Abstract
Guidance receptor signaling is crucial for steering migrating cells. Despite this, we generally lack direct measurements of such signaling. Border cells in Drosophila migrate as a tightly associated group, but dynamically, with front and rear cells exchanging places. They use the receptor tyrosine kinase (RTK) PDGF/VEGF receptor (PVR) as a guidance receptor, perceiving the attractant Pvf1. Here we determine the spatial distribution of PVR signaling by generating an antibody that specifically detects activated PVR in situ. PVR activity is very low in migrating border cells, due to strong activity of cellular phosphatases. Measurements of signal at the cell cortex show variability but a strong bias for both total active PVR and specific activity of PVR to be elevated at the front versus side of the leading cell, often with several-fold difference in signal levels. This polarized active PVR signal requires the E3 ubiquitin ligase Cbl and the recycling regulator Rab11, indicating a dependency on receptor trafficking. The endogenous ligand gradient contributes to shaping of signaling by increasing the specific activity of PVR toward the source in front cells. Surprisingly, signaling is also elevated at the back versus the side of rear cells. This distally polarized distribution of active PVR is ligand independent. Thus the actual guidance signal transmitted in border cells appears to integrate perceived ligand distribution with cell polarity or cell orientation with respect to the cluster. A general implication is that both group configuration and extrinsic cues can directly modulate guidance receptor signaling during collective cell migration.
Keywords: collective migration, directionality, Drosophila, guidance signal, receptor tyrosine kinase
Direct analysis of signal perception is key to understanding how cells interpret guidance signals to direct their migration. This has been difficult to achieve, however, because it is technically difficult to detect and measure signaling-active receptors—even more so when analyzing cell migration in its natural context of a 3D tissue. Important insights into guidance signaling have been obtained using local PIP3 accumulation as a proxy for activity of G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs) via PI3 kinase in different cell types (1–4). Differences have been observed between systems, which may either be due to differential signal perception or to differential downstream signal processing. Analysis of guidance receptor activation itself would improve our understanding of this crucial step in directed cell migration.
Apart from using different guidance receptors, such as GPCRs and RTKs, cells also differ in whether, under physiological conditions, they migrate individually (5) or collectively, as part of a group (6). When migrating individually, each cell is expected to detect and respond to spatial cues directly and independently. This may also be true for cells migrating as a group, but other possibilities exist. Interactions between cells could modulate perception of the external cue or guidance may be fully collective and require signal perception by the group (7).
Border cells undergo directed migration as a cell group in the Drosophila ovary (8). The system is genetically tractable and allows live imaging in the tissue, making it a good model for studying collective cell migration (9, 10). Once specified, border cells delaminate from the follicular epithelium, invade the underlying germ line tissue, and migrate between the giant nurse cells to the oocyte. They use the nurse cells as substratum for their migration with substrate adhesion mediated by E-cadherin (11). Border cells migrate as a tight group, always attached to one another. This attachment provides an inherent polarity for each cell, as intracluster cell interactions are distinguishable from interactions with the nurse-cell substrate. Live imaging has shown border cell migration to be very dynamic, with cells exchanging places frequently (9, 10). Border cells are directed by guidance cues from the oocyte and use two RTKs (PVR and EGFR) to read these cues (12, 13). Here we analyze guidance receptor activation in migrating border cells with focus on PVR because it has one essential ligand in this context, the secreted molecule Pvf1 (13, 14), whereas for EGFR, multiple ligands may be involved (12, 15).
Results
A Tool to Detect Active PVR in Vivo.
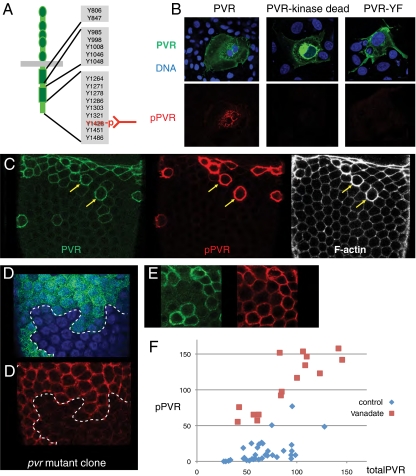
The fact that active RTKs autophosphorylate can be used to identify activated receptors as they display specific phosphoepitopes. To find an antibody that would specifically and with spatial resolution allow detection of active PVR in tissues, we generated a broad panel of monoclonal antibodies (mABs) against many potential tyrosine autophosphorylation sites in PVR and their peptide context (Table S1). One mAB, directed against pY1428 (Fig. 1A) was able to detect PVR overexpressed in COS cells but did not detect overexpressed kinase-inactive PVR or a PVR Y1428F mutant (Fig. 1B and Table S1), indicating this antibody detects active PVR, autophosphorylated on tyrosine 1428. Overexpressed, active EGFR and tyrosine-phosphorylated proteins in general were also not detected.
