Abstract
ErbB3/HER3 is one of four members of the human epidermal growth factor receptor (EGFR/HER) or ErbB receptor tyrosine kinase family. ErbB3 binds neuregulins via its extracellular region and signals primarily by heterodimerizing with ErbB2/HER2/Neu. A recently appreciated role for ErbB3 in resistance of tumor cells to EGFR/ErbB2-targeted therapeutics has made it a focus of attention. However, efforts to inactivate ErbB3 therapeutically in parallel with other ErbB receptors are challenging because its intracellular kinase domain is thought to be an inactive pseudokinase that lacks several key conserved (and catalytically important) residues—including the catalytic base aspartate. We report here that, despite these sequence alterations, ErbB3 retains sufficient kinase activity to robustly trans-autophosphorylate its intracellular region—although it is substantially less active than EGFR and does not phosphorylate exogenous peptides. The ErbB3 kinase domain binds ATP with a Kd of approximately 1.1 μM. We describe a crystal structure of ErbB3 kinase bound to an ATP analogue, which resembles the inactive EGFR and ErbB4 kinase domains (but with a shortened αC-helix). Whereas mutations that destabilize this configuration activate EGFR and ErbB4 (and promote EGFR-dependent lung cancers), a similar mutation conversely inactivates ErbB3. Using quantum mechanics/molecular mechanics simulations, we delineate a reaction pathway for ErbB3-catalyzed phosphoryl transfer that does not require the conserved catalytic base and can be catalyzed by the “inactive-like” configuration observed crystallographically. These findings suggest that ErbB3 kinase activity within receptor dimers may be crucial for signaling and could represent an important therapeutic target.
Keywords: dimerization, kinase inhibitor, catalytic mechanism, activation loop
Receptor tyrosine kinases (RTKs) from the EGF receptor (EGFR) or ErbB/HER family play important roles in animal development and disease (1) and are the targets of several important therapeutic agents used clinically to treat cancer. Each receptor contains a large extracellular ligand-binding region (targeted by therapeutic antibodies), a single transmembrane helix, and an intracellular tyrosine kinase domain (TKD) that is flanked by juxtamembrane and C-terminal regulatory regions and is targeted by specific small-molecule kinase inhibitors (1, 2). Ligand binding to the extracellular region promotes homo- or heterodimerization of ErbB receptors, leading to allosteric activation of their intracellular kinase domains through the formation of asymmetric dimers (3–5). Within an activated dimer, the C-terminal regulatory tail is trans-autophosphorylated on tyrosines and recruits downstream signaling molecules that contain phosphotyrosine-binding Src homology-2 (SH2) domains.
ErbB3/HER3 is unique among the mammalian ErbB receptors in being generally considered as kinase-inactive (6). When first cloned (7, 8), amino acid substitutions were noted at two particular sites that are conserved in other known kinases (9). A critical glutamate in helix αC of other kinases (indirectly involved in ATP binding) is replaced by a histidine in ErbB3 (H740). In addition, a conserved aspartate thought to function as a catalytic base in protein kinases (10)—deprotonating the substrate hydroxyl group—is replaced with asparagine in ErbB3 (N815). Initial biochemical studies concluded that ErbB3 is kinase-inactive (11, 12). The recombinant ErbB3 intracellular domain was reported neither to bind ATP nor to become autophosphorylated (12). Full-length ErbB3 expressed in insect cells could be affinity labeled with an ATP analogue (11), but only low-level autophosphorylation was seen that could not be increased by adding the ErbB3 ligand neuregulin and so was ascribed to endogenous insect cell kinases (11). Phosphorylation of ErbB3 (or chimerae containing its kinase domain) has been seen in immunoprecipitates from other cellular contexts (13–15) but may reflect trans-phosphorylation by other ErbB receptors (6, 11). The prevailing view, therefore, is that ErbB3 contains an inactive “pseudokinase” (6, 16).
ErbB2 and ErbB3 can be activated only through heterodimerization (6, 17), and both are tyrosine phosphorylated in neuregulin-induced heterodimers in cells that express only these two ErbB receptors (18). This observation presents a conundrum given current models for ErbB receptor activation, where one kinase domain in a dimer allosterically activates its neighbor and itself becomes trans-autophosphorylated as a consequence (3, 5). It is not clear how ErbB2 could be trans-phosphorylated in ErbB2/ErbB3 heterodimers through this mechanism unless ErbB3 has kinase activity.
There are additional reasons to suspect that ErbB3 might retain a low level of kinase activity. First, mutating the aspartate proposed to function as a catalytic base in the Csk tyrosine kinase (19) does not abolish (but does substantially reduce) kinase activity. Second, kinase activity has recently been reported for other presumed pseudo- or unusual kinases, including the Ca2+/calmodulin-activated Ser/Thr kinase (20), haspin (21), and Wnk (22)—although some other pseudokinases appear to be genuinely inactive (23). Here we show that the kinase domain of ErbB3 retains phosphoryl-transferase activity, as evidenced by trans-autophosphorylation of the purified ErbB3 intracellular domain. Kinase activity appears to be ∼1,000-fold weaker than that seen for EGFR but may be sufficient for receptor trans-phosphorylation in the context of an ErbB heterodimer. We demonstrate that the ErbB3 kinase domain binds ATP with an affinity similar to that of known active kinases. We also describe the crystal structure of the ErbB3 kinase domain, which provides a basis for understanding why kinase activity is reduced but not absent.
Results and Discussion
Autophosphorylation of the Purified ErbB3 Intracellular Domain.
