Then cells were prepared for ChIP analysis. See SI Materials and Methods for a detailed protocol.
Preparation of Mitochondria.
The mitochondrial fraction was prepared as previously described (36). A detailed protocol is described in SI Materials and Methods.
RNA Interference.
siRNA oligonucleotides whose sequences are listed in SI Materials and Methods were synthesized by Qiagen. HCT116 cells (p53+/+ and p53−/−), U2OS cells, and HEAC cells were plated at 50% confluence and transfected with the indicated siRNA oligonucleotide (50 nM) using Dharma-fect 1 (Dharmacon). Twenty-four hours later the cells were left untreated or exposed to daunorubicin, H2O2, or different drugs as indicated in the figure legends for 24 h before analysis.
Measurement of ROS.
Cells were incubated with 3 μM 2′,7′-dichlorodihydrofluorescein diacetate (DCF; Molecular Probes) for 15 min at 37 °C. After incubation, cells were washed with PBS, trypsinized, and resuspended in PBS solution, and fluorescence was measured using a FACScan flow cytometer (excitation at 488 nm, emission at 515–545 nm) and data analyzed with CELL Quest software.
Glutathione Assay.
Total GSH and GSSG were measured with the glutathione quantification kit (Dojindo). In brief, the deproteinated sample was used to determine GSH content via a standard enzymatic recycling procedure. To determine the GSSG content, an aliquot of the deproteinated supernatant was mixed with 2-vinyl-pyridine (Sigma-Aldrich) and triethanolamine (Sigma-Aldrich) and then assayed.
Determination of Glutamate and Glutamine Concentrations.
Concentrations of glutamate and glutamine in the medium were determined using a glutamine/glutamate determination kit (GLN-1; Sigma-Aldrich). The determination of L-glutamine was done in a two-step reaction: (i) deamination of L-glutamine to L-glutamate and (ii) dehydrogenation of the L-glutamate to α-ketoglutarate accompanied by reduction of NAD+ to NADH. The conversion of NAD+ to NADH was measured using a spectrophotometer at 340 nm. The amount of NADH is proportional to the amount of glutamate. A standard curve was determined for each day the samples were run to calculate the concentration of glutamate in the sample.
Supplementary Material
Acknowledgments
We are grateful to Professor Yasushi Saito and Naoko Hashimoto for helpful suggestions and discussions throughout this work. We thank Kayo Suzuki and Takako Hatada for expert technical assistance. This work was supported by National Institutes of Health Grants CA77742 and CA87497, the Global COE Program (Global Center for Education and Research in Immune System Regulation and Treatment); MEXT (Japan); Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (Japan) for Scientific Research on Priority Areas 17016010 and 20012010, Scientific Research (B) 21390147 and (C) 19659121, and Exploratory Research and Young Scientists (B) 20790367; the Tokyo Biochemical Research Foundation, the Sumitomo Foundation; the Mochida Memorial Foundation; the Takeda Science Foundation; the Sankyo Foundation of Life Science; and the Japan Diabetes Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002459107/DCSupplemental.
See Commentary on page 7117.
References
- 1.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 4.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 7.Kondoh H, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 8.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 9.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz-Lozano P, et al. p53 is a transcriptional activator of the muscle-specific phosphoglycerate mutase gene and contributes in vivo to the control of its cardiac expression. Cell Growth Differ. 1999;10:295–306. [PubMed] [Google Scholar]
- 11.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009;1787:328–334. doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamura S, et al. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol Res. 1999;11:281–285. [PubMed] [Google Scholar]
- 14.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 15.Bourdon A, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 16.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 17.Pérez-Gómez C, et al. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–542. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kvamme E, Nissen-Meyer LS, Roberg BA, Torgner IA. Novel form of phosphate activated glutaminase in cultured astrocytes and human neuroblastoma cells, PAG in brain pathology and localization in the mitochondria. Neurochem Res. 2008;33:1341–1345. doi: 10.1007/s11064-008-9589-9. [DOI] [PubMed] [Google Scholar]
- 19.Sablina AA, et al. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Sies H. Glutathione and its role in cellular functions. Free Radic Biol Med. 1999;27:916–921. doi: 10.1016/s0891-5849(99)00177-x. [DOI] [PubMed] [Google Scholar]
- 22.Matés JM, Pérez-Gómez C, Núñez de Castro I, Asenjo M, Márquez J. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. Int J Biochem Cell Biol. 2002;34:439–458. doi: 10.1016/s1357-2725(01)00143-1. [DOI] [PubMed] [Google Scholar]
- 23.Lora J, et al. Antisense glutaminase inhibition decreases glutathione antioxidant capacity and increases apoptosis in Ehrlich ascitic tumour cells. Eur J Biochem. 2004;271:4298–4306. doi: 10.1111/j.1432-1033.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 24.Ogunlesi F, Cho C, McGrath-Morrow SA. The effect of glutamine on A549 cells exposed to moderate hyperoxia. Biochim Biophys Acta. 2004;1688:112–120. doi: 10.1016/j.bbadis.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 26.Green DR, Chipuk JE. p53 and metabolism: Inside the TIGAR. Cell. 2006;126:30–32. doi: 10.1016/j.cell.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 27.Kovacevic Z, McGivan JD. Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol Rev. 1983;63:547–605. doi: 10.1152/physrev.1983.63.2.547. [DOI] [PubMed] [Google Scholar]
- 28.Watford M. Hepatic glutaminase expression: relationship to kidney-type glutaminase and to the urea cycle. FASEB J. 1993;7:1468–1474. doi: 10.1096/fasebj.7.15.8262331. [DOI] [PubMed] [Google Scholar]
- 29.Gómez-Fabre PM, et al. Molecular cloning, sequencing and expression studies of the human breast cancer cell glutaminase. Biochem J. 2000;345:365–375. [PMC free article] [PubMed] [Google Scholar]
- 30.Yudkoff M, et al. Glutathione turnover in cultured astrocytes: studies with [15N]glutamate. J Neurochem. 1990;55:137–145. doi: 10.1111/j.1471-4159.1990.tb08831.x. [DOI] [PubMed] [Google Scholar]
- 31.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 32.Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49:134–140. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 33.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 35.Chen K, Albano A, Ho A, Keaney JF., Jr Activation of p53 by oxidative stress involves platelet-derived growth factor-beta receptor-mediated ataxia telangiectasia mutated (ATM) kinase activation. J Biol Chem. 2003;278:39527–39533. doi: 10.1074/jbc.M304423200. [DOI] [PubMed] [Google Scholar]
- 36.Trounce A, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–494. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.