Fig. 1.
Detecting active PVR in vivo. (A) Schematic of PVR and target of the pPVR antibody (pY1428 and peptide context). (B) COS cells transfected with indicated GFP-fusion constructs. (Upper) GFP and DAPI signal. (Lower) pPVR antibody staining. The PVR-YF mutant has Y1428 changed to phenylalanine. (C) Follicular epithelium of a stage-10 egg chamber (slbo-Gal4 + UAS-PVR) stained with anti-PVR (green), anti-pPVR (red), and phalloidin (white). Two cells with high PVR expression indicated with yellow arrows. (D) Follicle epithelium [blue (DAPI) marking nuclei] with clone of Pvr/Pvr mutant cells marked by absence of GFP (green). Anti-pPVR (red) channel of same image shown below. (E) Epithelium with PVR overexpression as in C but after incubation with vanadate; setting for pPVR signal less sensitive than in C. (F) Plots of anti-PVR and anti-pPVR signals along the cortex of individual cells (after background subtraction), from multiple experiments, as shown in C and E, using the same settings. Arbitrary units.
To assess the specificity of this antibody in vivo, we analyzed cells of the follicular epithelium in the Drosophila ovary, which express a low level of PVR endogenously. Activated PVR, or pPVR, was detected at the cortex in follicle cells in which PVR was overexpressed at high levels (Fig. 1C). Cells with prominent pPVR labeling showed downstream PVR signaling effects such as F-actin accumulation (yellow arrows in Fig. 1C). pPVR was not detectable under normal conditions, but was detected in control cells, but not pvr−/− mutant cells, treated briefly with the phosphatase inhibitor vanadate (Fig. 1D). These findings suggest that this anti-pPVR antibody specifically detects active PVR, also in vivo. When phosphatases were inhibited, the pPVR signal increased linearly with PVR levels upon overexpression (Fig. 1 E and F). Without vanadate treatment, pPVR signal was very low and increased nonlinearly with increasing PVR expression (Fig. 1 C and F); however, the signal remained below that resulting from phosphatase inhibition, showing that overexpressed PVR can still be further activated. The vanadate experiments also indicated that PVR is continuously activated in normal cells, but effectively inactivated by cellular phosphatases.
Spatial Distribution of Active PVR in Border Cells Initiating Migration.
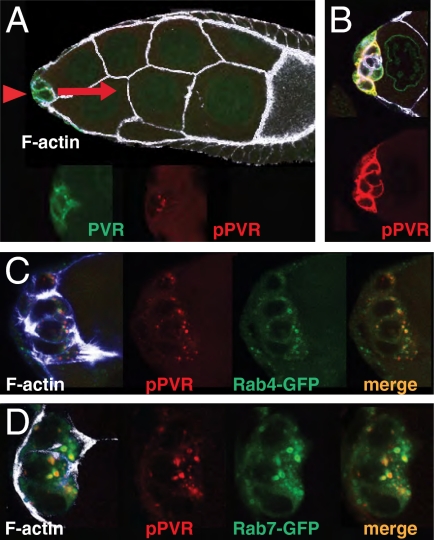
To analyze guidance signaling by PVR, we turned to border cells. The endogenous level of PVR in border cells is low and we had to moderately increase PVR levels to detect pPVR. At these levels, PVR activity was still stringently controlled by phosphatases, and we confirmed that directed migration was unperturbed. Indeed, elevated PVR ensured that directed migration was strongly dependent on the endogenous ligand Pvf1 (Fig. S1). The pPVR signal remained barely detectable at the border cell cortex (Fig. 2A). An estimated 30- to 100-fold increase in cortical pPVR was observed when phosphatases were inhibited (Fig. 2B). Thus phosphatases are very active in border cells and, consequently, only a small fraction of PVR molecules normally remain active at the plasma membrane. Without phosphatase inhibition, pPVR signal was enriched in large dots (Fig. 2A). pPVR colocalized with Rab4-YFP (Fig. 2C) as well as Rab7-GFP (Fig. 2D), indicating that this signal corresponds to endosomal compartments, and thus visualizes active PVR destined for recycling or for degradation. The endosomal signal was not obviously increased upon vanadate treatment (Fig. 2B), suggesting that this pPVR pool may be protected from cytosolic phosphatases, possibly present in internal structures of multivesicular bodies. The pPVR-rich dots were highly enriched toward the front of cells initiating migration, and may reflect active PVR endocytosed from the front plasma membrane.
Fig. 2.
