SUMMARY
Human embryonic stem cells (hESCs) share an identical genome with lineage-committed cells, yet possess the remarkable properties of self-renewal and pluripotency. The diverse cellular properties in different cells have been attributed to their distinct epigenomes, but how much epigenomes differ remains unclear. Here, we report that epigenomic landscapes in hESC and lineage committed cells are drastically different. By comparing the chromatin modification profiles and DNA methylomes in hESCs and primary fibroblasts, we find that nearly one-third of the genome differs in chromatin structure. Most changes arise from dramatic redistributions of repressive H3K9me3 and H3K27me3 marks, which form blocks that significantly expand in fibroblasts. A large number of potential regulatory sequences also exhibit a high degree of dynamics in chromatin modifications and DNA methylation. Additionally, we observe novel, context-dependent relationships between DNA methylation and chromatin modifications. Our results provide new insights into epigenetic mechanisms underlying properties of pluripotency and cell-fate commitment.
INTRODUCTION
Pluripotent embryonic stem cells (ES cells) possess the ability to differentiate into multiple cell lineages in the body (Thomson et al., 1998). The underlying molecular mechanisms of pluripotency and cell fate commitment have not been completely understood. At least two models account for differences between pluripotent stem cells and lineage-committed cell types. In the first, “master switches” activate distinct networks of transcription factors that govern the transcriptional program of each cell type and dictate cellular properties (Marson et al., 2008). For example, the OCT4/SOX2/NANOG network enables self-renewal properties of ES cells, and ectopic expression of these factors together with additional transcription factors reprogram somatic cells into pluripotent cells (iPS cells) (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). But the efficiency of reprogramming is low, suggesting the involvement of additional factors or mechanisms. Additionally, lineage-committed cells are stable over many cell divisions, after the initial “master switch” transcription factors are no longer expressed. Such a phenomenon, referred to as “cellular memory”, has been difficult to explain by the transcription factor network model (Ringrose and Paro, 2004).
Another model for cellular memory involves the cell’s epigenomic landscape consisting of covalent modifications to DNA or histones (Ringrose and Paro, 2004), which either enable or prevent parts of the genome to be active in different cell types. In stem cells, the epigenome is highly malleable and responsive (Meshorer and Misteli, 2006), unlike that of somatic cells. Multiple lines of evidence support this model. First, ES cells have a higher number of “bivalent” or “poised” promoters marking important developmental regulators compared to differentiated cells, as indicated by the repressive mark histone H3 lysine 27 trimethylation (H3K27me3) and the active chromatin modification H3K4me3 (Azuara et al., 2006; Bernstein et al., 2006; Pan et al., 2007). Second, immunofluorescent imaging showed that following differentiation, mouse ES cells display increased heterochomatin in the nucleus (Meshorer and Misteli, 2006). Additionally, depletion of the Jmjd1a and Jmjd2c demethylases for the heterochromatin modification H3K9me3 results in stem cell differentiation (Loh et al., 2007). Third, DNA methylation is found at the promoters of critical pluripotency genes such as Oct4 during differentiation, and is responsible to keep such genes silent in differentiated cells (Ben-Shushan et al., 1993; Deb-Rinker et al., 2005).
Recent large-scale analyses of DNA methylation and histone modifications revealed dynamic chromatin states and DNA methylation status at promoters and most CpG islands (Brunner et al., 2009; Meissner et al., 2008), showing that the methylation state of H3K4 is a good indicator of promoter DNA methylation levels in mammalian cells. This is consistent with prior studies indicating that H3K4 methylation disrupts DNA methylation by inhibiting contact of DNMTs with histones, whereas promoters marked only with H3K27me3 in mouse ES cells are more likely to exhibit de novo DNA methylation following differentiation (Meissner et al., 2008; Mohn et al., 2008). While these insights suggest potential mechanisms of epigenetic regulation at promoters, the epigenetic regulatory mechanisms outside of promoters remains largely unclear.
To obtain a better understanding of the epigenomic landscapes in the pluripotent and differentiated cell states, and explore the links between histone modifications and DNA methylation genome-wide, we conducted a comprehensive analysis of 11 histone modifications and a recently acquired genome-wide nucleotide resolution map of DNA methylation in H1 human embryonic stem cells (hESCs) and fetal lung fibroblasts (IMR90) (Lister et al., 2009). We show a large-scale expansion of H3K9me3 and H3K27me3 domains in differentiated cells relative to hESCs, which selectively affects genes related to pluripotency, development, and lineage-specific functions. We also find multiple epigenetic mechanisms by which key pluripotent transcription factors are silenced in somatic cells. Finally, our analysis of both cell types reveals context-dependent, complex relationships between chromatin modifications and DNA methylation.
RESULTS and DISCUSSION
Chromatin Modification Landscapes
We performed ChIP-Seq (Johnson et al., 2007) experiments to identify global histone modification patterns for H2BK5ac, H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K9me3, H3K18ac, H3K27ac, H3K27me3, H3K36me3, and H4K5ac. Antibodies were validated for specificity with peptide dot blot assays and Western blotting (Figure S1). ChIP-Seq experiments produced 5 to 40 million monoclonal, uniquely mapped tags per chromatin modification and input DNA for well correlated biological replicates in H1 hESCs and IMR90 fibroblasts (Figure 1, Supplemental Table S1, Figure S11). We chose acetylation marks, which are indicative of open chromatin, as well as H3K4 methylation marks featured at active enhancers and promoters (Heintzman et al., 2009; Heintzman et al., 2007). We contrasted this architecture with the repressive structures of H3K27me3 and H3K9me3 and the genic localization of H3K36me3, a transcription elongation-coupled factor (Krogan et al., 2003; Li et al., 2002).
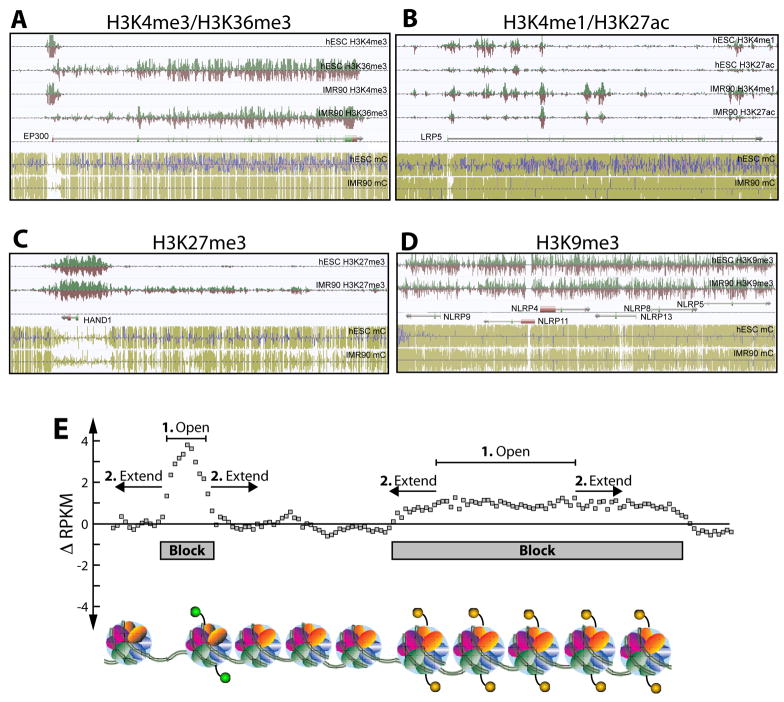
Figure 1. Histone Modifications form Block Structures.