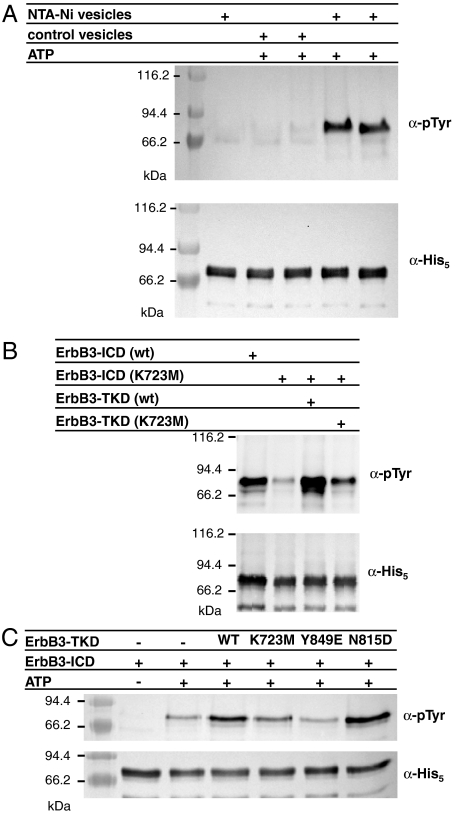
Our initial studies of the human ErbB3 intracellular domain (ErbB3-ICD665–1323) showed no evidence for autophosphorylation activity, consistent with previous reports (12). However, when ErbB3-ICD was clustered at the surface of vesicles by association of its N-terminal histidine tag with lipids bearing nitrilotriacetate (NTA)-Ni headgroups, robust tyrosine autophosphorylation could be seen in antiphosphotyrosine Western blots (Fig. 1A). This clustering method was used previously to promote assembly of histidine-tagged signaling receptor fragments on membrane surfaces (24) and was crucial for activating the EGFR kinase domain in recent studies by Kuriyan and colleagues (5). Autophosphorylation of ErbB3-ICD required both the presence of NTA-Ni lipid in the vesicles and ATP addition (Fig. 1A, two rightmost lanes). Mn2+ or Mg2+ ions were also required (Fig. S1A), with Mn2+ being slightly preferred. Consistent with the presence of all ErbB3 phosphorylation sites in the C-terminal tail, removing this region (leaving just the kinase domain: ErbB3-TKD665–1001) was accompanied by a loss of autophosphorylation.
Fig. 1.
Autophosphorylation of the ErbB3 intracellular domain in vitro. (A) ErbB3-ICD (3 μM) becomes autophosphorylated when incubated with ATP and vesicles containing 10% (mol/mol) NTA-Ni lipid (right two lanes), but not when either ATP or NTA-Ni lipid is absent. The upper panel is antiphosphotyrosine blot, and the lower is antipentahistidine loading control. Lanes 3/4 and 5/6 represent duplicate experiments. (B) ErbB3-ICD harboring a K723M mutation shows little autophosphorylation (lane 2) when treated with ATP and NTA-Ni vesicles as in A, whereas wild-type ErbB3-ICD autophosphorylation is robust. His-tagged ErbB3-TKD665–1001 added at 3 μM (lane 3) causes robust trans-phosphorylation of ErbB3-ICD (K723M), which is diminished by a K723M mutation in the TKD (lane 4). (C) Trans-phosphorylation of ErbB3-ICD by wild-type and mutated forms of ErbB3-TKD665–1001 was compared as described in the text.
To confirm that the autophosphorylation shown in Fig. 1A arises from ErbB3-ICD activity, and not from contaminating kinases, we analyzed autophosphorylation of a K723M-mutated variant under the same conditions. Lysine-723 in ErbB3 is equivalent to K721 in the ATP-binding site of the EGFR-TKD, at which mutations are known to abolish kinase activity (25). As shown in the second lane of Fig. 1B, a K723M mutation greatly diminished ErbB3-ICD autophosphorylation. However, K723M-mutated ErbB3-ICD could be trans-phosphorylated by adding isolated histidine-tagged ErbB3-TKD665–1001 (in the presence of NTA-Ni vesicles), as shown in lane 3 of Fig. 1B. Again, a K723M mutation diminished this activity (lane 4 in Fig. 1 B and C). Interestingly, mutating Y849 in the activation loop of ErbB3-TKD to glutamate impaired its ability to trans-phosphorylate ErbB3-ICD to an even greater degree (Fig. 1C), suggesting a possible role for Y849 in ErbB3 regulation. Taken together, these data show that the ErbB3 kinase domain retains sufficient activity for robust ICD autophosphorylation to be detectable in this simple assay. Key mutations at K723 and Y849 impair this activity. Introduction of aspartate at the position occupied by the catalytic base in other kinases (in the N815D mutant) did not significantly increase the trans-phosphorylation activity of ErbB3-TKD, consistent with a previous report (15).
Although these results demonstrate that ErbB3 can catalyze phosphoryl transfer, the catalytic efficiency is substantially lower than seen for activated EGFR-TKD. Indeed, we have not been able to detect significant phosphorylation of poly(Glu:Tyr) substrates or of peptides modeled on tyrosine phosphorylation sites within ErbB3 or ErbB2. By comparing rates of 32P incorporation when the ErbB3 and EGFR-TKDs trans-phosphorylate ErbB3-ICD (K723M), we estimate that ErbB3-TKD is approximately 1,000 times less active than fully activated EGFR-TKD under equivalent experimental conditions (Fig. S1B). Although the kinase activity of ErbB3 is modest, it could be sufficient to catalyze a physiologically relevant degree of receptor trans-phosphorylation in the context of ligand-induced ErbB receptor heterodimers as discussed below.
