Abstract
Class I phosphoinositide 3-kinases are enzymes that generate 3-poly-phosphoinositides at the cell membrane following transmembrane receptor stimulation. Expression of the phosphoinositide 3-kinase β (PI3Kβ) isoform, but not its activity, is essential for early embryonic development. Nonetheless, the specific function of PI3Kβ in the cell remains elusive. Double-strand breaks (DSB) are among the most deleterious lesions for genomic integrity; their repair is required for development. We show that PI3Kβ is necessary for DSB sensing, as PI3Kβ regulates binding of the Nbs1 sensor protein to damaged DNA. Indeed, Nbs1 did not bind to DSB in PI3Kβ-deficient cells, which showed a general defect in subsequent ATM and ATR activation, resulting in genomic instability. Inhibition of PI3Kβ also retarded the DNA repair but the defect was less marked than that induced by PI3Kβ deletion, supporting a kinase-independent function for PI3Kβ in DNA repair. These results point at class I PI3Kβ as a critical sensor of genomic integrity.
Keywords: cancer, genomic integrity
Class IA phosphoinositide 3-kinases (PI3Ks) are enzymes composed of a p85 regulatory and a p110 catalytic subunit that generate 3-poly-phosphoinositides (PIP3) following growth-factor receptor stimulation at the cell membrane (1, 2). p110α and p110β are expressed ubiquitously, and regulate cell division and embryonic development (3–9). p110α−/− mice show a developmental block at embryonic day (E) 9 to 10, as p110a is essential for vasculature formation (3, 9). Mice deficient in p110β die at E 2 to 3 (4); knock-in mice expressing a kinase inactive-p110β survive to adulthood, supporting a kinase-independent function for p110β in development (10). Nonetheless, the specific function of p110β in the cells remains elusive. We previously found that p110β is mainly nuclear and controls DNA replication (6, 7). Given the close connections between replication and repair, as well as the need for DNA repair mechanisms in development (11), we examined p110β involvement in the cell response to DNA double-strand breaks (DSB).
DSB are among the most deleterious lesions for genome integrity. Eukaryotic cells have developed two alternative DSB repair methods, direct ligation of the excised DNA ends (nonhomologous end joining, NHEJ) and homologous recombination (HR) (12–15). DNA repair is stimulated by a phosphorylation-based signaling cascade, the DNA damage response (DDR) (16–18). Current knowledge places three distal homologs of PI3K—DNA-PKcs, ATM, and ATR (termed class IV PI3K)—as key DDR regulators. DNA-PKcs regulates NHEJ, which is restricted mainly to the G1 phase, whereas ATM and ATR control HR (11, 13, 17).
The sequential process of DSB repair begins with formation of large protein complexes (foci) containing numerous repair proteins. Ku and DNA-PKcs initiate NHEJ; however, when NHEJ fails, cells proceed to HR (12). HR begins with binding of the MRN complex (Mre11-Rad50-Nbs1) to DNA; Nbs1 is considered the earliest sensor of DNA damage. MRN binding induces generation of single-strand DNA chains, which subsequently bind replication protein A. MRN also triggers ATM recruitment that activates the checkpoint mediator Chk2. Replication protein A supports recruitment of additional mediators, including ATR, which triggers Chk1 activation (13–16, 19). Chk1 and Chk2 stop the cell cycle for DNA repair; if DDR fails, cells undergo apoptosis (11, 20). In addition, when G2/M arrest fails, cells might continue to divide and accumulate DNA defects (21). Here we examine the contribution of PI3Kβ in DSB repair.
Results
p110β and PIP3 Localize to DNA Damage Foci.
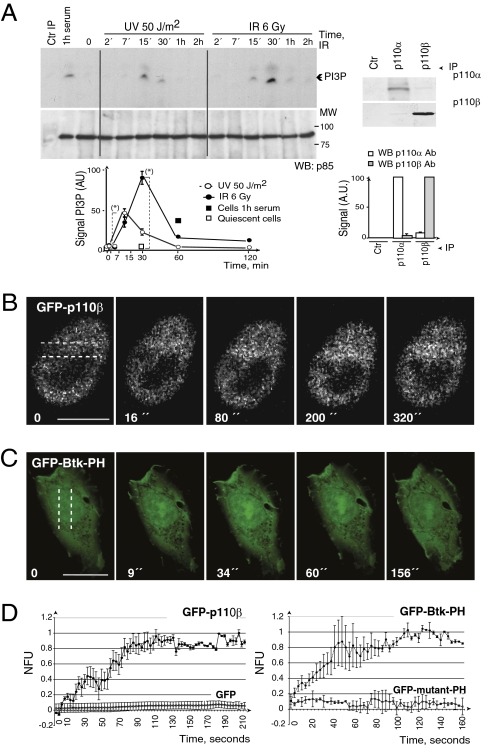
To determine whether p110β activity is stimulated by DNA damage, we exposed NIH 3T3 cells to UVC or IR, then immunopurified p110β using a specific antibody (Ab) (7), and perform an in vitro PI3K assay. Both treatments increased p110β activity (Fig. 1A). Considering that defects in the DDR often result in impaired cell-cycle checkpoints, we examined the consequences of interfering with p110β on DNA damage-induced G2 arrest. We determined the proportion of cells that cells progress into M phase by staining the cells with an S10-phospho-histone H3 (pH3)-specific Ab (22). We used shRNA to reduce p110β expression and the selective inhibitor TGX221 to inhibit p110β (7, 23). Inhibition of p110β moderately increased the proportion of pH3+ cells (Fig. S1A). Nonetheless, a larger proportion of p110β knockdown cells progressed into M phase after IR when compared with controls, indicating that p110β deletion inhibits G2 arrest (Fig. S1B). We also examined immortalized p110β−/− murine embryonic fibroblasts (MEF), and p110β−/− MEF reconstituted with WT- or with the kinase-dead (KR)-p110β (8). p110β deletion, but not kinase inactivation impaired G2 arrest, permitting entry into mitosis (Fig. S1C).