(A–D) AnnoJ Browser snapshots of representative examples of the relationship between histone modifications and DNA methylation. Strand-specific mapped tags are displayed above (green) and below the line (red). Strand-specific DNA methylation (mC): mCG (yellow-green bars), mCHG (blue bars), mCHH (pink bars). (A) The p300 (EP300) locus illustrating depleted mCG in the promoter containing H3K4me3, and H3K36me3 in the gene body of both cell types. (B) The NLRP gene cluster is encompassed by H3K9me3 in both cells types, a reduction in mCG is evident in IMR90. (C) H3K27me3 is present at the mCG depleted promoter of the developmental transcription factor HAND1. Spreading of H3K27me3 in IMR90 accounts for correlated differences with mCG. (D) H3K4me1 and H3K27ac occur at both the promoter of LRP5 and distal sites which show differences in mCG. (E) A schematic of the ChromaBlocks strategy for identifying chromatin domains from input-normalized ChIP-Seq data.
Visualization of these maps indicates that histone modifications show two distinct types of spatial distributions: small, punctuated peaks and large, spreading domains (Figure 1). Modifications such as H3K4 methylation and acetylation are typically found in peak-like structures (Figure 1A–B), while H3K36me3, H3K27me3, and H3K9me3 are usually seen in broad domains of varying widths (Figure 1A, C–D). It is possible that these two types of chromatin modification patterns correspond to distinct physical structures of chromatin fibers in vivo (Figure 1E).
Most peak-finding programs rely on the assumption of stranded distributions of tags at peaks, and as such cannot detect the broad domains typical of histone modifications(Kharchenko et al., 2008; Zhang et al., 2008). To systematically enumerate genomic regions enriched for chromatin modifications, we devised a computational method called ChromaBlocks, which is tailored to identify both peak-like structures and large chromatin domains from ChIP-Seq data (see Methods). This analysis yielded between 9,414 and 43,536 blocks for each chromatin modification in hESC or IMR90, occupying between 68 million and 510 million base pairs (Table S2). Analysis with permuted data indicates that this method is highly specific (median FDR = 0.97%, Table S2) and approaches saturation around 10 million unique monoclonal reads (Figure S2D-G). With this tool in place, we are able to assess varying chromatin structures and compare how the chromatin landscape of hESCs differs from that of differentiated cells.
Redistribution of Large Domains of Chromatin Modifications
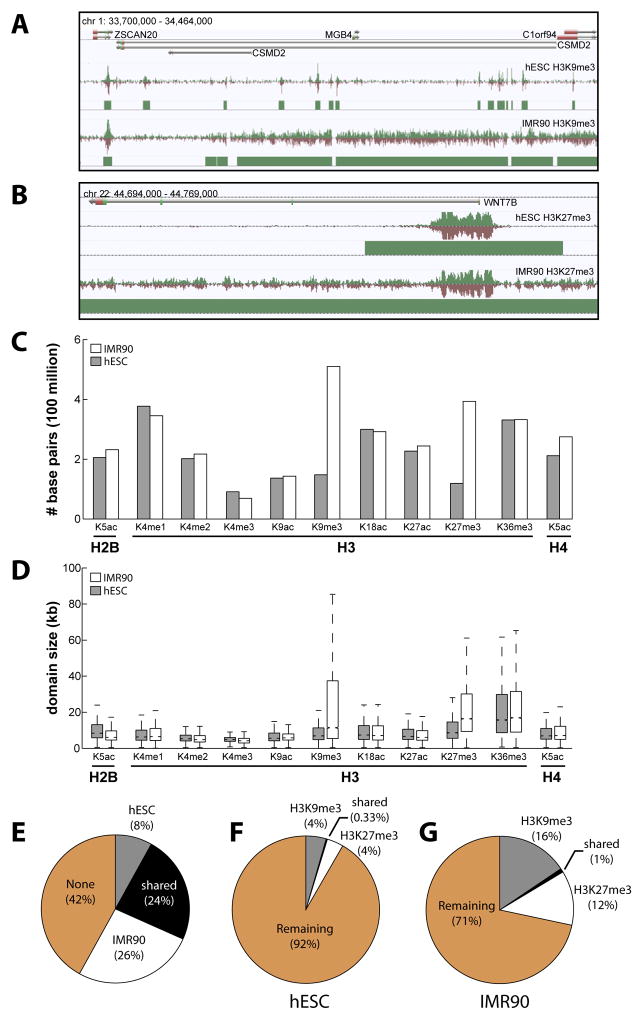
Comparing the domains in hESC and IMR90, we find that the chromatin architecture at a surprisingly large fraction of the genome is different (Figure 2A, B). Many of the H3K9me3 domains in hESCs appear small and interspersed (median length = 6.9 kb), but are expanded significantly in IMR90 cells (median length = 11.4 kb, p < 1E-15) (Figure 2A,C–D) (Table S3, S4). Correspondingly, the number of base pairs spanned by H3K9me3 in IMR90 (510 Mb) is 3.4 times larger than hESCs (148 Mb) (Figure 2C). Similarly, the number of base pairs spanned by H3K27me3 domains in IMR90 (394 Mb) is 3.3 times more than in hESCs (119 Mb) (Table S5, S6). The median size of an H3K27me3 domain in IMR90 (16400bp) expands to almost twice that of hESCs (8600bp, p < 1E-308), Wilcoxon rank sum test; Figure 2C), while coverage of H3K36me3 domains remains consistent (hESC median = 15700bp; IMR90 median size = 16900bp, p = 4.6E-7), (Figure 1A, 2C–D). In total, 50% of the IMR90 genome is spanned by chromatin domains from the 11 marks profiled, while only 32% of the hESC genome is in chromatin domains (Figure 2E). The difference is largely accounted for by H3K27me3 and H3K9me3 dispersal (Figure 2D, F–G). Each of these marks covers 4% of the hESC genome, but expands to 12% (H3K27me3) and 16% (H3K9me3) in IMR90 cells (Figure 2F–G).
Figure 2. Features of Chromatin Domains.
(A–B) Examples of (A) H3K9me3 and (B) H3K27me3 domains expanded in IMR90 relative to hESC. (C) The total number of base pairs spanned by domains and (D) the distribution of domain sizes for all mapped chromatin modifications. IMR90, white; hESC, grey (E) The fraction of the human genome spanned by the 11 chromatin modifications profiled in both cell types. The fraction of the (F) hESC and (G) IMR90 genomes spanned by H3K9me3 and H3K27me3 domains.
To confirm if expansion of repressive domains is a key characteristic of differentiated cell fate, we tested if expanded domains exist in other cells. We analyzed genome-wide profiles of H3K27me3 in CD4+ T cells (Barski et al., 2007; Cuddapah et al., 2009). Indeed, much of the CD4+ T cell genome (324 Mb) is spanned by H3K27me3 domains, nearly three times as large as in hESCs. Due to insufficient read depth for H3K9me3 in this cell type, we did not include H3K9me3 blocks in this analysis. The expansion of H3K27me3 holds true in various differentiated cell types. ChromaBlocks analysis of H3K27me3 shows expansion in non-transformed HUVEC (347Mb) and NHEK (312Mb) cells, as well as transformed GM12878 lymphoblasts (286Mb) and K562 leukemia cells (353Mb) (Figure S3A; data produced and released from the ENCODE Project (ENCODE, 2007)). All lineage-committed cell types examined, normal and cancer cell-types, have a clear increase in repressive chromatin structure compared to hESCs. In contrast, modifications associated with gene activity including H3K4me1, H3K4me2, H3K4me3, H3K27ac, and H3K36me3 are not expanded in these differentiated cell lines compared to hESC (data not shown).