ErbB3 TKD Fully Retains ATP-Binding Ability.
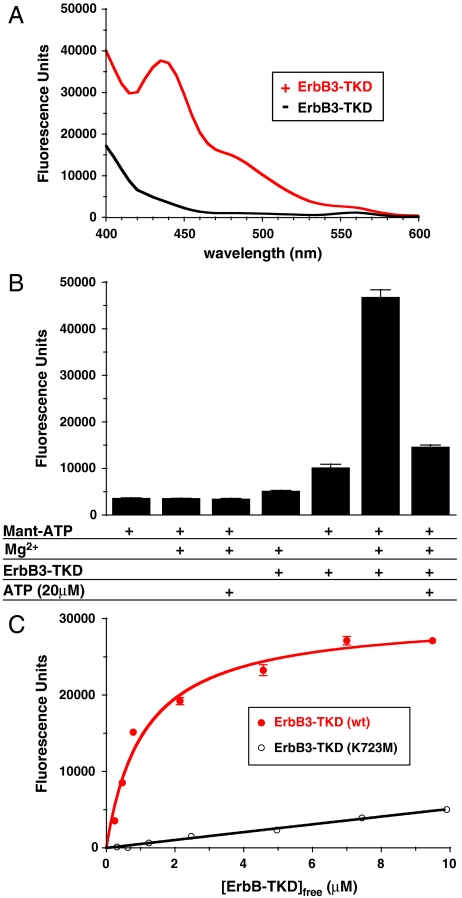
We next investigated the ATP-binding properties of ErbB3-TKD, because previous analyses yielded conflicting results (11, 12). We used mant [2′-(3′)-O-(N-methylanthraniloyl)]-ATP, a fluorescent analogue of ATP, for direct binding studies. As shown in Fig. 2A, a solution of 1 μM mant-ATP (plus 5 mM MgCl2) shows little fluorescence when excited at 280 nm, but a significant peak centered at 448 nm appears when 3 μM ErbB-TKD is added. This peak reflects FRET from tryptophans and/or tyrosines in the protein to the bound mant-ATP and is seen only when ErbB3-TKD, MgCl2, and mant-ATP are all present in the sample (Fig. 2B). Adding 20 μM ATP (a 20-fold excess over mant-ATP) substantially diminished mant-ATP binding by saturating the nucleotide-binding site of ErbB3-TKD. These results show that the ErbB3 kinase domain does bind ATP. To estimate the affinity of this binding, we monitored the FRET signal when titrating ErbB3-TKD into a solution containing 0.6 μM mant-ATP plus 5 mM MgCl2. A simple hyperbolic binding curve was observed (Fig. 2C), which suggested a Kd value of 1.1 μM for Mg2+/mant-ATP binding to ErbB3-TKD—arguing that the ErbB3 ATP-binding site will be saturated in vivo. This Kd value is smaller than apparent Km values of 4–47 μM for (Mg2+ or Mn2+) ATP measured with EGFR (26), ErbB2, or ErbB4 (27). Consistent with its reduced autophosphorylation activity (Fig. 1), K723M-mutated ErbB3-TKD also showed greatly diminished mant-ATP binding (Fig. 2C).
Fig. 2.
Mant-ATP binding to ErbB3-TKD648–1001. (A) Fluorescence emission spectra (with excitation at 280 nm) for 1 μM mant-ATP (plus 5 mM MgCl2) in the absence of ErbB3-TKD (Black) and with 3 μM added ErbB3-TKD648–1001 (Red). (B) Fluorescence emission of 1 μM mant-ATP at 450 nm under different conditions (with excitation at 280 nm). Protein-to-mant FRET is only seen when 1 μM mant-ATP, 5 mM MgCl2, and 3 μM ErbB3-TKD648–1001 are all present. The FRET signal is greatly reduced by adding 20 μM ATP to saturate the nucleotide-binding site in the kinase (far right). (C) Titrating ErbB3-TKD648–1001 into a 0.6 μM mant-ATP solution containing 5 mM MgCl2 yields a hyperbolic binding curve fit to give a Kd value of 1.1 μM for wild-type ErbB3-TKD. Mean ± standard deviation are shown for at least 3 independent experiments in B and C.
Crystal Structure of ErbB3-TKD Bound to an ATP Analogue and Mg2+.
To gain insight into how ErbB3-TKD retains activity despite substitutions at key residues, we determined its crystal structure. Crystals of ErbB3-TKD665–1001 grew only in the presence of 5 mM MgCl2 and the nonhydrolyzable ATP analogue adenylyl-imidodiphosphate (AMP-PNP) and diffracted x-rays to 2.8 Å resolution (Table S1). The structure was solved by molecular replacement (see Materials and Methods). The asymmetric unit contains two almost identical ErbB3-TKD665–1001 molecules, which pack in the crystal lattice in an N-lobe to N-lobe, C-lobe to C-lobe manner (Fig. S2).