Fig. 1.
p110β and PIP3 localize to damaged DNA foci. (A) NIH 3T3 cells were exposed to UV or IR and harvested at different times. p110β was immunoprecipitated (IP) (800 μg) from cell extracts and tested by in vitro PI3K assay. Control IP was with protein A. Graphs show PI3P signal intensity quantitation in arbitrary units (AU) (mean ± SD, n = 3). (*), P < 0.05. (Right) Western blot analysis of p110α or p110β IP (300 μg). Graphs show signal intensity quantitation in arbitrary units compared to maximum (mean ± SD, n = 3). (B) NIH 3T3 cells expressing GFP-p110β were microirradiated with an UV laser. We examined real-time GFP-p110β translocation to the DNA damage region. Dotted lines indicate laser paths. (C) GFP-Btk-PH-transfected U2OS cells (24 h) were microirradiated with an UV laser and examined as in B. (Scale bars in B and C, 15 μm.) (D) The graphs show normalized fluorescence units (NFU) of the mean of n = 3 experiments performed as in B and C and plotted as a function of time. Controls are GFP and GFP fused to the R25C mutant of the Akt PH domain.
DSB repair begins with formation of large protein complexes (foci) that contain many repair proteins (12). A large fraction of p110β localizes in the nucleus (7); we tested whether endogenous p110β formed foci after DNA damage. IR induced p110β localization in large nuclear foci (after 1 h) (Fig. S2 A and B). Moreover, simultaneous staining of p110β and γ-H2AX showed partial colocalization of endogenous p110β with γ-H2AX at DSB (Fig. S2C). To confirm p110β localization at DNA damage sites, we examined GFP-p110β translocation to DSB. Cells were irradiated with an UV laser that generates DSB in defined nuclear volumes (24, 25). GFP-p110β concentrated at laser tracks early and remained associated for the entire recording period (∼300 s) (Fig. 1B, Fig. S2D, and Movie S1).
To determine whether the PI3K product PIP3 concentrates at the site of DNA damage, we performed immunofluorescence analysis using anti-PIP3 Ab. This Ab stained the cell membrane, endomembranes, and the nuclei in exponentially growing NIH 3T3 cells (Fig. S3A). Ly294002 (pan-PI3K inhibitor) reduced the cellular PIP3 signal, whereas TGX221 inhibitor was more potent in reducing nuclear PIP3 (Fig. S3A). γ-irradiation of NIH 3T3 cells induced formation of foci (30 min) that were PIP3-positive; some were also γH2AX-positive (Fig. S3B). DSB foci also concentrated the enzyme substrate PI(4,5)P2 (Fig. S3C). To confirm that PIP3 concentrates at damage sites, we used the GFP-Btk-PH domain, which binds selectively to PIP3 (26). We microirradiated cells with a UV laser and examined real-time Btk-PH translocation to laser tracks; Btk-PH concentrated very early and remained in this region for the entire recording period (Fig. 1 C and D); GFP fused to the R25C mutant of Akt-PH domain (27) remained invariable (Fig. 1D and Fig. S3D). Therefore, PIP3 localizes at damaged DNA.
p110β Deletion Inhibits ATM and ATR Repair Pathways.
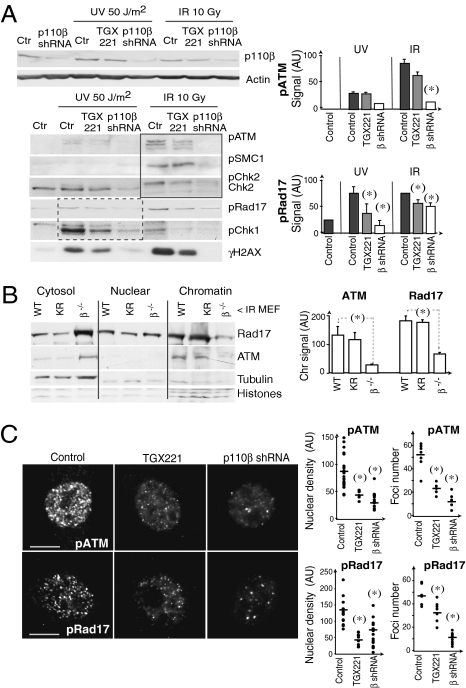
We next analyzed the impact of p110β depletion on the activity of ATM and ATR kinases. We examined phosphorylation of different substrates of ATM (pSMC1, pChk2) and ATR (pRad17, pChk1). γ IR induces more markedly ATM activation while UVC triggers principally the ATR route (Fig. 2A) (19, 24). Whereas reduction of p110β levels markedly diminished ATR and ATM pathways, p110β inhibition only partially reduced ATR route (Fig. 2A).
Fig. 2.
