Abstract
The establishment of agricultural economies based upon domestic animals began independently in many parts of the world and led to both increases in human population size and the migration of people carrying domestic plants and animals. The precise circumstances of the earliest phases of these events remain mysterious given their antiquity and the fact that subsequent waves of migrants have often replaced the first. Through the use of more than 1,500 modern (including 151 previously uncharacterized specimens) and 18 ancient (representing six East Asian archeological sites) pig (Sus scrofa) DNA sequences sampled across East Asia, we provide evidence for the long-term genetic continuity between modern and ancient Chinese domestic pigs. Although the Chinese case for independent pig domestication is supported by both genetic and archaeological evidence, we discuss five additional (and possibly) independent domestications of indigenous wild boar populations: one in India, three in peninsular Southeast Asia, and one off the coast of Taiwan. Collectively, we refer to these instances as “cryptic domestication,” given the current lack of corroborating archaeological evidence. In addition, we demonstrate the existence of numerous populations of genetically distinct and widespread wild boar populations that have not contributed maternal genetic material to modern domestic stocks. The overall findings provide the most complete picture yet of pig evolution and domestication in East Asia, and generate testable hypotheses regarding the development and spread of early farmers in the Far East.
Keywords: Asian colonization, mtDNA, phylogeography
The transition from hunting wild animals to stock-raising occurred in many places independently across the globe. In many cases, the combination of herding and agriculture spurred an increase in population size brought about by the advent of sedentary living, which in turn spurred a demographic migration outward from centers of domestication and agricultural origin (1). An understanding of the locations, timing, and processes of domestication are therefore essential to understanding not only the roots of modern civilization, but also the migratory trajectories that have shaped the modern geography of human languages and cultures (2).
The study of domestication has been transformed by biomolecular data that have provided new insights, not least of which has been the general conclusion that animal domestication occurred more often and in more places than had been suggested by traditional archaeological evidence (3, 4). Recent publications have sought to further explore the domestication of pigs using both archaeological (5 –8) and genetic approaches (9 –14). These studies confirmed long-suspected separate domestications of differentiated subspecies of European and Asian wild boar, and revealed that wild boar from regions such as Italy (10) and India (10, 14) were also either domesticated or at least incorporated into domestic stock originally derived from regionally differentiated wild populations. A clear correlation between phylogenetic signals and geographic provenance also allows pigs to be important proxies of human dispersal. As such, they have already revealed the movement of domesticated Near Eastern pigs into Neolithic Europe (9), as well as an unexpected (possibly Austronesian) trajectory connecting Southeast Asia to Oceania (11).
Recent studies in East Asia have highlighted the antiquity of the origins of agriculture and the domestication of plants and animals. Among current views are that early agricultural activities practiced by seasonally mobile cultivators focused on millets in northern China were well established along the Yellow River and Inner Mongolia by ~8,000 (cal) B.P. (5, 8, 15, 16), and domestication may have even begun 2,000 years earlier (17). In southern China, available evidence can be interpreted to suggest that it was sedentary hunter-gatherers (18) who first began cultivating rice along the Yangtze about 9,000–8,000 B.P., a process that culminated in the dependence upon domesticated rice agriculture by ~6,000 B.P. (19). Although dogs were likely the earliest domesticated animal in these regions, available zooarchaeological evidence has been interpreted to indicate that domestic pigs were prevalent in both northern and southern China by at least 8,000 B.P. (6, 7, 20). In both regions, however, pigs make up a small percentage of the earliest mammal bone assemblages that are instead dominated by remains of hunted deer (7, 21).
The extent to which pig domestication in each region was independent or connected by diffusion from a single origin remains to be established, although recent research based on complete mitochondrial genomes of East Asian pigs suggests that Chinese wild boar may have been domesticated independently in the Mekong and in the middle to downstream Yangtze regions (13), although the geographic boundaries of these regions were not defined. Current archaeological evidence suggests that once established, intensive, sedentary agriculture (including rice, millet, and pigs) expanded across various regions from Northeast Asia and Central China (see Table 1 for regional definitions) about 6,500–5,000 B.P., and southward from the Yangtze about 5,000–4,000 B.P. during the Late Neolithic (22, 23). The current understanding is that domesticated pigs and rice are also present in Thailand no earlier than ~4,000 B.P. (24), although the evidence for pigs is based upon traditional metrical analyses. A re-evaluation of the data using more sophisticated morphological techniques (e.g., (25, 26) is desirable because it may reveal an earlier appearance of domestic pigs in that region.