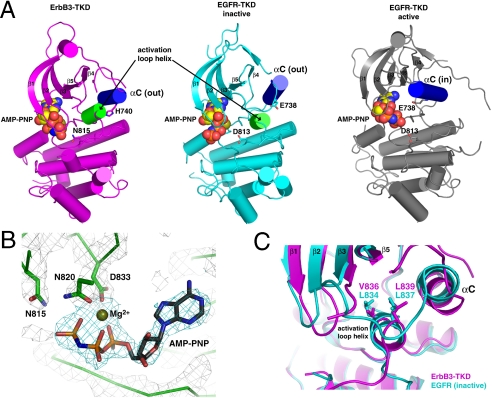
As shown in Fig. 3A, the overall structure of ErbB3-TKD665–1001 closely resembles inactive conformations of the EGFR, ErbB4, and Src kinases (5, 27). Helix αC of ErbB3-TKD665–1001 is displaced from the active site, in the “out” position typically seen in structures of inactive kinases (28). The activation loop of ErbB3-TKD665–1001 also retains the short α-helix that abuts helix αC in inactive EGFR and ErbB4 (green in Fig. 3A) and is the site of key mutations that activate EGFR in non-small cell lung cancer (NSCLC) (29). One particularly notable difference between ErbB3-TKD665–1001 and the other ErbB kinases is the length of helix αC, the amino-terminal half of which (∼1.5 turns in EGFR) is unraveled in ErbB3-TKD665–1001. This region of ErbB3-TKD forms a well-defined loop (residues 732–739) that appears to be anchored by backbone-mediated interactions with the β4/β5 loop and by projection of the F734 side chain into a hydrophobic pocket formed by side chains from β3, β5, αC, and the activation loop.
Fig. 3.
Crystal structure of ErbB3-TKD665–1001. (A) Cartoon representations are shown for ErbB3-TKD665–1001 (Left: Magenta), inactive EGFR-TKD (Middle: Cyan), and active EGFR-TKD (Right: Gray). EGFR-TKD structures are from PDB codes 2GS7 (5) and 2ITX (48). All structures have bound AMP-PNP and Mg2+ (shown in spheres). Helix αC is colored blue in each structure and is denoted as out (ErbB3 and inactive EGFR) or in (active EGFR). The short activation loop helix in ErbB3 and inactive EGFR is colored green. H740 in helix αC of ErbB3 and the similarly located E738 in EGFR are shown as sticks, as are ErbB3 N815 and EGFR D813. β-strands in the N lobe are labeled. (B) Close-up view of Mg2+-AMP-PNP in the ErbB3-TKD665–1001 active site, with electron density shown as a 2Fo - Fc map calculated at 1.8σ by using phases from a model with no nucleotide. Coordination of bound Mg2+ by the N820 and D833 side chains (from the DFG motif) and the AMP-PNP α- and β-phosphates is depicted. N815 (close to the γ-phosphate) is also marked. (C) Close-up of the short activation loop helix seen in ErbB3-TKD665–1001 (Magenta) and inactive EGFR-TKD (Cyan), with the two kinase domains superimposed. Mutations at L834 or L837 in EGFR activate the receptor in NSCLC. These residues overlay with V836 and L839 in ErbB3-TKD665–1001 and interact with the hydrophobic pocket that contributes to stabilization of helix αC in the out position. A V836A mutation inhibits ErbB3-TKD activity.
The ErbB3-TKD665–1001 structure confirms the similarity of its C lobe to that of EGFR-TKD, supporting previous arguments (from sequence comparisons) that the ErbB3 TKD should be able to function as an “activator” of the EGFR kinase domain in an asymmetric dimer (5). However, structural differences in the N lobe—including the shortened αC-helix—reinforce the argument that ErbB3-TKD cannot be activated (as “receiver”) by the same types of interactions thought to activate EGFR, ErbB4, and ErbB2 in heterodimeric complexes. Jura et al. (30) very recently reported an ErbB3-TKD674–1001 structure [Protein Data Bank (PDB) ID 3KEX] that is essentially identical to that reported here and have confirmed the ability of ErbB3-TKD674–1001 to function as an allosteric activator of EGFR-TKD in vitro. If the ErbB3-TKD does function as a receiver kinase in ErbB receptor dimers, it must do so through N-lobe interactions distinct from those described for EGFR (5) and ErbB4 (27).
Consistent with the mant-ATP-binding studies shown in Fig. 2, clear electron density for both AMP-PNP and Mg2+ was seen in the ErbB3-TKD665–1001 structure (Fig. 3B). The bound Mg2+ ion is coordinated by the side chains of N820 and D833 (from the DFG [Asp-Phe-Gly] motif), plus the α- and β-phosphates of AMP-PNP. The ϵ-amino group of the highly conserved K723 residue (in strand β3) interacts with both the α-phosphate of AMP-PNP and the D833 side chain, thus playing a key role in nucleotide binding as seen with inactive EGFR and indicated by the loss of kinase activity (Fig. 1) and mant-ATP binding (Fig. 2) in K723M mutants.
The activation loop configuration in ErbB3-TKD665–1001 resembles that of inactive EGFR-TKD and ErbB4-TKD (5, 27), characterized by a short α-helix (green in Fig. 3A) that contains several aliphatic side chains (L834, L837, and L838 in EGFR). These residues project into a hydrophobic pocket formed by side chains from both the β-sheet in the N lobe and helix αC. Mutations at L834 and L837 in the activation loop that disrupt these interactions have been found to activate EGFR in NSCLC (5, 29), and ErbB4 can be activated similarly in vitro (27). ErbB3 contains a conserved motif (V836, L839, and L840) in its short activation loop helix that makes a very similar set of interactions, apparently stabilizing the “inactive-like” position of helix αC. As shown in Fig. 3C, residues V836 and L839 in ErbB3 overlay almost exactly with L834 and L837 in EGFR (key sites of NSCLC mutations). Surprisingly, whereas mutating L834 in EGFR (or L839 in ErbB4) causes kinase activation, a V836A mutation in ErbB3 instead abolishes mant-ATP binding (Fig. S3) and—accordingly—the autophosphorylation activity seen for wild-type ErbB3-ICD in Fig. 1. Thus, mutations that destabilize the inactive state of EGFR and ErbB4 activate these kinases, whereas a mutation designed to destabilize the apparently inactive ErbB3 structure has the opposite (inactivating) effect.