Detection of active PVR in border cells initiating migration. (A) Early stage-9 egg chamber, with border cells (arrowhead) initiating migration. In this and all subsequent images, direction of migration is to the right (red arrow), the genotype is slbo-Gal4 + UAS-PVR, and white/blue is phalloidin signal. Total PVR (green); pPVR (red) signals are shown separately in the enlargement of the border cell cluster below. (B) Border cells as in A but after incubation with the phosphatase inhibitor vanadate for 10 min. (C) Border cell cluster from females expressing Rab4-YFP ubiquitously (green), showing overlap with the pPVR signal (merge). (D) Border cell cluster from females carrying UAS-Rab7-GFP. Note that overexpression of Rab proteins may change the size of targeted (endosomal) compartments, disallowing quantitative comparisons between genotypes (see, e.g., ref. 27).
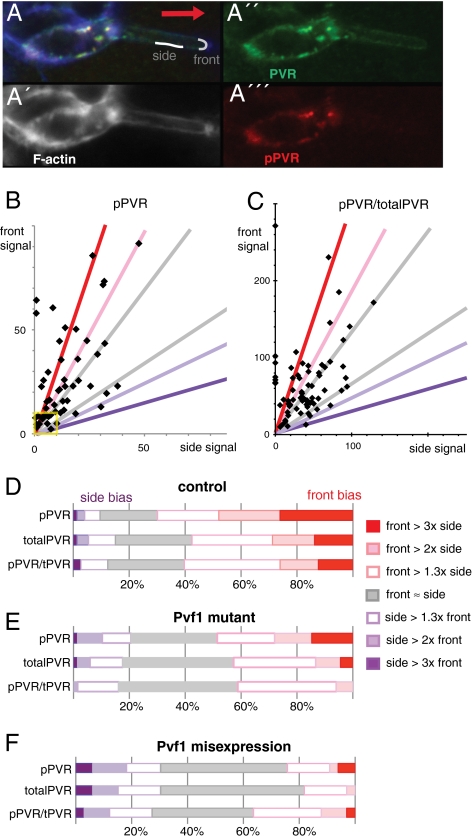
To directly evaluate the incoming guidance signal, we measured the low cortical pPVR and total PVR signals in border cells initiating migration. The pPVR signal indicates the amount of signal transmitted intracellularly, whereas pPVR divided by total PVR reflects what fraction of PVR is activated, or specific activity of PVR. Measurements were performed on the front border cell, averaging along a region of the front and of the side membrane in direct contact with the germ line substrate (Fig. 3A). The back of a border cell initiating migration abuts other border cells, and pPVR measurements here could reflect contributions by two cells. Where singular back membranes could be identified, they were similar to side membranes. The first noticeable feature was that the cortical signals of pPVR were quite variable (Fig. 3B), both in terms of signal levels and signal polarization, the ratio between front and side signals. A similar variable distribution was found for the specific activity of PVR (pPVR/totalPVR; Fig. 3C), indicating there is no fixed relationship between pPVR and total PVR levels. As images analyzed show snapshots of PVR activity, one interpretation of the variable signal is that the signaling output changes dynamically. The second noticeable feature was that despite the variation, there was a very pronounced front bias of active pPVR (fraction of snapshots with higher front signal; Fig. 3B and Fig. 3D Top). The bias was seen both for low levels of pPVR signal (yellow box in Fig. 3B) and moderate levels, and was highly statistically significant (P < 0.001). This front bias in pPVR output reflected two cooperating underlying biases: a tendency for total PVR protein to show front enrichment (Fig. 3D, second bar) and a tendency for the specific activity of PVR to be higher at the front (Fig. 3D, third bar, and Fig. 3C; P < 0.001 for both). To confirm these findings, we performed an independent set of experiments, with similar sample size but slight differences in how measurements were done. We obtained similar distributions (Fig. S2). Also, control experiments with an antibody detecting a general membrane associated protein showed no front bias (Fig. S3). Overall, the data indicate variable, likely dynamic, but clearly front-biased PVR signaling in the front cell.
Fig. 3.
Active PVR at the cell surface in border cells initiating migration. (A) Part of a border cell cluster initiating migration, focused on the front cell. Cortical traces typically used for quantitation of side and front signal are shown in gray line, drawn in phalloidin channel (A′), measured in anti-PVR channel (A′′), and pPVR channel (A′′′). (B and C) Plot of (average) pPVR signals and of pPVR/totalPVR signal at front versus side membranes of a front border cell initiating migration as in A, where background is subtracted from all signals first. Each point represents measurements from one border cell cluster; n = 76. In a few samples, side measurements were slightly below background and then set to 0. Colored lines correspond to limits for classifications used in D–F. Yellow box marks cell values close to background levels; excluding these does not noticeably change ratios depicted in D. (D) Distribution of signal ratios in front versus side measurements of pPVR (pPVRfront/pPVRside), totalPVR (PVRfront/PVRside), and pPVR/totalPVR (pPVRfront/pPVRside)/(PVRfront/PVRside) in front border cells initiating migration (as in B and C). Front ≈ side means front values ±30% of side values; front > 1.3× side means a front value 1.3–2× higher than side value; front > 2× side a front value 2–3× higher than side value, etc., expressed as fraction of clusters with indicated signal bias; n = 76. (E) Distribution of signal ratios in border cells initiating migration as for D, but in Pvf1/Pvf1 mutant females; n = 82. Corresponding scatter plots shown in Fig. S3. (F) Signal ratios in border cells initiating migration as for D, but also misexpressing Pvf1 (slbo-Gal4, EPgPvf1); n = 20.