Table 1.
Geographic definitions of regions discussed in the text
| Region | Areas region includes |
| Central China | Modern central China, except the southern portion of Yunnan province, Guangxi, and Guangdong provinces, and the northeast provinces of Liaoning, Jilin, Heilongjiang, and the northern portion of Inner Mongolia |
| Indo-Burma Biodiversity Hotspot (IBBH) (32) | Modern Vietnam, Laos, Cambodia, Myanmar, Thailand to the Kra Isthmus, and the southern portions of Yunnan province, Guangdong province, and Guangxi Zhuang Nationality Autonomous Region |
| South Asia | Modern India, Bangladesh, Bhutan, and Nepal |
| Northeast Asia | The northeast Chinese provinces of Liaoning, Jilin, Heilongjiang, and the northern portion of Inner Mongolia, modern North and South Korea, and the region of Southeast Russia that borders these regions |
| Oceania | The islands of Sumatra, Java, Borneo, and all of the islands to the east extending into the remote Pacific |
Genetic studies of pigs and wild boar from mainland East Asia have thus far been limited to analyses of modern samples, the conclusions of which have relied upon an assumption that past distributions can be extrapolated from present-day distributions. This assumption is risky given that throughout the Holocene, the geographic distribution of wild and domestic animals has varied dramatically as the result of numerous factors. Climatic fluctuations have resulted in significant range alterations among several large wild mammal species throughout Europe (27), and human hunting has created a modern patchwork of a formerly continuous brown bear population (28). Domestic populations have also experienced radical changes in their distributions. Frequencies of modern European cattle Y chromosome haplotypes, for example, bear little relation to Y haplotype frequencies derived from populations just a few hundred years old (29).
Multiple waves of human-mediated dispersal of other domesticates have created palimpsests of distinct mtDNA haplotypes, often rendering all but the most recent layer invisible. Chicken mtDNA haplotypes from Pacific islands have revealed a likely pattern of turnover resulting from successive waves of introduction (30). For pigs, an analysis of ancient material revealed an initial introduction of Near Eastern pigs into Europe, and their subsequent replacement by pigs domesticated from separate European wild boar populations—a pattern that would have remained invisible in an analysis that only made use of modern pigs (9). These examples show the power of natural processes and the human legacy of intervention to fundamentally alter the geographic distributions of wild and domestic animals, and they underscore the importance of including ancient specimens to obtain a more robust understanding of early patterns of domestication and human migration.
Results
Phylogenetic Analyses of East Asian Pigs.
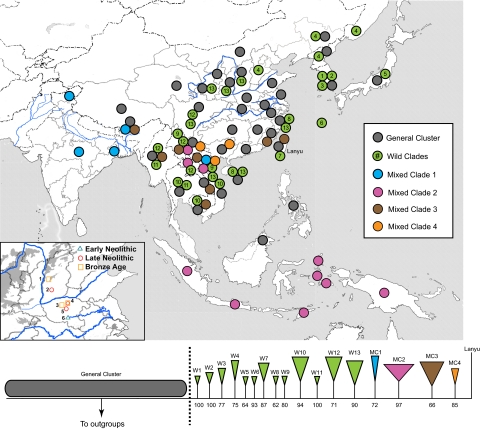
Despite the numerous additional samples and haplotypes included in this study, the general shape of the consensus Bayesian tree (drawn using an alignment of 662 bp of mtDNA control region sequence) is similar to that derived from previous studies (10, 11). The indigenous wild boar samples from ISEA and western Eurasia form well-supported clades that, relative to the group of East Asian samples focused upon here, are located in basal and derived positions respectively. The middle portion, defined by a large polytomy of clades and individual branches shown in Fig. 1, depicts samples from East Asia and forms the focus of this study.
Fig. 1.