Expansion of H3K27me3 and H3K9me3 Selectively Affects Developmental Genes
We investigated the extent to which genes are silenced in fibroblasts compared to hESCs by broad domains. Despite expansion of H3K27me3 domains, we observed similar numbers of promoters covered by H3K27me3 in hESCs (4736) and IMR90 (4279) cells (Figure S3B–C). However, 83% of H3K27me3 marked promoters are H3K4me3/H3K27me3 bivalent promoters in hESC, compared to 50% in IMR90 (Figure 3A). Of the 2485 promoters commonly marked by H3K27me3 in hESC and IMR90, 1512 show greater than 50% expansion in IMR90. The median H3K27me3 domain spanning a promoter in hESC is 10 kb, compared to 28 kb in IMR90 and 22 kb in CD4+ T cells, suggesting that expansion of H3K27me3 domains is a common feature of differentiated cells (Figure S3).
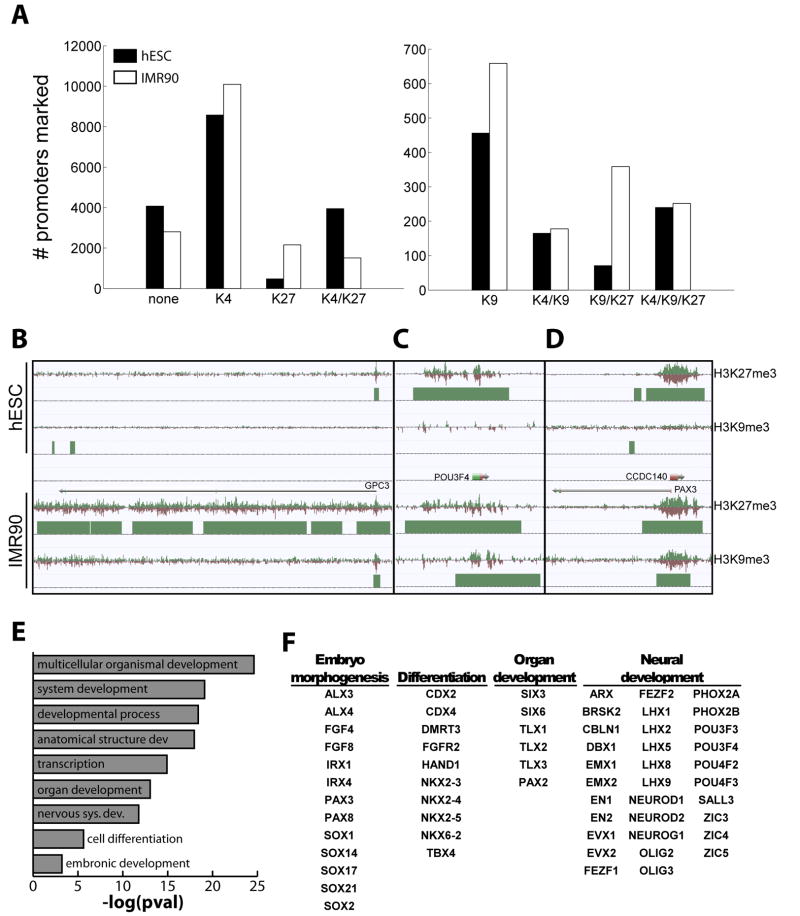
Figure 3. Expansion of Distinct Chromatin Blocks Marks Developmental Genes.
(A) The number of promoters marked by various combinations of chromatin modifications in hESC (black) and IMR90 (white). Exact combinations including H3K4me3, can be found for RefSeq genes in Supplemental Table S7. Abbreviations: K4, H3K4me3; K9, H3K9me3; K27, H3K27me3; none, lacking K4/K9/K27. (B–C) Snapshots of domains and chromatin structure around promoters which gain H3K9me3 and H3K27me3 in IMR90, for (B) GPC3, (C) POU3F4, and (D) PAX3. (E) Gene Ontology - biological processes enriched in promoters that are marked by both H3K9me3 and H3K27me3 in IMR90, along with (F) examples of developmentally relevant genes having this chromatin structure.
Gene Ontology (GO) analysis using DAVID (Dennis et al., 2003) of genes marked by H3K27me3 expansion in IMR90 reveals significant enrichment in developmental processes (Figure S3D), including genes from the BMP, FGF, FOX, SOX, and GATA families (Table S7). Additionally, GO analysis on the 314 promoters marked by H3K27me3 expansion in hESCs compared to IMR90 revealed significant enrichment of a small class of developmental proteins including brain-specific genes such as EMX2 and BAI1, as well as several HOX genes. The bivalent ESC-chromatin state often transitions to H3K27me3 and expands to cover the entire gene locus, and frequently neighboring gene loci. An additional 1742 promoters are marked by H3K27me3 only in IMR90 cells but not in hESCs, and are enriched for non-developmentally related biological processes including immune and defense response (Figure S3D). Interestingly, distinct genes are marked by expanded H3K27me3 domains in IMR90 and CD4+ T cells. There are 2138 promoters marked by H3K27me3 in hESCs that expand in IMR90 or CD4+ cells. Of these, 929 (44%) are IMR90-specific and 626 (29%) exhibit CD4+-specific expansion, while 583 (27%) expand in both cells. IMR90-specific expanded domains are enriched for T cell, B cell and lymphocyte related genes, while CD4+-specific expanded domains are enriched for extracellular matrix proteins, such as collagens. These results suggest that cell fate commitment is also a process of epigenetic gene repression that is unique to different lineages.
Similar to H3K27me3, there are 45% more H3K9me3-only marked domains in IMR90 (659) than in hESCs (456). However, a smaller fraction of gene promoters are spanned by H3K9me3 domains in hESCs (932, 5%) and IMR90 (1448, 8%) (overlap 456 promoters) (Figure 2C,F–G, Figure 3A). In several instances, gene family clusters are marked by H3K9me3 domains (Figure S5A; also see supplemental text). Interestingly, we observe 5 times as many promoters simultaneously marked by H3K9me3 and H3K27me3 in IMR90 (359) than hESCs (71) (Figure 3A). For example, the Simpson-Golabi-Behmel overgrowth syndrome gene, GPC3 (Pilia et al., 1996), is bivalent (H3K4me3/H3K27me3) and expressed in hESCs, yet is repressed by H3K9me3 and H3K27me3 in IMR90 (Figure 3B). Additionally, we observe key developmental transcription factors associated with both H3K9me3 and H3K27me3 in IMR90 cells. As examples, POU3F4 and PAX3 are marked by H3K27me3 in hESCs, and gain H3K9me3 in IMR90 to become dually marked for repression (Figure 3C–D). POU class transcription factors establish cell-type specific gene expression and cell fate decisions (Ryan and Rosenfeld, 1997), and PAX genes are critical for cell fate, organogenesis, and proliferation, leading to an important role in cancer (Loh et al., 2006). In light of this, we asked if H3K9me3/H3K27me3 marked promoters in IMR90 share similar functions. GO analysis revealed significant enrichment for developmental processes, including multicellular organismal development, system development, and anatomical structure development (Figure 3E), noting that many dual-marked promoters include key developmental regulators (Figure 3F), such as HOX genes (Figure S4; see supplemental text). The co-localization of H3K27me3 and H3K9me3 at genes was recently noted by others (Bilodeau et al., 2009), and confirms a link between PcG and H3K9me3. Not all H3K27me3 marked sites are associated with H3K9me3. This dual repression at specialized regulators may point to the importance of maintaining their silencing in differentiated cells. These genes serve as likely candidates for cell fate decisions.