Ligand Dependence of the Spatial Distribution of Active PVR.
The front of the front cell is closest to the oocyte, the source of the ligand Pvf1 (13). To assess ligand dependence of the pPVR signal, we analyzed border cells in a Pvf11624 mutant background. Pvf11624 is a strong loss-of-function allele and has the same phenotype as loss of PVR in the context of border cell migration (13). Live imaging confirmed that border cells were motile but much less directional in the Pvf1 mutant background (early stage of migration; n = 8). In the present experimental setup, migration was severely delayed for 95% of border cell clusters in the Pvf1 mutant (Fig. S1). Interestingly, active PVR was still detected in the Pvf1 mutant, with a distribution of signals for both pPVR and specific activity of PVR overlapping with that of the control situation (scatter plot in Fig. S4). The average signal was only slightly decreased. Some front bias was also retained for both pPVR and specific activity of PVR (significant at P < 0.05). However, the degree of signal polarization, or front bias, was reduced in the ligand mutant (Fig. 3E and Fig. S4). This was primarily seen as loss of cells with extreme front bias of PVR-specific activity (Fig. 3E Bottom), although there was also a reduction in total PVR front bias (Fig. 3E Middle). Uniform overexpression of the Pvf1 ligand resulted in 40% increased pPVR signal on average, and complete absence of front bias (Fig. 3F). These findings show that the ligand Pvf1 and, apparently, its correct distribution, is important for correct pPVR signal distribution.
It was surprising that many cells retained not only apparently normal signaling levels, but also significant front bias of activated PVR in a Pvf1 mutant with severe reduction in ligand level. As mentioned previously, phenotypic analyses indicated that all border cell clusters in the Pvf1 mutant were adversely affected. Perhaps the remaining moderate bias in signal localization is not sufficient for guidance and the signal must either be extremely front biased or remain biased for a long period to elicit the correct directional response. It may also not be adequate to analyze the front cell to understand guidance. We therefore decided to investigate PVR signaling in other cells of the border cell cluster.
Spatial Distribution of Active PVR in Migrating Border Cells Clusters.
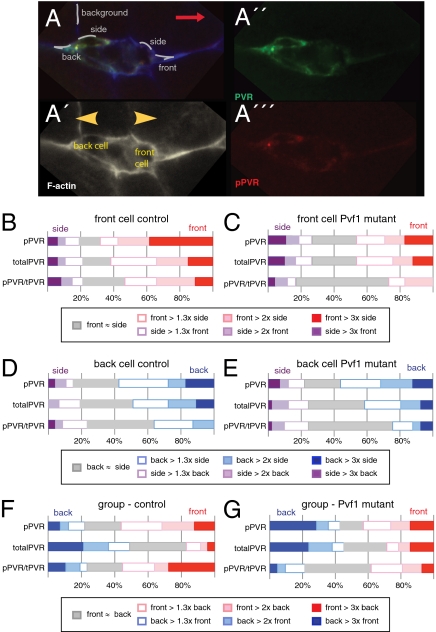
To investigate the spatial aspect of PVR activation and its ligand dependence in different border cells within a cluster, we analyzed them during migration. Specifically, we examined clusters during the first half of their migration toward the oocyte, as these generally have a clearly defined leading cell and a back cell oriented in the opposite direction (Fig. 4 A and A′). The orientation of the back cell relative to the cluster is then the mirror image of the front cell (yellow arrowheads point outward); ligand distribution is determined by orientation of the cluster in the tissue (red arrow points to oocyte). As for clusters initiating migration, the front cell had a robust front bias of activated PVR and of specific activity of PVR (Fig. 4B). Also, the front bias of active PVR was reduced in the Pvf1 mutant (Fig. 4C), in particular specific activity of PVR. Interestingly, the back cell also showed a bias, but less strong, and with opposite polarity: A higher fraction of clusters showed more pPVR signal at the back than at the side of the back cell (Fig. 4D; significant at P < 0.001). In the Pvf1 mutant, the back bias of the back cell (Fig. 4E) was similar to the front bias of the front cell (Fig. 4C) and back bias of control (Fig. 4D), indicating that this outward signaling bias is ligand independent. Most, but not all, of this outward bias is due to bias in total PVR distribution (compare top and middle bars in Fig. 4 C–E). Finally, we compared PVR activation in the front of the front cell with that in the back of the back cell in the same migrating cluster. Note that PVR expression levels may differ between cells of the group. Higher specific activity of PVR was more frequently observed in the front of the cluster than the back (Fig. 4F; P < 0.05), and this difference was ligand dependent (Fig. 4G). This finding suggest that the Pvf1 ligand also affects the relative level of PVR signaling between cells of the group.