Defective ATM and ATR pathways activation in p110β-deficient cells. (A) NIH 3T3 cells were transfected with p110β or control shRNA (48 h); other cells were treated with the p110β inhibitor TGX221 (30 μM, 4 h). Cells were irradiated (UVC or IR) and extracts collected after 1 h. Lysates were examined by Western blot using indicated Ab. ATM effectors are grouped with a square; ATR effectors are grouped with a dashed-line square. Graphs show band signal intensity (AU) (mean ± SD n = 3). (B) p110β−/− immortalized MEF alone or reconstituted with WT- or KR-p110β were exposed to IR (10 Gy), incubated (1 h), and fractionated. Extracts were examined by Western blot with indicated Ab. The graph shows the quantitation of the signal in the chromatin fraction (mean ± SD, n = 3). (C) NIH 3T3 cells transfected with p110β shRNA or treated with TGX221 were exposed to IR (3 Gy) and processed 15 min later for immunofluorescence (IF) using anti-phospho-Rad17 or -phospho-ATM Ab. Graphs show the integrated nuclear fluorescence intensity (nuclear density, Left) and the number of foci with signal intensity > 40 AU (Right) on a representative set of cells of n > 100 examined. (Scale bars, 15 mm.) (*), Student t test P < 0.05.
To examine the consequences of interfering with p110β expression or activity on ATM chromatin loading, we γ-irradiated cells, fractionated them as in ref. 7, and determined ATM content in the chromatin fraction; for the ATR pathway we analyzed Rad 17. ATM was present in the chromatin fraction of WT- and KR-p110β MEF, but was severely reduced in p110β−/− MEF; similarly, Rad17 loading onto chromatin was greatly impaired in p110β−/− MEF (Fig. 2B). Thus, p110β regulates ATM and ATR-pathway members binding to chromatin.
Immunofluorescence studies confirmed defective activation of ATM and ATR pathways in cells with reduced p110β expression upon irradiation. p110β inhibition reduced the signal intensity of pATM+ and pRad17+ foci, whereas p110β knockdown greatly diminished pATM+ and pRad17+ foci number (Fig. 2C). Histone H2AX is a substrate for ATM and ATR (as well as for DNA-PK) (14, 28). p110β inhibition reduced γH2AX signal intensity in DSB foci and p110β deletion nearly eliminated γH2AX signal (Fig. S4A). We also examined 53BP1, an ATM pathway effector that regulates chromatin structure at DSB (24). p110β inhibition delayed—but p110β knockdown virtually blocked—53BP accumulation at laser tracks (Fig. S4B and Movie S2). These results show that p110β expression is critical for the association of DDR proteins (ATM, Rad 17, γH2AX, and 53BP1) to DSB foci.
Endogenous p110β Associates to Nbs1.
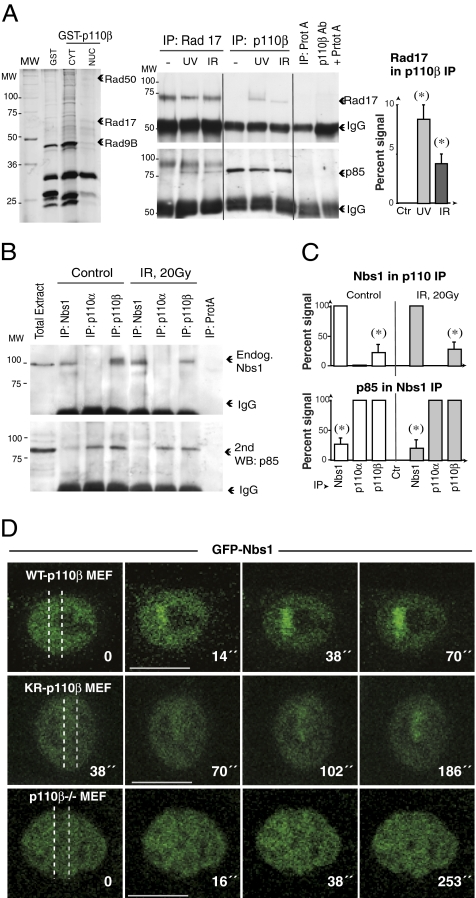
We used mass spectrometry to detect potential DDR proteins that interact with p110β. We transfected cells with GST-fused-p110β and performed a pull-down assay to identify nuclear proteins that might associate with p110β. We identified Rad50 (an MRN complex component), Rad17 (an ATR effector), and Rad9B (Fig. 3A), in complex with GST-p110β. We confirmed the association of Rad17 with p110β in intact cells following UVC or IR (Fig. 3A). In agreement the MRN component Rad50 association with p110β (Fig. 3A), recombinant p110α and -β were found to associate to exogenous human (h)Nbs1 (29). We tested whether endogenous p110β and Nbs1 formed a complex. Endogenous p110β, but not endogenous p110α (cytosolic, ref. 6), associated constitutively with endogenous Nbs1 (Fig. 3 B and C) and vice versa (Fig. 3C).