A map of East Asia showing modern political and Chinese and Indian province boundaries, and a phylogenetic tree depicting the relationships between clades of wild and domestic pigs in the region. Colored circles within each Chinese or Indian province, each country (other than China or India), and each island indicate the presence of at least a single sample belonging to the colored regions on the tree below. Each of the 13 individual clades made up solely of wild boar are colored green on both the tree and the map. The numbers below the clades represent the posterior probability support values. The branch leading to the unique Lanyu pig haplotype is labeled on the tree, and the word Lanyu is placed on the map. Four additional clades, labeled MC1–MC4 and colored blue, purple, brown, and orange, respectively, possess both regionally restricted wild boar populations and domestic pigs (Fig. S3). The left-hand side of the tree is a polytomious cluster made up of individual branches of both wild and domestic pigs (and nine clades made up solely of domestic pigs) collectively referred to as the general cluster and colored gray. The two sides of the tree are separated by a dashed black line also shown in Fig. 2. Specific location information for each sample can be found in Tables S1–S5 and Figs. S1–S3. The Indus, Ganges, Yellow, and Yangtze Rivers are highlighted in blue. The inset on the bottom left of the map shows the locations and relative ages of the archaeological sites from which the ancient pig bones were retrieved, and the numbers 1–6 correspond to the sites Gaohong, Taosi, Guchengzhai, Wangchenggang, Wadian, and Jiahu, respectively.
Of the short unaffiliated branches, 45 haplotypes (representing 167 samples) are found only in wild specimens, 92 haplotypes (339 samples) are found only in domestic specimens, and 21 haplotypes are found in both 87 wild and 582 domestic pigs. One exceedingly long branch leads to a haplotype sequenced in a population of domestic pigs originally discovered on the small island of Lanyu near the southeastern coast of Taiwan (31). Seventeen clades consist of at least two haplotypes made up of either wild samples or a mix of wild and domestic samples. The 13 clades consisting only of wild samples are labeled W1–W13, and the four mixed clades are labeled MC1–MC4 (Fig. 1 and Fig. S1).
Contrasting the phylogenetic position of these samples with their geographic provenance reveals several interesting patterns, even though some GenBank samples do not posses specific geographic information (Tables S1–S5). First, the domestic specimens within the general cluster are from throughout East Asia, and many of them are the most common Asian pig haplotypes found in globally distributed modern breeds (11). In contrast, the wild and mixed haplotype specimens in the general cluster are found mostly in Central China, or in countries and provinces (e.g., Bhutan and Yunnan Province) that border Central China.
The geographic patterning of the specimens found in the 13 clades with only wild boar, together with those in the four mixed specimen clades, show a different pattern. Of the wild clades, four (W5–W8) are restricted to islands, including Japan, Okinawa, Taiwan, and the southern Chinese island of Hainan (although two samples in this specific clade are from the mainland). Three clades are found only in South Korea (W1–W3). One clade (W4) is restricted to Northeast Asia (not including South Korea), one is restricted to Central China (W13; although two samples are also found in northern Vietnam), and five clades (W9–W12) are found only in samples from the region known as the Indo-Burma Biodiversity Hotspot (32) (IBBH). Of those, several clades are found only in smaller-scale regions.
Three of the four clades that possess haplotypes found in both wild and domestic samples have been previously identified, and additional samples presented here have bolstered support for and increased the distribution of them all. The first clade (MC1), found in South Asia, includes both native wild boar and domestic pigs from India and Nepal (10, 14). The second clade (MC2), referred to elsewhere as the Pacific Clade (11), now includes six samples found in northern Vietnam, Yunnan Province, and Laos. Despite additional sampling of domestic pigs from Central China and the IBBH, the only domestic or feral pigs that belong to this clade are found in Oceania. The third clade (MC3), referred to previously as MTSEA given their mountainous and Southeast Asian distribution (14), consists of both wild and domestic samples found almost exclusively in the IBBH. The fourth clade (MC4) is also restricted to the IBBH and consists of a single wild boar from Vietnam and 13 domestic samples from two southern Chinese provinces.