Fig. 4.
PVR signaling in front and back cells of border cell clusters during migration. (A) A border cell cluster during the first half of its migration toward the oocyte, overlay in A, phalloidin only in (A′), as well as anti-PVR (A′′) and anti-pPVR (A′′′) channels shown. Red arrow indicates direction of migration; yellow arrowheads (in A′) indicate front and back cell orientations relative to the rest of the cluster. Typical traces used for measurements are indicated in A. (B and C) Distribution of signal ratios for pPVR, totalPVR, and pPVR/totalPVR at front versus side in the front border cell of a migrating cluster in control (B) and Pvf1 mutant (C) background. Scoring and classification done as in Fig. 3D and expressed as fraction of clusters with indicated signal bias; n = 59 (control), n = 44 (Pvf1). (D and E). Distribution of signal ratios for pPVR, totalPVR, and pPVR/totalPVR at the back versus the side of the back border cell in migrating clusters (see, e.g., in A and A′) in control (D) and Pvf1 (E) mutant background. Scoring and classification as in B and C, except back signal replaces front signal. (F and G) Signal ratios of pPVR, totalPVR, and pPVR/totalPVR at the front of the front cell versus at the back of the back cell of migrating border clusters in control (F) and Pvf1 (G) mutant background. Scoring and classification as in B and C, except back of the back cell replaces side signal.
In summary, the amount of active PVR, and to a lesser extent the total PVR level, is influenced by a border cell's orientation relative to the rest of the cluster and is outward, or distally, polarized during migration. The pronounced front-biased PVR activation in front cells reflects the combined effects of this distal polarization and effects of the extrinsic guidance cue, Pvf1. In the absence of Pvf1, front and back cells are equally polarized in their PVR signaling, but in opposite directions; also, the usual increased signaling in the front versus back cell is not observed. Both of these changes may contribute to the cluster not showing properly guided movement without ligand.
Cellular Requirements for Basic Polarized Distribution of Active PVR.
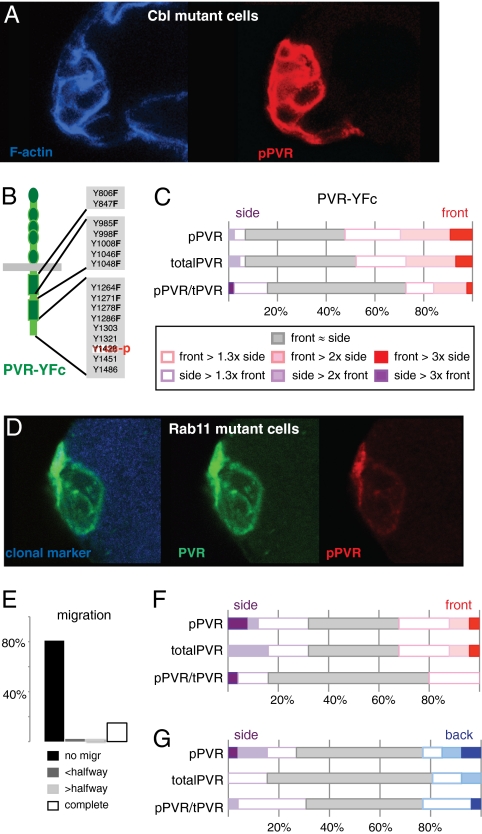
We next wanted to explore the basis for the observed ligand-independent, distally polarized bias of PVR signaling. We had previously found indications that regulators of RTK endocytosis, including the E3 ubiquitin ligase Cbl, contribute to guidance of border cells and to the distribution of total phosphotyrosine signal (16). The ability to directly detect active PVR allowed us to now test the effect of Cbl on distribution of active PVR. Cbl mutant clones showed two dramatic changes in the pPVR signal (Fig. 5A): a robust increase in signal levels (average of 10-fold) and delocalized signaling with no front bias (n = 19). The delocalized pPVR signal in Cbl mutant clones could be an indirect consequence of the increased signaling level. To address this, we analyzed a PVR mutant that cannot directly recruit Cbl but that has reduced overall signaling output, as 11 of the potential docking tyrosines are mutated to phenylalanine (PVR-YFc, Fig. 5B; see ref. 16). For PVR-YFc, the degree of front bias of pPVR signal was likewise markedly reduced (Fig. 5C, compare with Fig. 3D), in particular in terms of specific activity of PVR. This finding supports the idea that recruitment of Cbl to PVR affects the distribution of active PVR. Together, these data provide direct evidence that Cbl and receptor endocytosis affect the spatial distribution of the active RTK.