Fig. 3.
p110β associates with Nbs1 and Rad17. (A) NIH 3T3 cells were transfected with GST or GST-p110β-NLS (48 h). GST fusion proteins were purified, resolved in SDS/PAGE, and silver-stained. Band slices were examined by mass spectrometry (Left). NIH 3T3 cells were also irradiated with UVC (50 J/m2) or IR (10 Gy). After 1 h, endogenous p110β (1,500 μg) or Rad17 (300 μg) were IP from nuclear extracts (Right). Western blot was used to test for Rad17 in p110β IP and for p110β in Rad17 IP. The graph shows the percentage of p110β-associated Rad17 compared to the maximal Rad17 signal (in Rad17 IP from an equivalent protein amount) (mean ± SD, n = 3). (B) NIH 3T3 cells were irradiated with IR (10 Gy). After 1 h, endogenous p110β (500 μg) or endogenous Nbs1 (200 μg) were immunoprecipitated from nuclear extracts, resolved in SDS/PAGE and examined by Western blot using anti-Nbs1 Ab. The membrane was also blotted with p85 Ab to confirm equal loading. (C) The graphs show the percentage of p110β associated with Nbs1 compared to the maximal Nbs1 signal (Nbs1 IP from an equivalent protein amount) and quantitation of the reciprocal assay (mean ± SD, n = 3). (D) p110β−/− MEF reconstituted or not with WT or KR-p110β were transfected with GFP-Nbs1, laser microirradiated, and examined by real-time video microscopy. Dotted lines indicate laser paths. Scale bars, 15 mm.) (*), Student t test P < 0.05.
To define whether p110β regulates Nbs1 recruitment to damaged DNA, we examined translocation of GFP-murine-Nbs1 (30, 31) to laser tracks in p110β−/− MEF. In WT-p110β-MEF, Nbs1 accumulated early (at ∼15 s) and remained associated throughout the recording period (∼270 s); in p110β-KR cells, Nbs1 accumulated more slowly and in smaller amounts; in contrast, p110β deletion nearly abrogated Nbs1 accumulation at laser tracks (Fig. 3D and Movie S3). Indeed, 50% of p110β−/− MEF showed no Nbs1 accumulation and ∼50% showed very low intensity and unstable Nbs1 binding at laser tracks. Results were similar in NIH 3T3 cells. p110β expression is thus necessary for Nbs1 recruitment to DSB, whereas p110β activity enhances or stabilizes Nbs1 recruitment to these sites.
p110β Association Is Required for Nbs1 Binding to Damaged DNA.
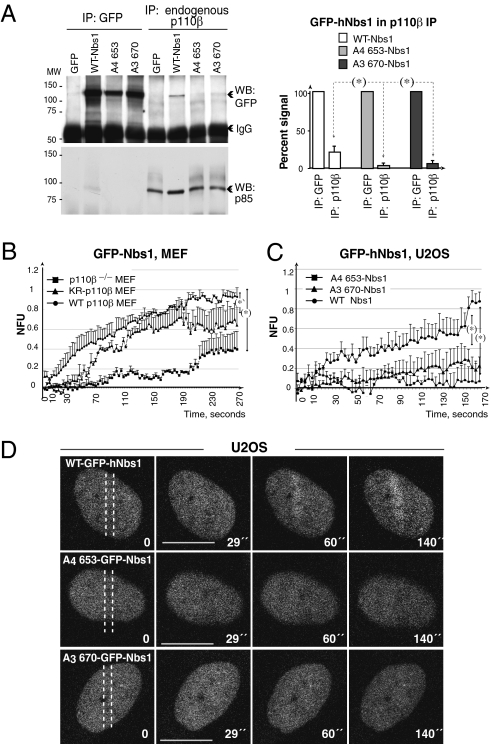
No viable Nbs1 mutant has yet been reported to disrupt the initial recruitment of MRN to DNA (15). To test whether p110β/Nbs1 complex formation is necessary for Nbs1 binding to DSB, we assayed which residues in Nbs1 mediate association with p110β. We examined residues 653 to 669 of hNbs1, which mediate interaction of recombinant p110α and Nbs1 (29). We transfected cells with WT GFP-hNbs1, or A4653-hNbs1, or with A3670-hNbs1, and examined Nbs1/endogenous p110β association. Endogenous p110β associated efficiently with WT, but very poorly with mutant forms of Nbs1 (Fig. 4A). p110b expression is required for Nbs1 translocation to laser tracks (Figs. 3D and 4B). In addition, GFP-WT-hNbs1 but not A4653-Nbs1 or A3670-Nbs1 concentrated at laser tracks (Fig. 4 C and D and Movie S4). These results show that p110β/Nbs1 association is necessary for Nbs1 recruitment to damaged DNA.
Fig. 4.
Nbs1 mutations that do not bind p110β are not recruited to DSB. (A) U2OS cells expressing GFP, GFP-hNbs1, -A4653-hNbs1, or -A3670-hNbs1 (36 h) were irradiated with IR (20 Gy). After 1 h, endogenous p110β (600 μg) or GFP-Nbs1 fusion proteins (250 μg) were immunoprecipitated from nuclear extracts, and examined by Western blot. The graph shows the percentage of GFP-Nbs1 associated to p110β, compared with the maximal GFP-Nbs1 signal (mean ± SD, n = 3). (*), P < 0.05. (B) MEF were irradiated with an UV laser, and examined by real-time video microscopy (as in Fig. 3C). The graphs show NFUs (mean ± SD, n = 7) plotted as a function of time. (C and D) U2OS cells expressing GFP-hNbs1, A4653-hNbs1 or A3670-hNbs1 (36 h) were examined as in B. Assembly curves (C) represent the mean ± SD (n = 4). (Scale bars, 15 μm.)