Quantum Mechanics/Molecular Mechanics (QM/MM) Simulation of Phosphoryl-Transfer by ErbB3.
From the observations described above, we hypothesized that the weak kinase activity of ErbB3 might utilize a mechanism distinct from that of other kinases and might be carried out by the inactive-like state depicted in Fig. 3A. This mechanism would explain why the V836A mutation prevents ATP binding and ErbB3-ICD autophosphorylation rather than activating the kinase further. We therefore undertook QM/MM simulations to delineate the energy landscape of phosphoryl transfer from ATP to a tyrosine substrate catalyzed by the ErbB3 structure from Fig. 3A and compared the outcomes with those seen in parallel computational studies of EGFR (see SI Text for details).
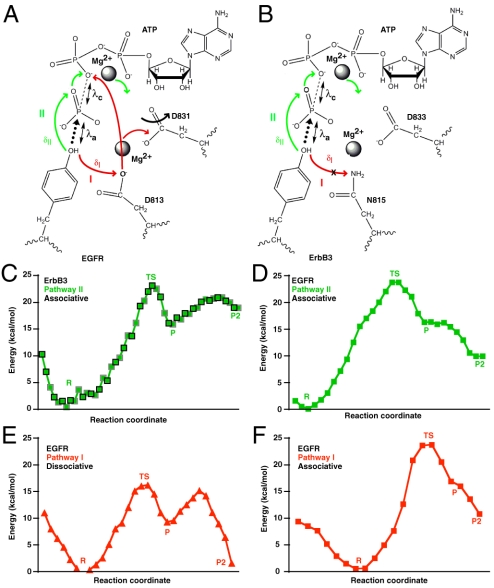
Experimental and theoretical studies have demonstrated that, in phosphoryl transfer by kinases and polymerases, nucleophilic attack on the target phosphate proceeds through a conformation that resembles a trigonal-bipyramidal transition state (31) and that phosphoryl transfer can occur through either an associative or a dissociative mechanism (31–33). A conserved aspartate (D813 in EGFR, D166 in protein kinase A) is proposed to function as a base acceptor for proton transfer from the hydroxyl group of the substrate, as depicted by the red arrows in Fig. 4A (pathway I). The presence of an asparagine at this location in ErbB3 (N815) precludes this pathway, requiring that proton transfer occurs via an alternative mechanism (Fig. 4B). Our QM/MM simulations reveal an alternative pathway in ErbB3 and EGFR (pathway II) that is characterized by migration of the substrate tyrosyl -OH proton to the O1γ oxygen of ATP and subsequently to the ATP O2β oxygen (green arrows in Fig. 4 A and B). Phosphoryl transfer in ErbB3 coincides with this pathway II proton transfer step and occurs exclusively through an associative mechanism (Fig. S4), with characteristic values of 1.9 Å for the formation (λa) and cleavage (λc) of bonds in the transition state and an estimated activation energy (Ea) of 23 kcal/mol (Fig. 4C). Parallel QM/MM simulations of EGFR in its active configuration showed that pathways I and II (which are mutually exclusive) can both be utilized for transfer of the tyrosyl -OH proton and compete with one another (Fig. S5). EGFR-catalyzed phosphoryl transfer concomitant with proton migration through pathway II occurs only through an associative transition state, with an estimated Ea of 24 kcal/mol (Fig. 4D)—equal to that seen for ErbB3. When proton transfer occurs through pathway I in EGFR, which involves proton abstraction by D813, the estimated Ea is significantly lower for the dissociative mechanism (Fig. 4E), at 16 kcal/mol, but is 24 kcal/mol for the associative mechanism (Fig. 4F).
Fig. 4.
QM/MM simulations of phosphoryl transfer. Schematic of the phosphoryl-transfer pathway in (A) EGFR-TKD and (B) ErbB3-TKD, as captured in QM/MM simulations. Mg2+ ions are marked, as are the catalytic aspartates (D831 in EGFR, D833 in ErbB3), the proposed catalytic base in EGFR (D813), and its replacement in ErbB3 (N815). Two potential pathways for proton migration (concomitant with phosphoryl transfer) are marked as described in the text: pathway I (Red) and pathway II (Green). The distance from the substrate -OH proton to the Oδ2 oxygen of D813 in EGFR is labeled δI, and its distance to the O1γ oxygen of ATP is δII (see Fig. S5 A and B). The nucleophilic attack distance λa (distance between the tyrosine oxygen and the ATP Pγ) and the bond cleavage distance λc (distance between the ATP Pγ and ATP O2/3β) are marked (see Fig. S5). (C–F) Energy changes along the reaction pathway in QM/MM simulations for the noted mechanisms involving pathway II (Green) or pathway I (Red). States correspond to “R” (reactant); “TS” (transition state with trigonal-bipyramidal geometry around Pγ); “P” (product after phosphoryl transfer with proton bound to ATP O1γ); and “P2” (product with proton transferred to ATP O2β); see Figs. S4–S6 for details. (C) Energy changes along the simulated ErbB3 reaction pathway (in the structure shown in Fig. 3A). Symbols bounded by black squares represent the forward scan, and those bounded by gray squares represent the reverse scan. Only pathway II is utilized, and estimated Ea for phosphoryl transfer is 23 kcal/mol. (D) Energy changes along the EGFR reaction pathway (active configuration) utilizing pathway II and the associative mechanism: Estimated Ea is 24 kcal/mol. (E) Phosphoryl transfer catalyzed by EGFR via the dissociative mechanism (utilizing pathway I for proton migration—via D813) has the lowest Ea value, at 16 kcal/mol. (F) Associative phosphoryl transfer concomitant with pathway I for EGFR has an estimated Ea of 24 kcal/mol.