Fig. 5.
Effects of perturbing Cbl or Rab11 on active PVR in border cells. (A) Image of Cbl homozygous mutant border cell clone, showing elevated anti-pPVR (red); compare with controls in Figs. 2A and 3A. (B) Schematic of PVR-YFc, a PVR mutant that does not bind Cbl-SH2 directly (16) but retains the pPVR epitope. (C) Distribution of signal ratios for pPVR, totalPVR, and pPVR/totalPVR at front versus side in the front border cell expressing PVR-YFc at initiation of migration; calculated and scored as in Fig. 3D; n = 53. (D) Image of Rab11 homozygous mutant border cell clone (mutant cells marked by absence of blue) at early stage 9, stained with anti-PVR (green) and anti-pPVR (red). (E) Quantification of border cell migration at stage 10 from Rab11 mutant border cells with slbo-Gal4 + UAS-PVR; n = 47; for control, see Fig. S1. (F) Distribution of signal ratios for pPVR, totalPVR, and pPVR/totalPVR at front versus side of front cells of Rab11 mutant clusters initiating migration as in E, calculated and scored as in Fig. 3D; n = 25. (G) Distribution of signal ratios for pPVR, totalPVR, and pPVR/totalPVR at back versus side of back Rab11 mutant cells during migration; n = 26. Measurements are from mixed clusters in which some cells are homozygous mutant for Rab11. Control genotype and scoring details in Fig. 4D.
RTK endocytosis could contribute to front-biased signaling either by primarily allowing removal of active RTK from the side of a cell or by promoting enrichment of active (and bulk) RTK at the front of a cell, possibly by recycling. The presence of active PVR in endosomal compartments primarily near the front of migrating cells (Fig. 2) provides some support for the latter hypothesis. Also, that several-fold differences in levels of active PVR levels can be observed in areas only a few microns apart—notably the front versus the side of many front cells (Figs. 3D and 4B)—could indicate an active enrichment process. To determine if there is a requirement for recycling, border cell clusters mutant for Rab11 were analyzed. Neither active PVR nor total PVR signals showed front bias in Rab11 mutant cells (Fig. 5 D and F), indicating PVR signaling was completely delocalized in this background. Correspondingly, a strong defect in directional migration was observed in cells with elevated PVR and reduced Rab11 (Fig. 5E). These measurements were performed on front cells initiating migration. pPVR signal was also found to be delocalized in migrating clusters in which the back cells were mutant for Rab11 (Fig. 5G, compare with Fig. 4D). Note that this happens without the increase in PVR signaling that was observed in Cbl mutant cells; in Rab11 mutant cells, the average cortical pPVR signal was similar to control levels. We conclude that the processes of endocytosis and recycling, likely of the receptor itself, are required for proper spatial distribution of PVR and active PVR in border cells.
Discussion
By direct detection of active PVR in migrating border cells, we have started to uncover what guidance receptor signaling actually looks like in vivo and note some interesting features. First, the endogenous ligand, Pvf1, is not required for signaling as such but significantly contributes to the shape of the response, promoting a prominent front-signaling bias in the front cell. The ligand primarily affects the front bias in specific activity of PVR. Correct ligand distribution is not sufficient for front-biased signaling, however; receptor trafficking is also required. Also, there is a general bias for active and total PVR to be enriched distally, at the region of the outer border cell membrane furthest from other cells of the group. The lack of polarization for both active PVR and total PVR in mutants affecting receptor trafficking suggests that the distal enrichment is active and trafficking dependent. Finally, these two aspects of spatial control of guidance receptor activity, effects of the extrinsic directional cue and cellular requirements, are interrelated and reinforce one another: In the Pvf1 mutant or upon overexpression, not only the specific activity of PVR but also the total PVR distribution is less polarized. Conversely, when trafficking is affected, the front bias in specific activity of PVR is also absent or strongly reduced. One possible explanation for this relationship is that trafficking of ligand-bound PVR is more effective or more polarized than trafficking of nonbound PVR. Given that PVR was found to be somewhat active without ligand, biased receptor trafficking could also provide the basic distal bias in PVR activity, with or without ligand. However, other factors, such as localized activity-dependent interactions or sequestering, could also contribute.