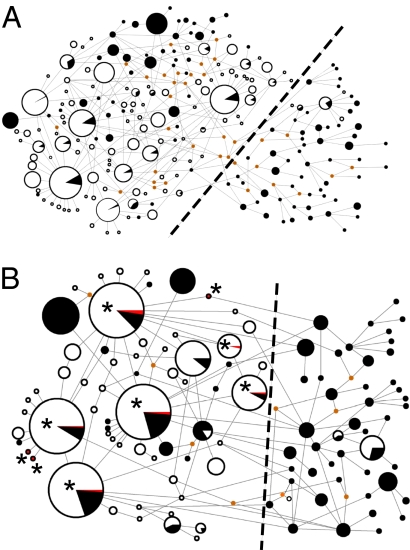
In addition to the phylogenetic tree, a network was drawn using only the modern samples (Fig. 2A and Fig. S2). Because missing sequence data cannot be incorporated into a network analysis, the alignment for the network consisted of fewer base pairs (378) to incorporate the maximum number of novel and GenBank samples. Despite this reduction, the pattern revealed in the tree and the network is consistent. The clades that consist solely of wild specimens are depicted as clusters of haplotypes on the right-hand side of the network, connected to the complex web of interconnected haplotypes on the left-hand side by inferred nodes. The sequences that make up the haplotypes on the left are the unaffiliated branches and differ from one another by a few frequently occurring mutations.
Fig. 2.
Two median-joining networks in which node sizes are proportional to haplotype frequencies depicting (A) the relationships among 1,540 modern wild and domestic samples in a 378-bp alignment and (B) the relative position of the ancient haplotypes after the archeological samples were added to a reduced 185-bp alignment. Wild, domestic, and ancient samples are shown in black, white, and red, respectively, and asterisks also mark the positions of the ancient samples. Inferred haplotypes are represented by small orange dots. The relative position of the Lanyu pig and mutations along the branches are shown in Fig. S2. The dashed black line dividing the networks correlates with the line shown in Fig. 1 demonstrating the consistent distinction between the general cluster and the wild and mixed haplotypes.
To incorporate the results of the 18 ancient samples from which we successfully generated reproducible DNA, a second network was constructed using a reduced alignment of 185 bp (Fig. 2B and Fig. S2). Two results from this second network based upon the smaller alignment are evident. First, although the truncation of the sequence length decreases the total number of haplotypes present in the network, the differentiation between the right- and left-hand sides of the network remains intact (Fig. 2B). Second, although three of the nine ancient haplotypes are novel, representing either previously inferred or new haplotypes positioned 1 bp from an existing haplotype, the six remaining haplotypes are identical to the most common haplotypes found within all modern East Asian pigs. In fact, the ancient samples derived from archaeological sites spanning 5,000 years possess five of the seven most common haplotypes shared by modern wild and domestic East Asian pigs. Despite this shared identity between many of the ancient and modern samples, the variability present within samples taken from the same site (Table S3) and the independent reproduction of the results (see Materials and Methods) suggests the sequences were generated from endogenous DNA.
Discussion
Natural Dispersal of Wild Boar.
The modern distribution of wild boar in East Asia reflects the natural dispersal of Sus scrofa out of ISEA onto the Asian continent (10). The spread of wild boar into the modern islands of Taiwan, Lanyu, Japan, and the Ryukyu chain was likely facilitated by the formation of land bridges between the East Asian mainland and these islands during periods of low sea level during the Pleistocene (33). The reformation of the East China Sea disrupted gene flow, and these populations became genetically divergent from their mainland ancestors.
Gene flow restriction can also result from a complex topography, which may explain the high degree of endemism in the IBBH (34), a region that possesses a diverse range of plants (35), animals (32), and languages (36). More specifically, Yunnan Province has previously been shown to be the only place where all seven mtDNA clades that consist of both wild and domestic chickens currently exist (37), a result that suggests not only that the significant number of geographically distinct wild boar lineages in the IBBH is not unusual, but also that this region may harbor additional, undiscovered populations of genetically distinct wild suids.
The overall pattern of wild boar distribution suggests that it is unusual for a single region to play host to more than one mitochondrially defined population (Fig. S3). The presence of two distinct wild boar populations in India is therefore unusual, as is the presence in Central China of both W13 wild boar and those from the general cluster. Although the distributions and sizes of global wild boar populations have clearly been affected by humans, the unexpectedly strong phylogeographic signal demonstrated for wild boar (10) suggests that wild boar populations have proved more resilient than other wild progenitor species at maintaining their natural ranges. As a result, it is reasonable to suppose that the process of pig domestication in different regions began with the local population of wild boar.