Our QM/MM simulations therefore argue that phosphoryl transfer can occur in ErbB3 (and indeed EGFR) without abstraction of the substrate tyrosyl -OH proton by the catalytic base aspartate. The proton can instead migrate to the O1γ oxygen of ATP, through pathway II in Fig. 4B. Phosphoryl transfer concomitant with this pathway can be catalyzed by ErbB3 in the inactive-like conformation shown in Fig. 3A, although it is predicted to be several orders of magnitude slower than the most favorable reaction channel in EGFR (i.e., pathway I through a dissociative mechanism)—consistent with our experimental observations. The finding in our QM/MM studies that this mechanism also operates in EGFR further suggests that mutating D813 in this receptor may not completely abolish its kinase activity. Indeed, a D813A-mutated EGFR variant was reported to retain the ability to promote EGF-dependent DNA synthesis and MAP kinase activation despite exhibiting greatly reduced receptor autophosphorylation (34). A low level of autophosphorylation in D813A-mutated EGFR, similar to that shown for ErbB3 in Fig. 1, may therefore be sufficient to mediate certain key aspects of its signaling.
Conclusions
Although the kinase domain of ErbB3 has long been assumed to be inactive (6–8) and has been classified as a pseudokinase (6, 16), the data presented here show that it binds ATP and promotes trans-autophosphorylation of the receptor’s intracellular domain when clustered at a membrane surface. QM/MM simulations also argue that the inactive-like ErbB3-TKD crystal structure presented here and recently by Jura et al. (30) can catalyze phosphoryl transfer. Thus, ErbB3 does appear to have an active tyrosine kinase in its intracellular domain. Our data suggest that it is ∼1,000-fold less active than EGFR-TKD, although it is quite possible that we have failed to activate ErbB3-TKD fully in our assays. A key question is whether or not the kinase activity evidenced in Fig. 1 is sufficient to explain cellular functions of ErbB3. There are several reasons to suspect so.
Stimulation of most RTKs involves initial “activating” trans-autophosphorylation events in response to ligand-induced dimerization that increase the catalytic efficiency of the kinase (assessed by using peptide substrates) by ∼150–1,000-fold overall (35–38). “Full” kinase activation by autophosphorylation at multiple sites is often required for the receptor to phosphorylate downstream signaling or docking molecules such as FGF receptor substrate-2 (FRS-2) and phospholipase-Cγ (PLCγ) (37) and to phosphorylate exogenous peptides efficiently in vitro. However, much lower levels of activity are clearly sufficient for the initial activating trans-autophosphorylation steps (which are mediated by the unphosphorylated kinase). For example, the unphosphorylated FGFR1 kinase catalyzes autophosphorylation of Y653, with which activation of this receptor begins. Prior to phosphorylation, the FGFR1 kinase domain is 500–1,000-fold less active than its “final” fully autophosphorylated counterpart that phosphorylates FRS-2 and PLCγ in normal signaling (37)—yet this weak activity is crucial for receptor function. The kinase activity we see for ErbB3 is similar to that of unphosphorylated FGFR and may well also be necessary (and sufficient) for receptor trans-autophosphorylation within ErbB receptor dimers (such as ErbB2/ErbB3 heterodimers)—even though ErbB3-TKD cannot phosphorylate peptide substrates efficiently (neither can unphosphorylated FGFR1-TKD). Indeed, trans-phosphorylation of ErbB2 within an ErbB2/ErbB3 heterodimer would be satisfyingly explained if ErbB3 kinase activity is responsible. EGFR (or ErbB2) may only require its “excess” kinase capacity (∼1,000-fold higher than seen for ErbB3) for phosphorylating exogenous substrates such as PLCγ as an additional component of signaling. This ability to phosphorylate exogenous substrates may be the only kinase function that ErbB3 lacks.
The weak kinase activity of ErbB3 could be clinically relevant in cases where this receptor drives resistance to agents that target other ErbB family members (2). ErbB3 signaling is important in resistance of NSCLC to the EGFR inhibitor gefitinib through amplification of the gene for Met, the hepatocyte growth factor receptor (39). ErbB3 also plays a key role in the ability of ErbB2-overexpressing cells to escape growth inhibition by the EGFR/ErbB2 dual-specific kinase inhibitor lapatinib (40). Efforts to inhibit ErbB3 in these (and other) settings—and thus combat this resistance—have focused on therapeutic antibodies (2). Our data suggest that evaluating the signaling importance of ErbB3’s weak kinase activity is also worthwhile and could represent a valuable therapeutic target. Indeed, ErbB3 autophosphorylation in vitro is not inhibited by lapatinib or by the EGFR inhibitors gefitinib or erlotinib (Fig. S7), leaving open the possibility that ErbB3 kinase activity promotes resistance to growth inhibition by EGFR/ErbB2-targeted drugs. If this is the case, selective inhibition of the ErbB3 kinase could be an important additional therapeutic strategy.
Materials and Methods
Plasmid Construction and Protein Preparation.