Border cells migrate as a tight and free (nontethered) group of cells, and their directional movement is guided by external cues. Our findings suggest that the group structure may directly affect the perception of guidance information, in addition to the external ligand distribution: The distal enrichment of active and total PVR indicates that PVR activity reflects the position and orientation of the cell relative to other cells of the cluster, not just relative to the tissue. This, in turn, suggests that cell–cell contact within the group directly or indirectly affects guidance signaling. Such a cell contact-dependent shaping of signaling could in principle allow polarized guidance receptor activation that is spatially controlled exclusively by the organization of a cell group, and not by ligand distribution. Uniform ligand, or no ligand, could therefore give directional signaling output if a group is not free but organized in an inherently polarized manner. This effect could be responsible for, or contribute to, directional cell movement in other types of collective migration—for example, sheet or slug migration in tissue culture models or in vivo (6, 17).
The combined effects of cellular features and extrinsic cues on guidance receptor activity observed for border cells is possibly advantageous for directional migration—it may aid detection of low ligand levels or it may help cells maintain the same response as they move up the ligand gradient. In other systems, guidance receptor activity may directly reflect the external guidance cue concentration, for example for GPCR-based detection of external cAMP in Dictyostelium (18, 19), one popular model of chemotaxis in eukaryotic cells. Amplification of signaling differentials also occur in such systems, but likely further downstream of the guidance receptor (20, 21). The latter type of signal perception strategy may allow a more acute response to changing external stimuli. Though intriguing, these ideas remain very speculative as we generally lack direct information on guidance receptor signaling status. Future efforts to measure guidance receptor activation as well as downstream outputs in different types of directionally migrating cells may allow us to better appreciate how different guidance signal perception strategies are chosen and used.
Methods
Monoclonal antibodies were generated by immunizing mice with the tyrosine phosphorylated peptides (Peptide Specialty Laboratories) listed in Table S1 and selecting clones by ELISA analysis that responded to the phosphorylated peptide over 2-fold better than the identical but nonphosphorylated peptide. Mouse immunization, production of monoclonals, and initial screening and testing was performed at the monoclonal facility, EMBL Monterotondo. Subsequent testing of positive clones was done by detecting autophosphorylated PVR in COS cells. COS cells were transiently transfected with CMV-PVR plasmids encoding wild-type, kinase dead, or PVR-YF mutants and processed for immunofluorescence (standard protocols). Thirteen different clones, representing five epitopes, were positive and proceeded for testing in vivo, on ovaries from slbo-Gal4 + UAS-PVR females and slbo-Gal4 + UAS-EGFR females (Table S1). Antibodies with no/weak staining, background staining, or cross-reactivity with EGFR were eliminated, leaving only one specific clone, anti-PY7-14D6, directed against phosphorylated Y1428 of PVR in the peptide context. Supernatants from mAB-producing hybridomas were used (undiluted).
Optimized protocol for anti-pPVR staining: Ovaries were finely dissected in Grace's cell insect medium (Sigma) for a maximum of 20 min and immediately fixed in Grace's + 4% paraformaldehyde for 20 min. Ovaries were rinsed twice in wash buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.05% Nonidet P-40, 1 mg/mL BSA], washed for 30 min and manually dissociated into single egg chambers. Blocking was performed for 30 min in wash buffer with 5 mg/mL BSA (block buffer) and incubated overnight at 4 °C with primary antibody diluted 1:5 (mAB for pPVR) and 1:200 (rat polyclonal antibody (13) for total PVR) in block buffer. The remaining procedure was standard, with secondary antibodies (Jackson Immunoresearch Laboratories) and rhodamine-coupled phalloidin (Molecular Probes). In case of vanadate treatment, 0.5 mM vanadate was prepared freshly from a stock solution and added to a modified dissection medium appropriate for live imaging (10).
To quantify cortical signals, the central, or most extended, sections of migrating cells were selected, and nonsaturating images were captured on a Leica confocal, quality controlled, and quantified in unmodified form. A 1.5- to 3.5-μm-long line was drawn based in phalloidin (F-actin) channel (side or front/back), expanded to eight pixels (corresponding to 430 nm), and average pixel intensity measurements were performed in channels detecting pPVR and total PVR. Background was similarly measured along a nearby nurse cell membrane for each sample. Statistical significance of differences observed (bias for front over side, for example) was determined using the Wilcoxon signed-rank test. To allow a rough comparison of signal levels between different genetic backgrounds, we used the average of pPVRsignal/pPVRbackground as a measure of average signal strength.