Proliferating Cell Nuclear Antigen Concentrates at DNA-Damaged Zones Downstream of Nbs1.
Some of the proteins controlling DNA replication also regulate DNA repair. MRN complexes localize at replication forks (32), and proliferating cell nuclear antigen (PCNA), which controls DNA replication (33), also regulates NHEJ and HR in yeast (34, 35). We described that p110β associates with PCNA, controlling DNA replication (7). We explored whether PCNA also localizes to DSB in mammals, and whether this process is controlled by p110β. Although the net amount of chromatin-bound PCNA did not increase after irradiation, DNA damage induced RFP-PCNA translocation to laser tracks (Fig. S5 A–C). PCNA chromatin binding diminishes by p110β inhibition, and more markedly upon p110β knockdown (Fig. S5A). In addition, p110β inhibition and, more strikingly, p110β deletion reduced PCNA localization at laser tracks (Fig. S5 and Movie S5). Thus, PCNA localizes to laser tracks in a p110β-dependent manner.
Because both PCNA and Nbs1 binding to DSB were controlled by p110β, to define the primary event regulated by p110β in DDR, we simultaneously examined the translocation of Nbs1 and PCNA to DSB in MEF (Movies S6 and S7). p110β deletion affected both Nbs1 and PCNA loading. Nonetheless, Nbs1 translocated slightly earlier than PCNA to damaged DNA in control cells (Fig. S5). In addition, in some p110β-deficient cells in which GFP-Nbs1 did not mobilize to laser tracks, we detected PCNA translocation; finally, p110β inhibition induced a more pronounced defect in Nbs1 than PCNA traslocation (Fig. S5). These findings show that interference with p110β affects the earliest sensor of DNA damage (Mre/Rad50/Nbs1) more severely than PCNA loading and that these processes might be independent.
p110β Deletion Induces Genomic Instability.
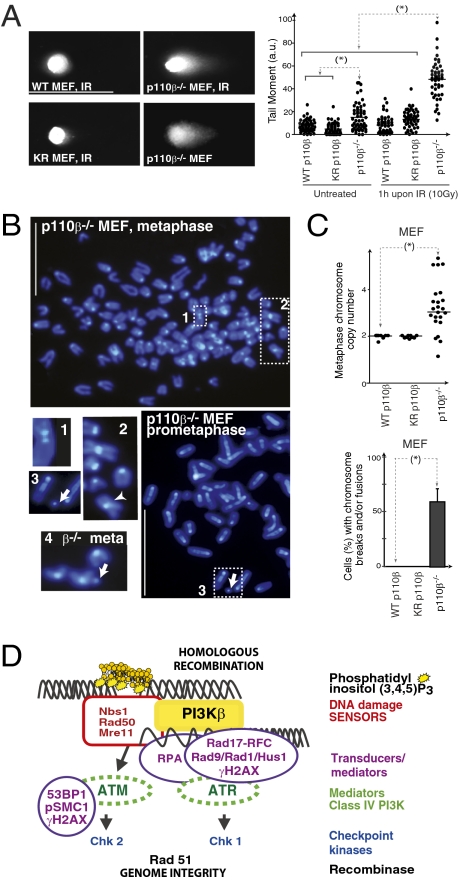
We focus our study on DSB repair, as p110β associates with HR components (Fig. 3). To demonstrate defective DSB repair in p110β−/− cells, we searched for the presence of DSB in untreated cells. In addition, we irradiated these cells (γ-IR, 10 Gy) and incubated them for 1 h to permit DNA repair; we quantitated DSB using the neutral comet assay (36). This assay showed that p110β−/− MEF, but not WT or KR-p110β MEF, already had spontaneous comets in untreated cultures (∼20% of cells), and showed a large proportion of comets (indicative of DSB) 1 h after IR (Fig. 5A). Accordingly, p110β-depleted NIH 3T3 cells were radiation-sensitive, as they showed a higher rate of DNA damage-induced apoptosis than controls (Fig. S6). Because p110β-deficient cells fail to repair DSB and do not stop at G2/M following damage (Fig. S1), they might accumulate DNA defects. We examined chromosome breaks and chromosome numbers by DAPI staining of MEF metaphases. p110β−/− MEF, but not WT or KR-p110β MEF, had abnormal chromosome numbers, chromosome breaks, and disjunction figures (Fig. 5 B and C), showing that p110β deletion causes genomic instability.
Fig. 5.
p110β deletion induces radiation sensitivity and genomic instability. (A) Representative p110β−/− immortalized MEF, alone or reconstituted with WT- or KR-p110β, were exposed to IR (10 Gy) and incubated (1 h), or were untreated; all were tested in neutral comet assays. The graph shows the tail moment for these MEF (mean ± SD, n = 50 cells). Tail Moment = (%DNA in tail × tail length)/100. (Scale bar = 125 μm.) (B) DAPI staining of representative p110β−/− MEF in metaphase or prometaphase (indicated). p110β−/− MEF showed supernumerary chromosomes and fused chromosomes (two chromosomes with one centromere, inset 2, arrowhead). Control adjacent chromosomes (with two centromeres) are shown in inset 1. Chromosome breaks are shown in insets 3 and 4 (arrows). Representative images of n = 50 examined. (Scale bar = 15 μm.) (C) Percent-MEF with the indicated phenotypes. (D) Repair of DNA DSB is stimulated by a phosphorylation-based signaling cascade termed the DNA damage response. The earliest DSB sensor for HR is the MRN complex, which binds to DNA and activates the class IV PI3K ATM. ATM permits binding of replication protein A, which assists subsequent recruitment of Rad17-RFC, Rad1/Rad9/HUS1 and ATR/ATRP complexes. ATR activates Chk1 and ATM activates Chk2; both checkpoint kinases contribute to arresting the cell cycle while cells repair DNA. PI3Kβ also binds to DSB where it generates PIP3 and helps recruiting Nbs1, in a kinase-independent manner, regulating DNA repair. *, P < 0.05.