DNA fragments encoding the human ErbB3 intracellular domain (ErbB3-ICD665–1323) or TKD (ErbB3-TKD665–1001 or ErbB3-TKD648–1001) with N-terminal hexa-histidine tag were subcloned into the pFastBac1 vector (Invitrogen), and proteins were expressed in Spodoptera frugiperda Sf9 by using the Bac-to-Bac expression system (Invitrogen). Cells were lysed in 20 mM Hepes pH 8.0, 300 mM NaCl, 5% glycerol, 5 mM 2-mercaptoethanol with protease inhibitor cocktail (Roche), and proteins purified by using Ni-NTA beads (Qiagen) followed by anion exchange and size exclusion chromatography.
Analysis of Autophosphorylation by Western Blotting.
Vesicles were prepared as described (5) containing 5–10% (mol/mol) 1, 2-dioleoyl-sn-glycero-3-{[N(5-amino-1-carboxypentyl) iminodi-acetic acid]succinyl} nickel salt (DOGS-NTA-Ni, from Avanti Polar Lipids) in dioleoylphosphatidylcholine (Sigma). Vesicles (125 μM total lipid) were incubated with ErbB3 proteins at the noted concentrations in 100 mM MOPS pH 7.4, 200 mM NaCl, 5% glycerol, 1 mM ATP, 5 mM MgCl2, 2 mM MnCl2, and 0.1 mM DTT. After 30 min at 25 °C, reactions were stopped by adding 50 mM EDTA and SDS-PAGE gel-loading buffer. Proteins were analyzed by immunoblotting with pY20 phosphotyrosine antibody (BIOMOL) and anti-His5 antibody (QIAGEN), by using HRP-conjugated anti-mouse (Amersham) and enhanced chemiluminescence for detection.
Mant-ATP-Binding Assay.
Mant-ATP [2′-(3′)-O-(N-methylanthraniloyl)adenosine 5′-triphosphate] was purchased from Invitrogen. Binding assays were carried out in 20 mM Tris pH 8.0, 200 mM NaCl, 5% glycerol, 1 mM DTT, 5 mM MgCl2, and 0.6 μM Mant-ATP, and varying amounts of ErbB3648–1001. Fluorescence measurements employed a Tecan SAFIRE II plate reader, with excitation at 280 nm and emission detection at 450 nm (5 nm bandwidth).
Crystallization and Structure Determination.
Purified ErbB3-TKD665–1001 was concentrated to 6 mg/mL and incubated with 5 mM MgCl2 and 1 mM AMP-PNP (Sigma). Crystals were obtained by using the hanging-drop method at 21 °C by mixing protein with an equal volume of reservoir solution [50 mM MOPS pH 7.4, 27% (wt/vol) PEG8000, 0.17 M (NH4)2SO4, and 15% glycerol]. Crystals were frozen in liquid nitrogen directly from the mother liquor. Diffraction data were collected at the Argonne Advanced Photon Source (beamline GM/CA CAT) and processed with HKL2000 (41). The structure was solved by molecular replacement using Phaser (42), with the inactive EGFR (V924R) TKD structure (PDB code 2GS7) (5) as a search model. Coot (43) was used for model building, and refinement employed both REFMAC (42) and CNS (44). Translation/libration/screw refinement (45) was employed in the later stages of refinement, by using REFMAC (42), with anisotropic motion tensors refined for each lobe of the kinase domain. Structure figures were generated with PyMol (46).
QM/MM Simulations.
Models for the ErbB3 and EGFR kinase domains with bound Mg2+ and substrate were constructed (see SI Text for details), and the ground state of each ternary complex was obtained from energy minimizations (Fig. S6) to ensure uniformity in reactant states across all systems prior to QM/MM simulations. Resulting models were prepared for QM/MM simulations (see SI Text for details) in which the quantum region contained the two Mg2+ ions, water molecules within 5 Å of the Mg2+ ions, segments of the ATP and peptide substrate, and two catalytic residues (N815 and D833 in ErbB3, D813 and D831 in EGFR). The system was subjected to 1,200 steps of the adopted basis Newton–Raphson minimization. For QM/MM molecular dynamics simulations, the system was heated to 300 K and then subjected to constant temperature dynamics by using a Langevin thermostat for 10 ps with a 1-fs integration step. We describe reaction pathways in terms of simple artificial reaction coordinates χj (47). For phosphoryl transfer through the associative mechanism, χ1 is the distance between the reactive tyrosyl oxygen and the ATP γ-phosphate, χ2 is the distance between the tyrosyl O- and ATP O2/3β, and χ3 is the ATP Pγ- ATP O2/3β distance. For phosphoryl transfer through dissociative mechanisms, an additional distance χ4 includes the coordinate for proton abstraction (the distance between the tyrosyl OH- and D830∶Oδ2). Restrained minimization as well as restrained sampling simulations were performed along χ by using the QM/MM Hamiltonian with a potential bias term (see SI Text for details). Reaction paths were computed by energy minimizations in the presence of restraints and recalculating single-point energies in their absence.
Supplementary Material
Acknowledgments.
We thank members of the Lemmon, Ferguson, and Radhakrishnan laboratories for valuable discussions and advice. This work was supported in part by grants from the National Cancer Institute (R01-CA079992-11 to M.A.L.) and National Science Foundation (CBET-0730955 and CBET-0853539 to R.R.). S.E.T. was supported by a National Science Foundation Graduate Research Fellowship and a Graduate Assistance in Areas of National Need Award from the Penn Bioengineering Department. Computational resources were provided in part by the National Partnership for Advanced Computational Infrastructure under the allocation grant MRAC MCB060006. Crystallographic data were collected at the GM/CA Collaborative Access Team at the Advanced Photon Source (APS) that has been funded by the National Cancer Institute (Y1-CO-1020) and National Institute of General Medical Sciences (Y1-GM-1104). Use of APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-ENG-38.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates for the ErbB3 kinase domain bound to AMP-PNP have been deposited in the Protein Data Bank (www.rcsb.org/pdb) with the identification number 3LMG.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002753107/DCSupplemental.