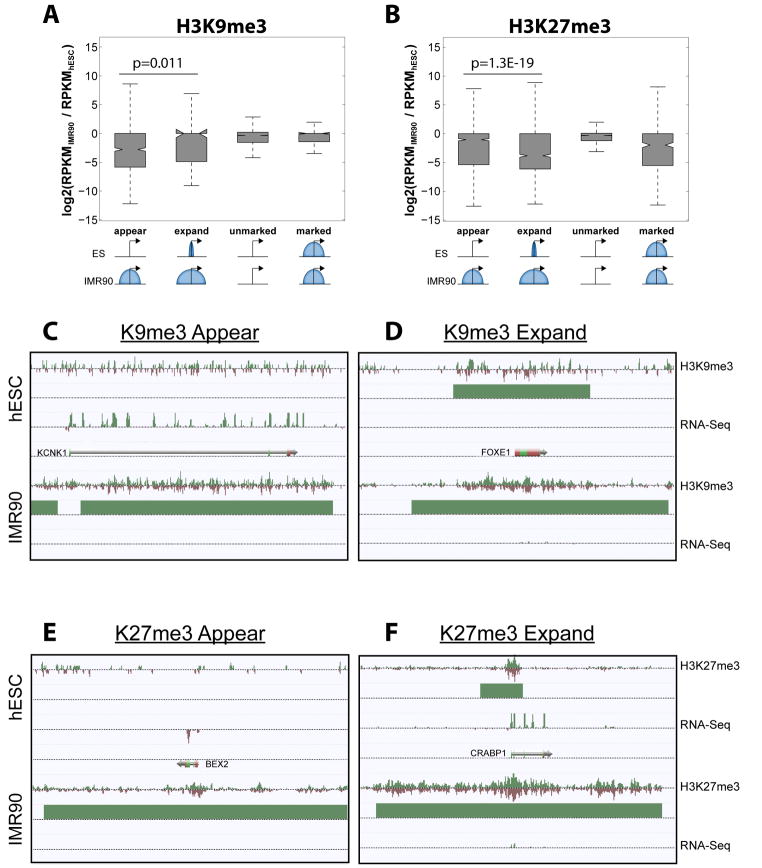
With distinct spatial modes of H3K9me3 and H3K27me3 marking genes, we asked how these modes affect gene repression. We considered four cases: 1) promoters not marked in hESCs but marked in IMR90 (appear), 2) promoters covered by blocks in hESCs but expanded by at least 50% in IMR90 (expand), 3) promoters unmarked in both cells (unmarked), and 4) promoters marked by similarly sized blocks in both cells (marked). Table S7 shows the H3K4-, K9-, and K27-me3 states for all genes and expression changes. The acquisition of H3K9me3 at promoters showed the most significant decrease in gene expression (p = 0.011) compared to the other H3K9me3 scenarios (Figure 4A, C–D). In contrast, the greatest repression for H3K27me3 cases occurred upon domain expansion (Figure 4B, E–F). This may imply that the presence of H3K9me3 alone is enough to suppress gene expression and expansion of domains has a smaller effect on gene repression. By contrast, the presence of narrowly marked H3K27me3 promoters is frequently insufficient to silence a gene (Figure 4F), as they are often in a bivalent state, but the expansion seems to maintain stable silencing of gene expression in the differentiated cells (p = 1.3E-19).
Figure 4. Expression of Genes in Different Types of Chromatin Domains.
(A) Change in gene expression for promoters where H3K9me3 appears, expands, remains unmarked, or remains similarly marked in IMR90 compared to hESC. (B) As in (A), but for H3K27me3. (C–F) Snapshots of genes exhibiting (C) H3K9me3 appearance, (D) H3K9me3 expansion, (E) H3K27me3 appearance, and (F) H3K27me3 expansion, as observed in IMR90 relative to hESC.
Global analysis of associations between Chromatin Modifications and DNA Methylation
DNA methylation is a critical component of the epigenome that represses gene expression through promoter CG methylation, in addition to localization at heterochromatin and repetitive elements in the genome. This mark is thought to maintain long-term repression and to be less dynamic than histone modifications. There is also evidence that some crosstalk between these epigenetic mechanisms exists, suggesting a direct link between H3K9 methyltransferases (HKMT) and DNA methyltransferases (DNMTs) (Fuks et al., 2003; Jackson et al., 2002; Lehnertz et al., 2003; Li et al., 2006; Tamaru and Selker, 2001). However, mutations in the SET domain of HKMT G9a do not alleviate DNA methylation (Dong et al., 2008; Tachibana et al., 2008). To date, it remains to be determined if other chromatin structures or regulatory elements have an association with DNA methylation.
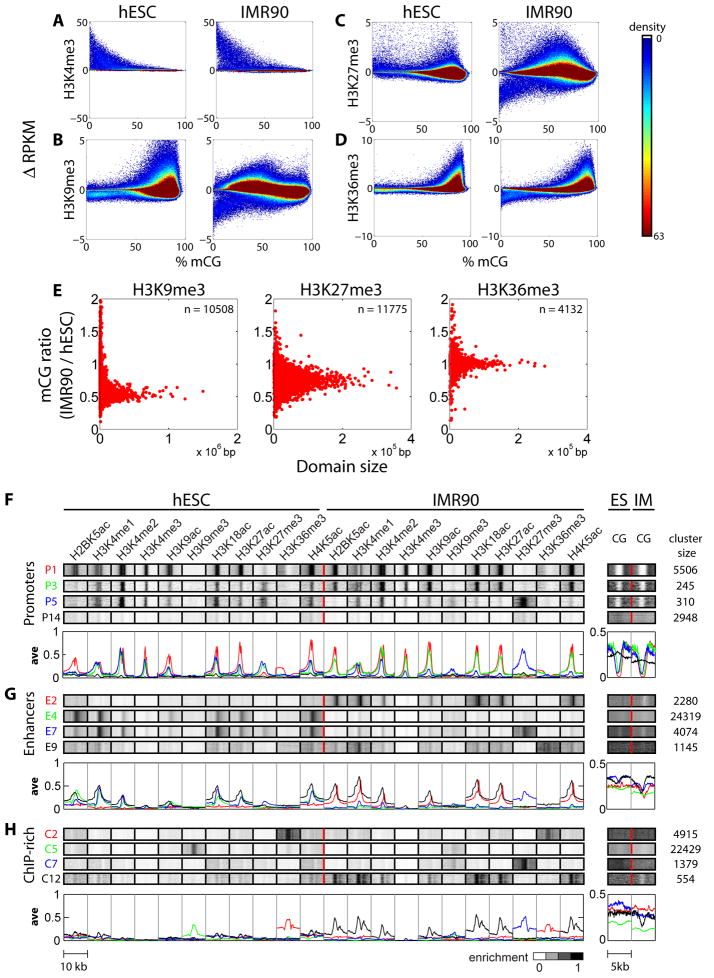
Using genome-wide, nucleotide resolution maps of DNA methylation (mC) generated in H1 hESCs and IMR90 cells (Lister et al., 2009), we systematically investigated the relationships between the 11 chromatin modifications described above and DNA methylation. Using a sliding window approach, we plotted the presence of ChIP-Seq tags for each histone modification normalized to input, against the average percentage of CG methylation (mCG) in the same window to determine the association of the two epigenetic components in hESCs and IMR90 (Figure 5A–D; Figure S6). Consistent with previous findings, we observe that H3K4me3 is inversely correlated with mCG (Figure 5A; Figure 1A). Similarly, H3K27me3 is associated with hypomethylation of DNA at promoters, consistent with previous observations that bivalent promoters are typically hypomethylated (Brunner et al., 2009; Meissner et al., 2008). Outside of promoters, H3K27me3 is also positively associated with DNA methylation across a broad range of mCG. Interestingly, the degree of association between H3K9me3 and DNA methylation is less prevalent than that between mCG and H3K36me3, which is always associated with fully methylated DNA in both hESCs and IMR90 (Figure 5B,D; Figure 1A). These results suggest cell-type specific relationships between DNA methylation and histone modifications (Figure 1C; also see Figures 6 & S8–S9).