Discussion
Class I PI3K were thought to act mainly by increasing PIP3 production at the cell membrane. Here we report a unique function for PI3Kβ in the control of DSB repair. PI3Kβ was required for the binding of the first DNA damage sensor protein Nbs1 to double-strand breaks. Indeed, endogenous PI3Kβ bound to endogenous Nbs1, and this complex was necessary for efficient concentration of Nbs1 at DSB. Both PI3Kβ deletion and mutation of Nbs1 at the site of association with p110β resulted in highly defective Nbs1 localization at DSB. Because of this function in DNA damage sensing and subsequent DDR (Fig. 5D), p110β deletion resulted in defective G2 arrest, radiation sensitivity, DSB accumulation, and genomic instability.
These findings and our description on the role of p110β in DNA replication (7) suggest that there is a fundamental difference between the function of the other ubiquitous class IA PI3K, p110α, which is mainly cytosolic (6, 7), and p110β. Whereas p110α regulates cell growth, as well as G0 > G1 and G1 > S phase transitions, p110β cooperates with p110α in G1 progression (6) but also concentrates in the nucleus, where it regulates DNA homeostasis.
We show that p110β is activated following DNA damage and concentrates at DSB. Both p110β kinase activity and a kinase-independent p110β function regulate DNA repair. Whereas PI3Kβ inhibition delayed activation of the DDR, PI3Kβ deletion almost abrogated it. The kinase-independent capacity of p110β to associate Nbs1 is critical for its recruitment to damaged DNA and, in turn, for amplification of the DDR. In the absence of p110β, Nbs1 was not recruited to laser tracks and, after IR, p110β knockdown cells had a very small number of pATM and pRad17-containing foci and showed defective ATM and ATR pathway activation. These defects explain the failure in G2 arrest of p110β-deficient cells, as well as their radiation sensitivity, DSB accumulation, and DNA instability.
In contrast to the consequences of deleting p110β, its inhibition delayed, but did not abrogate Nbs1 association to laser tracks; the number of IR-induced foci was roughly 50% that of normal cells and showed reduced pATM and pRad17 intermediate frequency (IF) signal intensity. In vivo imaging of cells with inhibited p110β showed unfocused, delayed, and unstable concentration of Nbs1, PCNA, and 53BP1 in laser tracks, suggesting that p110β activity stabilizes or facilitates protein recruitment to DSB. Given the high negative charge of PIP3, the reported local increase in PIP3 at DSB might help to maintain DNA (also negatively charged) in an open conformation by repelling electrostatic forces. Alternatively, PIP3 might sequester histones (positively charged), contributing to stabilization of chromatin in an open conformation at DSB. PIP3 localization at DSB sites might also recruit PH domain-containing proteins, such as PKBα/Akt1, which concentrate at DSB, associates to DNA-PK, and promotes cell survival (37). PTEN, a phosphatase that dephosphorylates PIP3, also controls DSB repair by regulating Rad51 expression (38). The potential crosstalk between PI3Kβ, PKBα/Akt1 and PTEN action in DDR requires further analysis.
We conclude that whereas p110β activity (PIP3) facilitates DNA repair, p110β expression is required for this process, supporting the concept that p110β contribution in DDR is mainly kinase-independent. The kinase-independent function of p110β was also seen in KR-p110β knock-in mice; these mice were born at lower numbers (50%) than expected. Moreover, at E 13.5, two groups of pik3cbKR/KR littermate embryos were found; 70% appeared normal and 30% appeared small and moribund (10). In MEFs obtained from the latter, KR-p110β content was smaller than in the apparently normal ones, supporting the existence of p110β kinase-independent functions (10). Tissue-specific p110β-deletion mouse models have been reported; whereas PTEN−/− prostates develop tumors, p110β-deletion in PTEN−/− prostate impeded tumorogenesis (8). p110β−/− prostates did not show apparent morphological defects (8). The observation that prostate develops in conditional p110β−/− mice (8) contrasts with the requirement of p110β for DNA replication and repair, suggesting that other proteins might replace or compensate p110β function in some tissues. Conventional p110β deletion, however, impairs embryonic development very early, at E 2 to 3 (4). Because DSB repair is critical for embryonic development (11), we hypothesize that the kinase-independent function of p110β in DNA repair might contribute to cause the embryonic lethality of p110β−/− mice.
Current knowledge classifies three distal homologs of PI3K, DNA-PKcs, ATM, and ATR, as key DDR regulators. DNA-PKcs controls NHEJ; ATM and ATR control HR (12–15). This model should be expanded to integrate the nuclear class I PI3K isoform p110β in DNA repair pathway activation at the DSB-sensing step (Fig. 5D). We have examined the function of p110β on HR DSB repair, as p110β associates with HR repair proteins (Fig. 3); however, we cannot rule out the involvement of p110β in additional repair processes. The involvement of p110β in nuclear PIP3 production and DDR increases the complexity of the mechanisms by which class IA PI3K pathway might regulate survival and tumorigenesis. p110β activity is essential for prostate cancer formation in the mouse (8), suggesting the use of interfering p110β-based therapy. Our results point to the importance of testing the status of the p53 gene (frequently mutated in cancer), because defects in apoptosis mechanisms in cancer cells might result in increased genomic instability following interference with p110β.