Pig Domestication in China.
Both genetic and archaeological evidence has demonstrated conclusively that pigs were domesticated in East Asia. The other possible instances of domestication discussed below thus far lack corroborating archaeological evidence, and we therefore refer to them as “cryptic domestication.” Despite the robust evidence in Central China, a lack of resolution has prevented any definitive conclusions to be drawn regarding the number of geographic origins and instances this process may have taken place (7, 21). The data presented in this study demonstrate that the most common modern domestic haplotypes found in Central China are also the most common Asian haplotypes found across East Asia, in Australian feral pigs, and in modern European and American breeds—the latter as a consequence of the 18th-century drive to improve European breeds by hybridizing them with imported Asian pigs (38, 39). The lack of fine geographical resolution of the mtDNA d-loop data across Central China (probably resulting from both a lack of genetic differentiation between wild boar populations, and a history of human-mediated movement) precludes any conclusions regarding multiple centers of domestication within Central China. The future use of techniques such as multiple nuclear markers and shape analyses may provide the requisite resolution to address this question.
The position of ancient DNA sequences among the most common haplotypes on the network, however, suggests that modern Chinese pigs are the direct descendants of the original populations of domestic pigs sampled along the Yellow River. This evidence does not rule out a separate domestication of pigs in other parts of Central China, but it does demonstrate a long-term continuity between early and modern domestic pigs. In addition, neither modern nor ancient pig samples in this study share any genetic affinity with a clade of wild boar (W13) that is also present across Central China (Fig. S3). The combination of a shared geographical distribution but maintenance of a strict genetic differentiation suggests not only that domestic haplotypes have not leaked into wild populations, but also that neither ancient nor modern keepers of domesticated pigs made a lasting effort to incorporate the females of separate wild populations into their domestic stock.
Cryptic Pig Domestication in South Asia.
A clade of wild and domestic pigs (MC1) made up of samples from across India and Nepal represents another regionally distinct input of wild boar into domesticated pigs (Fig. S3). At present, little is known about the history of pig domestication in India. Sus bones are a widespread but minor component of archaeological assemblages throughout India and Pakistan (40). An initial increase and subsequent rapid decrease of pig remains in successive periods at Mehrgarh in Pakistan raises the possibility that efforts were made to keep pigs during the late fourth millennium B.C., but were later abandoned (41). Because detailed morphometric evidence for separating wild and domesticated pigs in this region is not yet available (42, 43), the archaeological evidence is inconclusive. What the genetic evidence implies, however, is that modern Indian domestic pigs are derived from local wild boar. If pigs were introduced from East Asian or Near Eastern sources, the invading pigs must have mixed heavily with the indigenous wild populations.
Cryptic Pig Domestication in Southeast Asia and the East China Sea.
There is currently no indication of domestic pigs in peninsular Southeast Asia (the IBBH) until the end fifth millennium BP when they appear alongside the first evidence of sedentary agriculture (24, 44). The genetic evidence, however, demonstrates that some breeds of domestic pig share haplotypes with three clades of differentiated wild boar that are currently found only in the IBBH, suggesting that wild boar from this region have been involved in domestication (Fig. S3).
Of the three clades in this region, the Pacific Clade (MC2) was discussed in a recent publication that presented evidence for a route taken by domestic pigs from mainland Asia through ISEA and into the Near then Remote Oceania (11). The sole sequences that rooted the pathway in mainland Asia were reported from two pigs from northern Vietnam (45). The number of wild boar from the IBBH that fall into this clade has been extended by the discovery of one wild boar sample from Laos and three samples from Yunnan Province, China, all four of which possess Pacific Clade haplotypes. These samples expand the geographic range of wild boar possessing Pacific Clade signatures and add support to the hypothesis that the Pacific Clade is indigenous to peninsular Southeast Asia. This evidence supports the postulated Neolithic expansions of Austroasiatic language speakers along the major Southeast Asian rivers from Yunnan (46). The fact that no modern domestic pigs possessing Pacific Clade haplotypes have yet been found in mainland Asia is most likely a consequence of a replacement of native pigs by pigs introduced from Central China, a situation analogous to that seen in the Near East during later prehistory (9).