Figure 5. Genome-wide Correlation Analysis between Histone Modifications and Methylated CG (mCG).
(A–D) Change in RPKM for several histone modifications relative to input control were calculated within 2.5kb window for (A) H3K4me3 and 5kb for (B) H3K9me3, (C) H3K27me3, and (D) H3K36me3, and plotted against %mCG in the same window throughout the genome. Density of spots in the plot is displayed by the heatmap (right). (E) For H3K9me3 (left), H3K27me3 (middle), and H3K36me3 (right), spreading chromatin domains were identified as IMR90 domains having at most 20% overlap with hESC domains. Shown is the IMR90 to hESC ratio of %mCG for each of these domains, as a function of domain size. (F–H) Representative Punctuated Chromatin Signatures. ChromaSig was used to simultaneously cluster and align all 11 chromatin modifications mapped from both cell types at (F) promoters, (G) predicted enhancers, and (H) regions of ChIP-Seq enriched sequences. On the left are the chromatin signatures recovered for each cluster, and on the right are %mCG tracks appended after clustering. Enrichment of histone modifications and %mCG are indicated by the heatmap (bottom). The number of genomic loci in each cluster is indicated on the far right. Average profiles for each modification are plotted beneath the representative clustergrams. Profiles are color-coordinated with the cluster ID, e.g. P1 (red), P3 (green), P5 (blue), P14 (black).
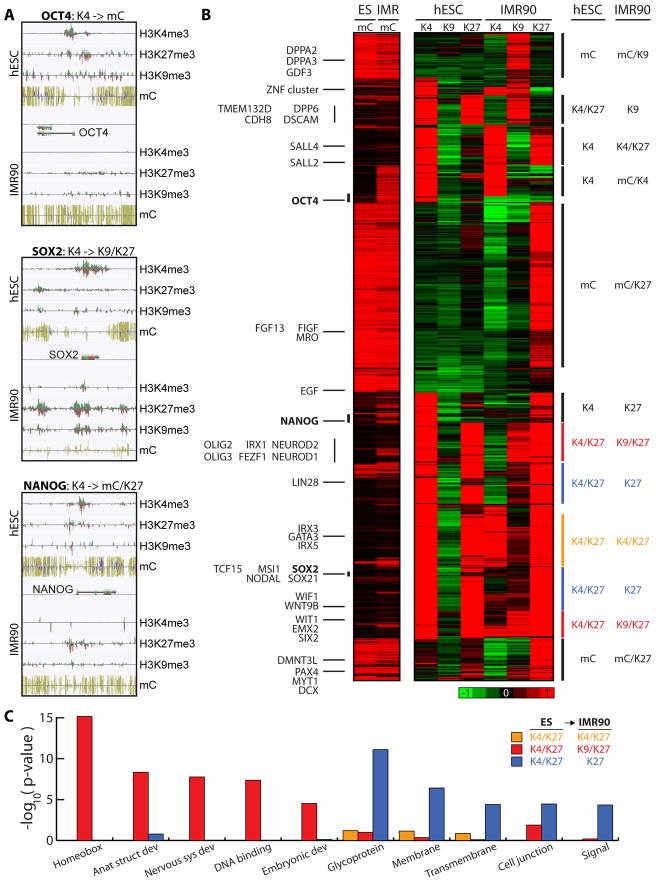
Figure 6. Distinct Modes of Epigenetic Repression at Developmental Genes.
(A) Snapshots of chromatin modifications and DNA methylation around OCT4, SOX2, and NANOG in hESC and IMR90, illustrating distinct transitions to repressive epigenetic states. Abbreviations: K4, H3K4me3; K9, H3K9me3; K27, H3K27me3; mC, DNA methylation. (B) Unsupervised clustering of H3K4me3, H3K9me3, H3K27me3, and mCG at promoters of genes down-regulated by 2-fold in IMR90 relative hESCs. Relevant genes are indicated to the left. Distinct clusters are noted on the right. The enrichment scale is shown below the heatmap. (C) GO analysis of genes whose promoters remain bivalent (H3K4/K27me3), acquire H3K9me3 (H3K4/K27me3 to H3K9/K27me3), or lose H3K4me3 (H3K4/K27me3 to H3K4me3) in IMR90.
H3K36me3 and DNA methylation have been described in exons of transcribed gene bodies (Ball et al., 2009; Hellman and Chess, 2007; Kolasinska-Zwierz et al., 2009). Examining the enrichment of H3K36me3 and mCG across 231,984 human exons, we observe that there is concordance between H3K36me3 and mCG (Figure S7A–B). However, H3K36me3 is more positively correlated with gene expression (Figure S7C–D), as most exons are marked with DNA methylation (Figure S7A–B). A possible mechanism may involve H3K36me3 in the recruitment of DNA methyltransferases to maintain DNA methylation at transcribed gene bodies. This would fit the “methylation paradox”, noting that while promoter CpG islands remain unmethylated, transcribed CpG islands show an increase in DNA methylation (Jones, 1999). However, DNA methylation is also found outside transcribed regions. Therefore, while H3K36me3 predicts DNA methylation, the converse is not true.
To assess how changes in chromatin blocks correlate with changes in DNA methylation, we analyzed the genomic regions that were differentially associated in IMR90. Regions of the genome uniquely marked by H3K9me3 in IMR90 cells contain roughly half as much mCG as in hESC (Figure 5E, Figure S7E–G). This depletion is most evident for larger domains, while small domains have a wide variance of mCG. Similarly, H3K27me3 domains unique to IMR90 are also depleted of mCG relative to hESCs, though at levels intermediate to H3K9me3 and H3K36me3 blocks, which exhibit equivalent levels of DNA methylation in each cell type (Figure 5E–F, Figure S7E–G). This intermediate level of methylation is not due to the contribution of promoters (Figure S7H). These relationships hold when comparing domains marked in both hESCs and IMR90. Together, these results suggest that, on a global scale, gain of the repressive modifications H3K27me3 or H3K9me3 is associated with a corresponding decrease in DNA methylation level in the differentiated IMR90 cells. This observation suggests a much more complex relationship between H3K9me3 or H3K27me3 and DNA methylation than previously proposed (Cedar and Bergman, 2009; Suzuki and Bird, 2008).