Materials and Methods
Reagents and cDNA.
β-actin antibody (Ab) was from Sigma, histone Ab from Chemicon Intern, p110β (IF) and Rad17 (IP) from Santa Cruz, p110β from Cell Signaling, Chk1 and Chk2 from Novocastra and Upstate; Rad17 (Western blot), pRad17, and p-SMC1 were from Abcam. Other antibodies used were PIP3 (Echelon Bioscience), PCNA (BD Transduction), p-histone H3 (Ser10; Beckman Coulter), p85 (Upstate), GFP (Roche), γH2AX (Millipore), p-ATM (1981; Rockland); p-Chk1 (Ser345), ATM, and Nbs1 were from Cell Signaling. p110α Ab was donated by A. Klippel (Merck, Boston, MA). [γ-32P]ATP was from Amersham. TGX221 (used at 30 μM) was from ACCC. p110β shRNA were from Origene and other shRNA were described (6). All remaining reagents were from Sigma. The construct encoding the PKB-mutant-PH domain in the pEGFP-C1 vector was a gift of J. Downward (Cancer Research, London, United Kingdom). pSG5-Myc-p110β (7), pEGFP-C1-53BP1 (24), murine Nbs1-2GFP (30), and pEN-mRFP-PCNAL2 (33) have been described. pEGFPC1-p110β and pEBG-GST-p110β were constructed by subcloning p110β from pSG5-myc-p110β into pEGFP-C1 and pEBG. GFP-Btk-PH domain was described (26). pEGFPC2-WT- hNbs1, pEGFPC2-A4653- hNbs1 and -A3670- hNbs1) were prepared by subcloning into pEGFP-C2 (mutants were kindly donated by Y. C. Chen, Yang-Ming University, Taipei, Taiwan) (29).
Cell Culture and Transfection.
Cell lines were maintained as reported (7). Immortalized p110β-deficient MEF were donated by J. Zhao and T. Roberts (Dana-Farber Cancer Institute, Boston, MA) (8). Cell synchronization at G1/S and metaphase arrest were described (6, 7). To examine the proportion of cells in mitosis, we stained the cells with an antibody recognizing S10-phospho-histone H3 (22). We used Jet Pei (Genycell) for transfection. Cells transfected with p110β shRNA were selected for 48 h in medium plus 2 μg/mL puromycin. To increase the nuclear localization of recombinant GFP-p110β, it was transfected with NLS-p85 (7).
Irradiation-Induced DNA Damage.
Cells were irradiated using UV or ionizing radiation (IR). UVC radiation was generated using a UV generator (λ = 254 nm; Vilber Laurmat) and IR was delivered by a γ-irradiator (MARK 1, Shephard and Associates) that uses a 137Cs probe. Real-time recruitment of DNA repair proteins to microlaser-generated DNA damage sites was as reported (24, 25). Before laser treatment, the cell medium was changed to a phenol red-free DMEM (Invitrogen). Immediately after microirradiation, repeated images of the same field were acquired in an integrated confocal unit operated by LCS software v2.61. Images were recorded at ∼3.2 s after DSB generation, with a gap of 3.2 s per image.
Cell Lysis, Immunoprecipitation, Western Blotting, PI3K Assay, and GST-p110β Pull-Down.
Total cell lysates were prepared in RIPA lysis buffer (7). For protein-protein interactions, cells were extracted as cytosolic and nuclear fractions (7). Triple fractionation (cytosol, nuclear soluble, and chromatin), IP, Western blot, and PI3K assays were as described (7).
For pull-down, NIH 3T3 cells transfected with pEBG-GST or -p110β-NLS (48 h) were fractionated as cytoplasmic and nuclear extracts. Nuclear lysates (1 mg) were incubated with glutathione beads (2 h, 4 °C). Beads were washed twice with lysis buffer and once with 50 mM Tris/HCl pH 7.5; bound p110β was resuspended in 2 × Laemmli buffer and resolved by SDS/PAGE. The gel was stained using a silver staining kit (Amersham). Stained protein bands were excised and the gel cut into small pieces and analyzed by mass spectrometry. p110β-associated proteins were identified by comparison with the peptide sequence database.
Immunofluorescence, Comet Assay, and Statistical Analyses.
Cells were fixed with 4% formaldehyde in PBS (10 min, room temperature), blocked using PBS buffer, and permeabilized with 0.3% TX-100 PBS (10 min). Cells were incubated with antibodies (1 h, room temperature), followed by three washes with staining buffer. Secondary antibodies were added to samples and incubated (1 h, room temperature), followed by three washes. Mounting medium containing DAPI (VectaShield) was added and cells were visualized in an AV100 Flow-view Olympus microscope or Leitz DMRB (Leica). Neutral comet assays were performed using the Comet Assay Kit (Trevigen) according to manufacturer's instructions.