There have been several demographic expansions of agricultural populations into the IBBH that could have brought domestic pigs from Central China with them, including Austronesian speakers through ISEA and parts of the mainland coastal regions (47), post-Neolithic expansions of Sino-Tibetan speakers (47, 48), and Austro-Tai or Miao-Yao groups from Southern China (49). Importantly, the replacement did not extend beyond the IBBH, thus leaving intact populations of domestic Pacific Clade pigs on New Guinea and the Pacific Islands. This pattern also suggests that people carrying Pacific Clade domestic pigs left the IBBH before Central Chinese domestic pigs arrived.
Like the Pacific Clade, the MC3 (MTSEA) clade also possesses wild boar with unique signatures that are restricted to the IBBH (although a single anomalous wild boar from Taiwan also clusters in this group). Unlike the Pacific Clade, MC3 contains 66 domestic pigs scattered across the IBBH, and two pigs from Bhutan (Fig. S3). This pattern suggests another instance of the incorporation of IBBH wild boar into regionally restricted domestic pigs, although neither the time frame nor the mode of incorporation can be deduced. In addition to the two clades described herein, the discovery of four additional clades made up solely of wild boar within the IBBH (W9–W12) speaks to the complexity of suid evolution and domestication in this region.
All 14 samples (13 domestic and one wild) found in the MC4 clade are also found only in the IBBH (Fig. S3). Curiously, like the Pacific Clade, the only wild boar is found in Vietnam, whereas the domestic samples are all from either the adjacent Chinese provinces of Yunnan and Guangxi. This clade adds further weight to the suggestion that wild boar from this region were either domesticated or somehow incorporated into introduced domestic pigs.
Further east, a genetic survey of pigs originally found on the island of Lanyu off the east coast of Taiwan discovered a highly unusual haplotype (31, 50) that, despite its genetic distance, still clusters alongside other East Asian signatures. There are currently no wild boar on Lanyu, suggesting either that the wild ancestor of these pigs was endemic to Lanyu and has since been exterminated, or that Lanyu domestic pigs were derived from an as-yet-undiscovered population of wild boar whose genetic differentiation may be explained by rising and falling sea levels discussed previously. Further genetic and morphological analyses of these pigs may reveal the first instance of domestication of an island endemic.
Conclusions
The evidence presented here suggests the following evolutionary history of pigs in East Asia. Having originally evolved in ISEA, wild Sus scrofa migrated (without human assistance) across the Kra Isthmus on the Malay Peninsula into Mainland Asia. From here, they spread across the landscape and, after traveling over land bridges, onto the islands of Japan, the Ryukyu chain, Taiwan, and Lanyu where they evolved unique mitochondrial signatures. After millennia of hunting and gathering, a major biocultural transition occurred early in the Holocene during which human populations in East Asia domesticated a variety of plants and animals, including pigs. This process took place at least once in the Yellow River drainage basin where millet may have been first domesticated as early as 10,000 B.P. (17), and may have also taken place independently in the downstream Yangtze River region where rice may have been domesticated (8, 19). Two things are clear from the ancient DNA evidence presented here. First, unlike Europe, modern Chinese domestic pigs are the direct descendants of the first domestic pigs in this region. Second, despite the occurrence of a genetically distinct population of wild boar throughout modern China, this population has neither been incorporated into domestic stocks nor exterminated as a result of hunting or introgression with feral pigs.
This genetic evidence also supports separate domestication pathways (however independent) of one population in India and three wild boar populations indigenous to Peninsular Southeast Asia. Given the relative geographic proximity of the Southeast Asian clades (Fig. S3), it is possible that the domesticated haplotypes were all present in a single population of wild boar, as is the case for modern Yaks (51). Regardless, only the Pacific Clade domestic pigs were transported out of Southeast Asia (to ISEA and the Pacific) before they were replaced in their homeland by domestic pigs derived from nonindigenous populations of wild boar. The domestic pigs derived from the second and third populations of wild boar are currently restricted to Peninsular Southeast Asia, and may have played a part in the local extermination of Pacific Clade domestic pigs. It is important to point out that these populations represent exceptions, and that many genetically distinct (and still extant) populations of wild boar exist throughout East Asia, the majority of which have never been part of a domestication process. More generally, these cryptic domestication processes in India and Southeast Asia are currently based only on genetic data, although new morphological techniques are available and should be used to test different domestication scenarios (26).