DNA Methylation at Potential Regulatory Sequences
A complex relationship also exists between mCG and H3K4me1, H3K4me2 and histone acetylation, which mark active regulatory sequences such as promoters and enhancers (Figure 5A–D, S6). To relate complex patterns of histone modifications with mCG at functional elements, we utilized an unbiased clustering algorithm, ChromaSig (Hon et al., 2008). Focusing on modifications with a punctuated footprint (Figure 5F–H, Figure S8 – all clusters), our analysis revealed 42 frequently occurring patterns, grouped into three categories corresponding to known promoters (Figure 5F, S8A, Table S8), H3K4me1-predicted enhancers (Figure 5G, S8B, Table S9) and other regions with enriched chromatin modification signals, ChIP-rich regions (Figure 5H, S8C, Table S10).
While most promoters have a strong inverse correlation between H3K4me3 and mCG, a subset of gene promoters lacking chromatin modifications are hypermethylated and typically silenced (Figure 5F, S8A & S9A,D). At enhancers (Table S11, S12) cell-specific relationships exist. Acetylated, IMR90-specific enhancers (Figure 5G: clusters E1–E3, E10–E11) show depletion of mCG in IMR90 cells, but not in hESCs (cluster E2). Non-acetylated IMR90-specific enhancers have minimal mCG depletion (cluster E9). At shared enhancers (E5, E7, E8), both cell types have mCG depletion (Figure 5G: E7) while hESC-specific enhancers (E4, E6 E12) show no obvious depletion of mCG (E4 in Figure 5G; S8B, S9B, E). Instead, hESC enhancers and transcription factor binding sites for OCT4, SOX2, NANOG and others are depleted of non-CG methylation (Lister et al., 2009). Therefore, the chromatin modifications at these elements are largely cell specific, and are inversely related to DNA methylation in a cell-specific manner. However, the mechanism for the role of CG versus nonCG methylation is unclear.
ChromaSig analysis of ChIP-rich regions outside of known promoters and predicted enhancers confirmed our block analysis, illustrating H3K36me3 regions enriched for mCG (Figure 5H cluster C2; S8C cluster C1–C3), and the presence of peak-like H3K9me3 sites in hESCs (Figure 5H, cluster C5). ChromaSig also detects patterns missed by peak-finding or prediction algorithms. Cluster C12 illustrates likely IMR90 cell-specific enhancers previously missed (Figure 5H), and clusters C6–C7 with H3K27me3 in hESCs are unannotated promoters enriched for DNA methylation, similar to promoter clusters P16–P17 (Figure 5H, cluster C7). These ChIP-rich regions outside of known annotations provide interesting avenues for future study. Much like promoters, ChIP-rich regions show little variation in DNA methylation across cell types (Figure S9C). Therefore, our analysis reveals that H3K4me1-predicted enhancer regions, which we previously showed to be cell-type specific (Heintzman et al., 2009), exhibit the most dynamic DNA methylation changes.
Distinct Mechanisms of Epigenetic Repression of Pluripotency Genes in Fibroblasts
The core pluripotent transcription factors OCT4, SOX2, and NANOG are each marked in IMR90 cells by distinct combinations of repressive modifications (DNA methylation, H3K9me3, and H3K27me3). The OCT4 promoter is repressed through DNA methylation, as previously demonstrated (Barrand and Collas, 2009; Ben-Shushan et al., 1993; Deb-Rinker et al.). The SOX2 promoter harbors both H3K27me3 and H3K9me3, and NANOG shows an increase in promoter mCG and acquires H3K27me3 (Figure 6A). To determine if other genes are regulated by these epigenetic modes of repression, we enumerated nearly 1400 genes with > 2-fold reduction in expression and that acquired at least one repressive modification in IMR90 relative to hESCs. From an unbiased hierarchical clustering of these genes, we observe that only small groups of genes follow the OCT4-, SOX2- and NANOG-like patterns (Figure 6B, Table S13). Due to their limited expression, it is not surprising that few genes are repressed in the same manner. However, these small, uniquely marked sets of genes may provide insight to stem cell biology or disease such as cancer. For example, the NANOG-like class contains KLF8, a known regulator of the reprogramming factor KLF4. The OCT4-like class contains the cell cycle regulator RAB25, which has a role in cell cycle regulation and tumor invasion (Caswell et al., 2007). Because a common mechanism of epigenetic repression exists, these genes may also be positively co-regulated in a tissue-specific manner. This is most evident with SOX2, a marker of neural stem cells. Many of the SOX2-like class of genes have known roles in neurobiology, such as CADPS, RIC3, ZIC5 and KIF1A.
Other combinations are more common. A large number of DNA methylated promoters gain H3K27me3 (mC to mC/H3K27me3), which may reflect a functional link between these marks for a sub-set of genes. Previous studies have suggested that H3K27me3 marked promoters are linked to DNA hypermethylation in cancer cells (Ohm et al., 2007; Schlesinger et al., 2007; Vire et al., 2006; Widschwendter et al., 2007). Here, we note the reciprocal relationship also occurs. Not all statically marked mC genes acquire H3K27me3, a smaller class gain H3K9me3 (mC to mC/H3K9me3) (Figure 6B), suggesting different mechanisms. hESC bivalent promoters, which are enriched for developmental regulators, are equally distributed amongst three states in IMR90 cells: remaining H3K4/27me3 or becoming H3K9/K27me3 or mC/H3K27me3 marked. GO analysis showed that the developmental regulators tend to be marked by H3K9/27me3 in IMR90 (Figure 6C). Moreover, genes dually marked for repression (mC/K27me3 and K27/K9me3) constitute the majority of the 1400 genes examined. Presumably, these added levels of epigenetic repression decrease the likelihood of escaping gene repression and altering cell fate. The multiple modes of repression suggest several mechanisms are in place to restrict de-differentiation in lineage-committed cells.