Statistical analyses were performed using StatView 512+. Gel bands, curve integration, and fluorescence intensity were quantitated with ImageJ software. Cell cycle profiles were analyzed with multicycle AV for Windows (Phoenix Flow Systems). Statistical significance was evaluated with the Student's t test and the χ2 test calculated using Prism5V.5.0 software. Fluorescence values in the irradiated region were annotated during recording. NFU were obtained by subtracting the fluorescence value of the first frame and comparing this value to maximal fluorescence intensity in control cells (considered 1). Tail Moment was calculated using Comet Assay IV software from Perceptive Instruments.
Supplementary Material
Acknowledgments
We thank Drs. M. C. Cardoso (Max-Delbrück-Centrum, Berlin, Germany) G. G. Poirier (University of Laval, Quebec, Canada), Y. C. Chen (Yang-Ming University, Taipei, Taiwan), J. Zhao and T. Roberts (Dana-Farber Cancer Institute, Boston, MA) for reagents. We also thank Dr. J. Santos (Centro Biología Molecular Severo Ochoa, Madrid), A. Suárez and V. Silió for support with different analyses, D. Megías (Confocal Microscopy Service, Centro Nacional de Investigaciones Oncologicas, Madrid) for help with laser irradiation, M. Marqués, J. L. Rodríguez, and V. Pérez for advice, and C. Mark for editorial assistance. This work was financed by grants from the Spanish Association Against Cancer and the Spanish Ministry of Science and Innovation (SAF2004-05955 and Network of Cooperative Research in Cancer RD07/0020/2020).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914242107/DCSupplemental.
References
- 1.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B, Ali K, Bilancio A, Geering B, Foukas LC. Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Bi L, Okabe I, Bernard DJ, Wynshaw-Boris A, Nussbaum RL. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J Biol Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- 4.Bi L, Okabe I, Bernard DJ, Nussbaum RL. Early embryonic lethality in mice deficient in the p110beta catalytic subunit of PI 3-kinase. Mamm Genome. 2002;13:169–172. doi: 10.1007/BF02684023. [DOI] [PubMed] [Google Scholar]
- 5.Foukas LC, et al. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 6.Marqués M, et al. Phosphoinositide 3-kinases p110alpha and beta regulate cell cycle entry, exhibiting distinct activation kinetics in G1 phase. Mol Cell Biol. 2008;28:2803–2814. doi: 10.1128/MCB.01786-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marqués M, et al. Specific function of phosphoinositide 3-kinase beta in the control of DNA replication. Proc Natl Acad Sci USA. 2009;106:7525–7530. doi: 10.1073/pnas.0812000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia S, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graupera M, et al. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–666. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 10.Ciraolo E, et al. Phosphoinositide 3-kinase p110beta activity: key role in metabolism and mammary gland cancer but not development. Sci Signal. 2008;1:ra3. doi: 10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown EJ. Analysis of cell cycle progression and genomic integrity in early lethal knockouts. Methods Mol Biol. 2004;280:201–212. doi: 10.1385/1-59259-788-2:201. [DOI] [PubMed] [Google Scholar]
- 12.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr Opin Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 15.Difilippantonio S, Nussenzweig A. The NBS1-ATM connection revisited. Cell Cycle. 2007;6:2366–2370. doi: 10.4161/cc.6.19.4758. [DOI] [PubMed] [Google Scholar]
- 16.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 17.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 18.Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupré A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 20.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 21.Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 22.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson SP, et al. PI 3-kinase p110beta: a new target for antithrombotic therapy. Nat Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 24.Murga M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruhlak MJ, Celeste A, Nussenzweig A. Monitoring DNA breaks in optically highlighted chromatin in living cells by laser scanning confocal microscopy. Methods Mol Biol. 2009;523:125–140. doi: 10.1007/978-1-59745-190-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito K, Scharenberg AM, Kinet JP. Interaction between the Btk PH domain and phosphatidylinositol-3,4,5-trisphosphate directly regulates Btk. J Biol Chem. 2001;276:16201–16206. doi: 10.1074/jbc.M100873200. [DOI] [PubMed] [Google Scholar]
- 27.Watton SJ, Downward J. Akt/PKB localisation and 3′ phosphoinositide generation at sites of epithelial cell-matrix and cell-cell interaction. Curr Biol. 1999;9:433–436. doi: 10.1016/s0960-9822(99)80192-4. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Capetillo O, Allis CD, Nussenzweig A. Phosphorylation of histone H2B at DNA double-strand breaks. J Exp Med. 2004;199:1671–1677. doi: 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YC, et al. Activation of phosphoinositide 3-kinase by the NBS1 DNA repair protein through a novel activation motif. J Mol Med. 2008;86:401–412. doi: 10.1007/s00109-008-0302-x. [DOI] [PubMed] [Google Scholar]
- 30.Lukas C, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haince JF, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 32.Maser RS, et al. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol Cell Biol. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sporbert A, Domaing P, Leonhardt H, Cardoso MC. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 2005;33:3521–3528. doi: 10.1093/nar/gki665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balajee AS, Geard CR. Chromatin-bound PCNA complex formation triggered by DNA damage occurs independent of the ATM gene product in human cells. Nucleic Acids Res. 2001;29:1341–1351. doi: 10.1093/nar/29.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 36.Lee RF, Steinert S. Use of the single cell gel electrophoresis/comet assay for detecting DNA damage in aquatic (marine and freshwater) animals. Mutat Res. 2003;544:43–64. doi: 10.1016/s1383-5742(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 37.Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 38.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.