For all experiments in border cells, the control genotype was slbo-Gal4 + UAS-PVR, and mutants were introduced into this background. The Pvf11624 mutant used is a strong loss-of-function mutant (13); in homozygous mutant ovaries, transcripts were barely detectable by RT-PCR (less than 2% of wild type). EPg11235 (13) was used for Pvf1 misexpression. The following alleles, recombined onto the appropriate FRT chromosome, were used for clonal analysis: Pvr1, a null allele (22), CblF165, a likely null allele (23), Rab11j2D1, a strong hypomorphic allele (24). tub-Rab4-GFP (25) and UAS-Rab7-GFP (26) were used as markers. PVR-YF mutants are described in ref. 16. PVR-YFa (Y1428F) construct is used in Fig. 1B.
Supplementary Material
Acknowledgments
We thank Temasek Life Sciences Laboratory and the European Molecular Biology Laboratory (EMBL) for support at various stages of this project. We specifically thank Alan Sawyer and the EMBL monoclonal facility, and Rainer Pepperkok for hosting K.J. Thanks to Minna Poukkula for live imaging of border cell samples, Zhang Rui for help with statistical analysis, Susan Eaton and Marcos Gonzalez-Gaitan for flies, Adam Cliffe, Darren Gilmour and Stefano De Renzis for helpful discussions, and Stephen Cohen for comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915075107/DCSupplemental.
References
- 1.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 2.Servant G, et al. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell. 2005;8:215–227. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Haugh JM, Codazzi F, Teruel M, Meyer T. Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J Cell Biol. 2000;151:1269–1280. doi: 10.1083/jcb.151.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halin C, Rodrigo Mora J, Sumen C, von Andrian UH. In vivo imaging of lymphocyte trafficking. Annu Rev Cell Dev Biol. 2005;21:581–603. doi: 10.1146/annurev.cellbio.21.122303.133159. [DOI] [PubMed] [Google Scholar]
- 6.Rørth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- 7.Rørth P. Collective guidance of collective cell migration. Trends Cell Biol. 2007;17:575–579. doi: 10.1016/j.tcb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Montell DJ. Border-cell migration: The race is on. Nat Rev Mol Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- 9.Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 10.Bianco A, et al. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- 11.Niewiadomska P, Godt D, Tepass U. DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J Cell Biol. 1999;144:533–547. doi: 10.1083/jcb.144.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duchek P, Rørth P. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science. 2001;291:131–133. doi: 10.1126/science.291.5501.131. [DOI] [PubMed] [Google Scholar]
- 13.Duchek P, Somogyi K, Jékely G, Beccari S, Rørth P. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- 14.McDonald JA, Pinheiro EM, Montell DJ. PVF1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with Taiman. Development. 2003;130:3469–3478. doi: 10.1242/dev.00574. [DOI] [PubMed] [Google Scholar]
- 15.McDonald JA, Pinheiro EM, Kadlec L, Schupbach T, Montell DJ. Multiple EGFR ligands participate in guiding migrating border cells. Dev Biol. 2006;296:94–103. doi: 10.1016/j.ydbio.2006.04.438. [DOI] [PubMed] [Google Scholar]
- 16.Jékely G, Sung HH, Luque CM, Rørth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Vitorino P, Meyer T. Modular control of endothelial sheet migration. Genes Dev. 2008;22:3268–3281. doi: 10.1101/gad.1725808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 19.Ueda M, Sako Y, Tanaka T, Devreotes P, Yanagida T. Single-molecule analysis of chemotactic signaling in Dictyostelium cells. Science. 2001;294:864–867. doi: 10.1126/science.1063951. [DOI] [PubMed] [Google Scholar]
- 20.Van Haastert PJ, Devreotes PN. Chemotaxis: Signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 21.Charest PG, Firtel RA. Feedback signaling controls leading-edge formation during chemotaxis. Curr Opin Genet Dev. 2006;16:339–347. doi: 10.1016/j.gde.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 22.Brückner K, et al. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev Cell. 2004;7:73–84. doi: 10.1016/j.devcel.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Pai LM, Barcelo G, Schüpbach T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61. doi: 10.1016/s0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- 24.Dollar G, Struckhoff E, Michaud J, Cohen RS. Rab11 polarization of the Drosophila oocyte: A novel link between membrane trafficking, microtubule organization, and oskar mRNA localization and translation. Development. 2002;129:517–526. doi: 10.1242/dev.129.2.517. [DOI] [PubMed] [Google Scholar]
- 25.Marois E, Mahmoud A, Eaton S. The endocytic pathway and formation of the Wingless morphogen gradient. Development. 2006;133:307–317. doi: 10.1242/dev.02197. [DOI] [PubMed] [Google Scholar]
- 26.Entchev EV, Schwabedissen A, González-Gaitán M. Gradient formation of the TGF-beta homolog Dpp. Cell. 2000;103:981–991. doi: 10.1016/s0092-8674(00)00200-2. [DOI] [PubMed] [Google Scholar]
- 27.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: A key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.