Repressive Chromatin Domains in Induced Pluripotent Cells (iPSC)
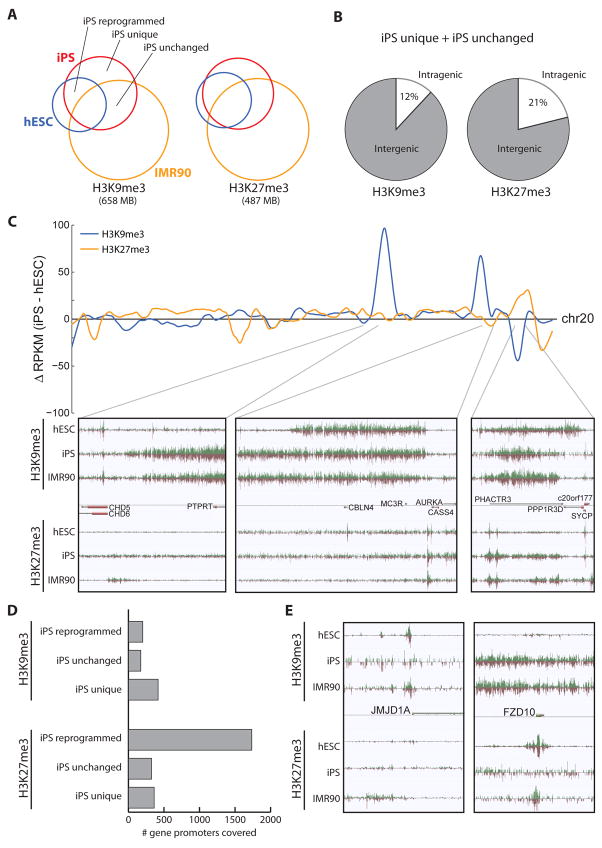
iPS cells and hESCs are both pluripotent and share the ability to self-renew. Furthermore, the global gene expression profiles of IMR90-derived iPS cells are more similar to hESCs than the original IMR90 cells (Yu et al., 2007). However, it is remains to be determined if this is true of repressive chromatin structure. Using iPS cells reprogrammed from IMR90 cells by Yu et al., we asked if repressive domains are a key feature of cell fate, and therefore are broad domains remodeled during reprogramming, especially outside a few key gene promoter regions. We observed that reprogramming results in a noticeably reduced distribution of repressive domains (Figure S10), with H3K9me3 reduced to 275 MB and H3K27me3 reduced to 195 MB, between the values observed in IMR90 (510 MB, 394 MB) and hESC (148 MB, 119 MB). Despite the wider coverage of H3K9me3 in IMR90, there is more overlap between iPS cells and hESC (H3K9me3 = 105 MB, H3K27me3 = 102 MB) than between IMR90 and hESC (H3K9me3 = 76 MB, H3K27me3 = 68 MB), indicating that reprogramming has resulted in an epigenome that is more similar to the hESC epigenome (Figure 7A, Figure S10), consistent with previous observations (Maherali et al., 2007; Mikkelsen et al., 2008; Sridharan et al., 2009).
Figure 7. Comparison of repressive chromatin structure in hESC and iPS cells.
Overlap of repressive chromatin domains in hESC (blue), iPS (red), and IMR90 (yellow) cells, for H3K9me3 (left) and H3K27me3 (right). The total coverage of these modifications is indicated below. Regions related to reprogramming are labeled: iPS reprogrammed regions, defined as overlap of iPS and hESC but not IMR90; iPS unchanged regions, defined as overlap of iPS and IMR90 but not hESC; and iPS unique regions. (B) The average genic distribution of iPS unchanged and iPS unique regions. (C) (top) Difference in RPKM of iPS profiles compared to hESC profiles for H3K9me3 (blue) and H3K27me3 (yellow) throughout the length of chr20. (bottom) Zoomed in views of genomic regions having differences between iPS and hESC cells. (D) For the three types of iPS regions, the number of gene promoters covered by H3K9me3 (top) or H3K27me3 domains (bottom). (E) Snapshots of repressive chromatin structure at JMJD1A (left) and FZD10 (right) promoters, illustrating differences in iPS and hESC cells.
On a global scale, the profiles of H3K9me3 and H3K27me3 in iPS cells are similar to those in hESCs (Figure 7C). However, regions of discordance exist, at times spanning genic regions (Figure 7C). To dissect these differences, we defined three interesting chromatin structures related to reprogramming. The first, denoted iPS reprogrammed, are domains shared between iPS and hESC. The second, denoted iPS unchanged, are marked in iPS and IMR90 cells, therefore not in a hESC-like state. The third, denoted iPS unique, are domains not marked in either hESC or IMR90 cells. The iPS unchanged and iPS unique groups have comparable genomic distributions, generally spanning intergenic regions of the genome, with only 12% of H3K9me3 and 21% of H3K27me3 iPS domains intragenic on average (Figure 7B). Thus, while there are subtle differences in repressive chromatin structure between iPS and hESC, most of these differences are confined to regions outside of genes.
iPS-reprogrammed H3K27me3 domains cover nearly 5 times as many gene promoters as the other groups (iPS reprogrammed, 1736; iPS unchanged, 325; iPS unique, 362) (Figure 7D). This indicates that reprogramming has generally re-established the same H3K27me3 structure as hESCs at promoters, and only a small fraction are incorrectly reprogrammed. We do not observe the same phenomenon for H3K9me3 with more genes in iPS unique (417) than iPS reprogrammed (201) or iPS unchanged (173). These iPS/hESC differences in H3K9me3 structure at gene promoters is evident at several important epigenetic and developmental genes, e.g. H3K9me3 demethylase JMJD1A is marked by H3K9me3 in both hESC and IMR90 but not iPSCs (Figure 7E). Also interesting, the WNT receptor FZD10 retains the incorrect repressive mark: H3K9/K27me3 in IMR90, H3K27me3 in hESCs, yet H3K9me3 in iPSCs (Figure 7E). Recently, Chin et al found 3947 genes differentially expressed between several iPS and hESC lines (Chin et al., 2009). We tested if H3K9me3 or H3K27me3 was differentially enriched at these gene promoters between IMR90-derived iPSCs and H1 hESCs. From the 3947 genes, 770 promoters are marked by H3K27me3 in iPS cells, of which only 14.5% are not marked in hESC (p = 1, Binomial, expectation is 15.2%). In contrast, 160 promoters are marked by H3K9me3 in iPS cells, 68% of which are not marked in hESC (p = 0.0023, Binomial, expectation is 56.8%). Thus, it appears that H3K9me3 contributes more than H3K27me3 to the differences in gene expression between iPS and hESC. Perhaps these observations indicate flexibility in gene repression during reprogramming.
Our findings suggest that expanded repressive domains are a key aspect of differentiated cell fates, perhaps reducing the plasticity seen in stem cells. Reprogramming of somatic cells involves remodeling of these domains to reflect the hESC epigenome. Therefore, it is likely that partially reprogrammed iPS (piPS) cells still harbor expanded chromatin domains that prevent them from achieving pluripotency. Our results call for future studies of these chromatin marks in additional hES, iPS and piPS cells. Moreover, expediting chromatin remodeling would likely improve reprogramming efficiency.
CONCLUSION
A fundamental question is how the identical genome sequence gives rise to a diversity of cell types with different gene expression profiles and cellular functions. By comparing the epigenome of pluripotent stem cells to that of a differentiated cell, we provide evidence that lineage-committed cells are characterized by significantly expanded repressive chromatin domains that selectively affect genes involved in pluripotency and development, suggesting that epigenetic mechanisms play a critical role in cellular differentiation and maintenance of differentiated cellular state. Recently, a similar conclusion was reached by examining H3K9me2 in mouse ES cells and differentiated cells (Wen et al., 2009). However, an independent analysis of the data using a different block finding method has reached at a different conclusion, leading to a debate on the genomic distribution of H3K9me2 domain in mammalian cells (Filion and van Steensel, 2010; Wen et al., 2010). In the present work, the expansion of H3K27me3 or H3K9me3-marked repressive domains was confirmed by several measures. First, the expansion of H3K27me3 was illustrated in independently derived data from multiple cell types. Second, expanded H3K27me3 and H3K9me3 show reduction in IMR90-derived iPS cells. Finally, we applied an independent block-finding algorithm, TileHMM (Humburg et al., 2008), used to reanalyze the Wen et al. H3K9me2 data (Filion and van Steensel, 2010), and confirm the expansion of H3K27me3 and H3K9me3 in IMR90 relative hESCs (Figure S3E). Our result provides additional support to the general model proposed by Wen et al. (2009).