References
- 1.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Baselga J, Swain SM. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 3.Jura N, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Red Brewer M, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: The biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 7.Kraus MH, Issing W, Miki T, Popescu NC, Aaronson SA. Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: Evidence for overexpression in a subset of human mammary tumors. Proc Natl Acad Sci USA. 1989;86:9193–9197. doi: 10.1073/pnas.86.23.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plowman GD, et al. Molecular cloning and expression of an additional epidermal growth factor receptor-related gene. Proc Natl Acad Sci USA. 1990;87:4905–4909. doi: 10.1073/pnas.87.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanks SK, Quinn AM, Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 10.Madhusudan, et al. cAMP-dependent protein kinase: Crystallographic insights into substrate recognition and phosphotransfer. Protein Sci. 1994;3:176–187. doi: 10.1002/pro.5560030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., III Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sierke SL, Cheng K, Kim HH, Koland JG. Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J. 1997;322(Pt 3):757–763. doi: 10.1042/bj3220757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraus MH, Fedi P, Starks V, Muraro R, Aaronson SA. Demonstration of ligand-dependent signaling by the erbB-3 tyrosine kinase and its constitutive activation in human breast tumor cells. Proc Natl Acad Sci USA. 1993;90:2900–2904. doi: 10.1073/pnas.90.7.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallasch C, et al. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prigent SA, Gullick WJ. Identification of c-erbB-3 binding sites for phosphatidylinositol 3'-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J. 1994;13:2831–2841. doi: 10.1002/j.1460-2075.1994.tb06577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Berger MB, Mendrola JM, Lemmon MA. ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett. 2004;569:332–336. doi: 10.1016/j.febslet.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Riese DJ, II, van Raaij TM, Plowman GD, Andrews GC, Stern DF. The cellular response to neuregulins is governed by complex interactions of the erbB receptor family. Mol Cell Biol. 1995;15:5770–5776. doi: 10.1128/mcb.15.10.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole PA, Grace MR, Phillips RS, Burn P, Walsh CT. The role of the catalytic base in the protein tyrosine kinase Csk. J Biol Chem. 1995;270:22105–22108. doi: 10.1074/jbc.270.38.22105. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee K, et al. CASK functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eswaran J, et al. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci USA. 2009;106:20198–20203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min X, Lee BH, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure. 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Kornev AP, Taylor SS. Pseudokinases: Functional insights gleaned from structure. Structure. 2009;17:5–7. doi: 10.1016/j.str.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrout AL, Montefusco DJ, Weis RM. Template-directed assembly of receptor signaling complexes. Biochemistry. 2003;42:13379–13385. doi: 10.1021/bi0352769. [DOI] [PubMed] [Google Scholar]
- 25.Honegger AM, et al. Point mutation at the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell. 1987;51:199–209. doi: 10.1016/0092-8674(87)90147-4. [DOI] [PubMed] [Google Scholar]
- 26.Qiu C, et al. In vitro enzymatic characterization of near full length EGFR in activated and inhibited states. Biochemistry. 2009;48:6624–6632. doi: 10.1021/bi900755n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu C, et al. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 29.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 30.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci USA. 2009;106:21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction. Science. 2003;299:2067–2071. doi: 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- 32.Valiev M, Yang J, Adams JA, Taylor SS, Weare JH. Phosphorylation reaction in cAPK protein kinase-free energy quantum mechanical/molecular mechanics simulations. J Phys Chem B. 2007;111:13455–13464. doi: 10.1021/jp074853q. [DOI] [PubMed] [Google Scholar]
- 33.Williams DM, Cole PA. Proton demand inversion in a mutant protein tyrosine kinase reaction. J Am Chem Soc. 2002;124:5956–5957. doi: 10.1021/ja025993a. [DOI] [PubMed] [Google Scholar]
- 34.Coker KJ, Staros JV, Guyer CA. A kinase-negative epidermal growth factor receptor that retains the capacity to stimulate DNA synthesis. Proc Natl Acad Sci USA. 1994;91:6967–6971. doi: 10.1073/pnas.91.15.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cobb MH, Sang BC, Gonzalez R, Goldsmith E, Ellis L. Autophosphorylation activates the soluble cytoplasmic domain of the insulin receptor in an intermolecular reaction. J Biol Chem. 1989;264:18701–18706. [PubMed] [Google Scholar]
- 36.Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Mol Biol. 2001;8:1058–1063. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- 37.Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 38.Till JH, et al. Crystal structure of the MuSK tyrosine kinase: Insights into receptor autoregulation. Structure. 2002;10:1187–1196. doi: 10.1016/s0969-2126(02)00814-6. [DOI] [PubMed] [Google Scholar]
- 39.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 40.Sergina NV, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Winn MD, Isupov MN, Murshudov GN. Use of TLS anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 47.Rosta E, Woodcock HL, Brooks BR, Hummer G. Artificial reaction coordinate “tunneling” in free-energy calculations: The catalytic reaction of RNase H. J Comput Chem. 2009;30:1634–1641. doi: 10.1002/jcc.21312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.