The eradication of Pacific Clade domestic pigs from Southeast Asia, like the eradication of Near Eastern domestic pigs from Europe, underscores the importance of ancient DNA in temporal narratives of domestication. Although domestic plants and animals form the basis of most sedentary societies, they also instigate demographic change, cultural expansion, and the formation of large-scale trade networks, all of which can significantly alter the distribution of biological organisms associated with them, often leading to a complete turnover of populations. The close mitochondrial affiliations of intraspecies populations also renders it difficult to identify phylogeographic patterning at a regional scale, thus limiting our ability to spot independent domestication events. Still, the evidence presented here strongly suggests an intriguingly complex pattern of local domestication and regional turnover, and underscores the need for further integrated archaeological and genetic research.
Materials and Methods
Ancient and Modern Samples.
A total of 48 ancient pig samples from six archaeological sites in the lower and middle Yellow River drainage basin of northern China (Fig. 1 Inset) were subjected to strict protocols for extraction and replication (see SI Text). The bones ranged in age from 9,000 to 3,100 B.P. based upon a stratigraphic association with 14C dates of the contexts from which the bones were extracted. Domestication is a process (3), and here we use the terms wild and domestic loosely. Given the continuous nature of domestication (among other factors), a definitive status determination has not been possible, although several lines of preliminary evidence suggest that all of the ancient samples were derived from pigs that were undergoing, or had undergone significant morphological change as a result of their close relationships with humans (SI Text). Of the 48 bones, 18 yielded a 185-bp fragment of mitochondrial control region DNA amplified in two fragments. Details regarding the archaeological sites, the contexts from which the bones were recovered, and the full results of the DNA analysis can be found in Table S3.
In addition, a 698-bp fragment was amplified from 151 domestic and wild pig samples across Central China, including 66 Chinese breeds (Table S1). Sequences from these samples were combined with 1,390 GenBank entries to generate a dataset comprising a total of 1,541 individual pigs representing at least 22 Chinese provinces and 15 additional countries in East Asia and the Pacific (Table S2).
Analysis of Sequence Data.
The 151 modern sequences generated as part of this study were combined with 1,390 sequences obtained from GenBank and aligned by eye using Se-Al (http://tree.bio.ed.ac.uk/software/seal/) to generate a 662-bp alignment. Collapse1.2 (http://darwin.uvigo.es/) was then used to identify 288 unique haplotypes (117 within wild boar, 141 within domestic, 30 shared by domestic and wild boar). The 18 ancient samples from which DNA was successfully amplified were aligned alongside the modern samples. Despite the shorter fragment length relative to the modern samples, there were still nine separate haplotypes identified among the 18 ancient sequences.
Phylogenetic analysis was performed with MrBayes 3.1.2 (52) using model parameters (GTR+G+I) identified by ModelTest (53). Parameter estimates and consensus trees resulting from 10 MrBayes runs were recorded and contrasted. The posterior probabilities listed on the tree in Fig. 1 represent the lowest recorded values among all of the runs. Trees were rooted using warthogs (Phacocheorus aethiopicus).
Median-joining networks were also created using Network 4.1 (54) to further elucidate the differences among the varying haplotypes. Because the software does not allow for missing data, we used a 378-bp fragment (of the 662 bp generated) to construct a median-joining tree of 1,541 East Asian pigs (Fig. 2A and Fig. S2). To incorporate the sequence data generated from the ancient samples, a second median-joining network was created after reducing the alignment to 185 bp (Fig. 2B and Fig. S2).
Supplementary Material
Acknowledgments
We thank Nu He (Institute of Archaeology, Chinese Academy of Agricultural Science, Beijing, China), Juzhong Zhang (University and Science and Technology of China, Beijing, China), Sheng Ma (Shanxi Provincial Institute of Archaeology, Taiyuan, China), Quanfa Cai, and Yanming Fang (Henan Provincial Institute of Archaeology, Zhengzhou, China) for provision of ancient samples. We also thank Changxin Wu (College Animal Science and Technology, China Agricultural University, Beijing, China) for helping establish the aDNA Laboratory at the China Agricultural University, Hui Zhou and Dawei Cai (Research Center for Chinese Frontier Archaeology, Jilin University, Changchun, China) for replication, and Charles Higham and Thomas Cucchi for assistance with the manuscript. This work was supported by grants from the National Basic Research Program of China (Grant 2006CB102100) and the National Key Technology R&D Program of China (Grant 2006BAK21B03). G.L. was supported by an EMBO post-doctoral Fellowship. All 151 modern and 18 ancient sequences were submitted to Genbank with reference numbers FJ601390–FJ601547.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ601390– FJ601547).