Knockout studies of H3K9 methyltransferases and H3K27 methyltransferases lead to differentiation or developmental defects (Dodge et al., 2004; Faust et al., 1998; O’Carroll et al., 2001; Pasini et al., 2007; Peters et al., 2001; Tachibana et al., 2002), suggesting that epigenetic mechanisms play a critical role in cell fate determination. The expansion of H3K9me3 and H3K27me3 domains in differentiated cells relative hESCs support this model. Reprogrammed cells re-organize their chromatin architecture to reflect the less repressive state of stem cells. This suggests inhibiting repressive chromatin structure would facilitate reprogramming either through inhibition of HKMTs or overexpression of demethylases. Early evidence supports this hypothesis (Shi et al., 2008a; Shi et al., 2008b; Wendt et al., 2008). Collectively, this suggests one aspect of epigenetic cell fate lies in the structure of repressive or compacted chromatin.
Methods
ChIP-Seq
ChIP was carried out as previously described with 500ug chromatin and 5ug antibody (antibodies are listed in Supplement) (Heintzman et al., 2007; Kim et al., 2007). ChIP libraries for sequencing were prepared following Illumina protocols with the minor modifications (see Supplement) (Illumina, San Diego, CA). Libraries were sequenced using the Illumina GAII machine as per manufacturer’s protocols. Following sequencing cluster imaging, base calling and mapping were conducted using the Illumina pipeline (Illumina, San Diego, CA). All data have been deposited to the Sequence Read Archive (SRA), accession SRP000941.
Cell growth
IMR90 cells were grown as previously described (Kim et al., 2007). H1 embryonic stem cells were grown as previously described using Matrigel (BD Biosciences) and mTeSR (Ludwig et al., 2006a; Ludwig et al., 2006b).
Data normalization
For each chromatin mark and the input control we divide the genome into 100 bp bins, count the number of reads falling into each bin i, and compute the number of reads per kilobase of bin per million reads sequenced (denoted RPKMmark,i), with the exception of the input where RPKM is computed over the 5 consecutive bins centered at i. Because input represents the entirety of the genome and is not sequenced to saturation like ChIP data, a 500bp bin is used to reduce noise that occurs in a 100bp window. Finally, normalized ChIP enrichment is computed as ΔRPKM = RPKMmark,i − RPKMinput,i.
ChromaBlocks
To identify both peak-like and domain-like enrichment, a block is started for 1) regions of 10 consecutive bins having average ΔRPKM ≥ 1.5 with at least 5 bins above this threshold or 2) regions of 50 consecutive bins having average ΔRPKM ≥ 0.5 with at least 25 above this threshold. Once a block is started, its boundaries are extended in both directions until reaching a region of 50 bins having average ΔRPKM < 0.1 (Figure 1E). A full description is described in the Supplement Methods.
Quantifying DNA methylation
For a given cytosine residue with CG context at position i in the genome, its measure of mCG Mi is defined as the fraction of methylated cytosines called out of all methylC-Seq reads spanning i. Given a region, we then use %mCG as a measure of DNA methylation enrichment, defined as the sum of the Mi for all CG dinucleotides in the region, divided by the total number of CG dinucleotides in the region. The only exception is when calling promoters enriched with DNA methylation for a given cell. Here, we use an absolute measure of mCG with Mi = 1, and define enriched promoters as those with %mCG ≥ 50% for a 1-kb window centered at the TSS.
Identifying punctuated chromatin signatures
RefSeq promoters are clustered by chromatin signature using ChromaSig as described (Hon et al., 2008) (motif width w = 4000 bp, wandering distance d = 1000 bp, σanother = 2.5, and pa = 0.01). Putative enhancers in hESC and IMR90 were predicted on the basis of chromatin signatures as described (Heintzman et al., 2007) and clustered by ChromaSig (w = 4000 bp, d = 2000 bp, σanother = 2.5, and pa = 0.001). Regions of significant ChIP enrichment of histone modifications were identified as described (ChromaSig) (p-value cutoff 1E-3), and filtered to remove promoters, predicted enhancers, gene 3′ ends, and CTCF binding sites, and finally clustered by ChromaSig (w = 4000 bp, d = 1000 bp, σanother = 2.5, and pa = 0.001). In all instances, ChromaSig performed the clustering on all 11 chromatin modification maps from both cell types simultaneously.
Supplementary Material
Acknowledgments
We acknowledge the Bradley Bernstein group at Harvard MGH East and the ENCODE consortium for generating the H3K27me3 datasets for HUVEC, NHEK, GM and K562 cell types, which were released to the public on 2009-10-07. R.D.H. is supported by an American Cancer Society Postdoctoral Fellowship. R.L. is supported by a Human Frontier Science Program Long-term Fellowship. This work was supported by the following: NIH Epigenomics Roadmap Project (to B.R., J.A.T, J.R.E and W.W.), the California Institute for Regenerative Medicine (B.R.), the Ludwig Institute for Cancer Research (B.R.), the Mary K. Chapman Foundation (J.R.E.), and the J.T.F. Morgridge Institute for Research (J.A.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrand S, Collas P. Chromatin states of core pluripotency-associated genes in pluripotent, multipotent and differentiated cells. Biochem Biophys Res Commun. 2009;391:762–767. doi: 10.1016/j.bbrc.2009.11.134. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Ben-Shushan E, Pikarsky E, Klar A, Bergman Y. Extinction of Oct-3/4 gene expression in embryonal carcinoma x fibroblast somatic cell hybrids is accompanied by changes in the methylation status, chromatin structure, and transcriptional activity of the Oct-3/4 upstream region. Mol Cell Biol. 1993;13:891–901. doi: 10.1128/mcb.13.2.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, Mills GB, Humphries MJ, Messent AJ, Anderson KI, et al. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell. 2007;13:496–510. doi: 10.1016/j.devcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb-Rinker P, Ly D, Jezierski A, Sikorska M, Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J Biol Chem. 2005;280:6257–6260. doi: 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schubeler D, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125:4495–4506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Steensel B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nat Genet. 2010;42:4. doi: 10.1038/ng0110-4. author reply 5–6. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol. 2008;4:e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humburg P, Bulger D, Stone G. Parameter estimation for robust HMM analysis of ChIP-chip data. BMC Bioinformatics. 2008;9:343. doi: 10.1186/1471-2105-9-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281:19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon GC, Tonti-Filippini J, Nery JR, Lee LK, Ye Z, Ngo QM, et al. Human DNA methylomes at single-base resolution reveal widespread cellspecific epigenetic signatures. 2009 doi: 10.1038/nature08514. Under Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006a;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, Crandall LJ, Daigh CA, Conard KR, Piekarczyk MS, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006b;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Pilia G, Hughes-Benzie RM, MacKenzie A, Baybayan P, Chen EY, Huber R, Neri G, Cao A, Forabosco A, Schlessinger D. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet. 1996;12:241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Ryan AK, Rosenfeld MG. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008a;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008b;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Reply to [ldquo]Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells[rdquo] Nat Genet. 2010;42:5–6. doi: 10.1038/ng0110-4. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.