This article contains supporting information online at www.pnas.org/cgi/content/full/0912264107/DCSupplemental.
References
- 1.Bellwood P. First Farmers: The Origins of Agricultural Societies. Oxford: Blackwell; 2005. [Google Scholar]
- 2.Diamond J, Bellwood P. Farmers and their languages: The first expansions. Science. 2003;300:597–603. doi: 10.1126/science.1078208. [DOI] [PubMed] [Google Scholar]
- 3.Dobney K, Larson G. Genetics and animal domestication: New windows on an elusive process. J Zool. 2006;269:261–271. [Google Scholar]
- 4.Zeder MA, Emshwiller E, Smith BD, Bradley DG. Documenting domestication: The intersection of genetics and archaeology. Trends Genet. 2006;22:139–155. doi: 10.1016/j.tig.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Barton L, et al. Agricultural origins and the isotopic identity of domestication in northern China. Proc Natl Acad Sci USA. 2009;106:5523–5528. doi: 10.1073/pnas.0809960106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J, Flad RK. Pig domestication in ancient China. Antiquity. 2002;76:724–732. [Google Scholar]
- 7.Flad RK, Yuan J, Li S. In: Late Quaternary Climate Change and Human Adaptation in Arid China. Madsen DB, Gao X, Chen FH, editors. Amsterdam: Elsevier; 2007. [Google Scholar]
- 8.Fuller DQ, Qin L, Harvey E. In: Past Human Migrations in East Asia. Matching Archaeology, Linguistics and Genetics. Sanchez-Mazas A, Blench R, Ross MD, Peiros I, Lin M, editors. London: Routledge; 2008. pp. 40–83. [Google Scholar]
- 9.Larson G, et al. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc Natl Acad Sci USA. 2007;104:15276–15281. doi: 10.1073/pnas.0703411104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson G, et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 11.Larson G, et al. Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in island southeast Asia and Oceania. Proc Natl Acad Sci USA. 2007;104:4834–4839. doi: 10.1073/pnas.0607753104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Megens HJ, et al. Biodiversity of pig breeds from China and Europe estimated from pooled DNA samples: Differences in microsatellite variation between two areas of domestication. Genet Sel Evol. 2008;40:103–128. doi: 10.1186/1297-9686-40-1-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu GS, et al. Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia. Genome Biol. 2007;8:R245. doi: 10.1186/gb-2007-8-11-r245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka K, et al. Mitochondrial diversity of native pigs in the mainland South and South-east Asian countries and its relationships between local wild boars. Anim Sci J. 2008;79:417–434. [Google Scholar]
- 15.Liu XY, Hunt HV, Jones MK. River valleys and foothills: Changing archaeological perceptions of North China's earliest farms. Antiquity. 2009;83:82–95. [Google Scholar]
- 16.Bettinger R, Barton L, Morgan C. The origins of food production in North China: A different kind of agricultural revolution. Evol Anthropol. 2010;19:9–21. [Google Scholar]
- 17.Lu HY, et al. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc Natl Acad Sci USA. 2009;106:7367–7372. doi: 10.1073/pnas.0900158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuller DQ, Qin L. Water management and labour in the origins and dispersal of Asian rice. World Archaeol. 2009;41:88–111. [Google Scholar]
- 19.Fuller DQ, et al. The domestication process and domestication rate in rice: Spikelet bases from the lower Yangtze. Science. 2009;323:1607–1610. doi: 10.1126/science.1166605. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y, Zhang J. Restudy of the pigs' bones from the Jiahu site in Wuyang County, Henen. Kaogu. 2008;1:90–96. (in Chinese) [Google Scholar]
- 21.Jing Y, Flad R, Luo YB. Meat-acquisition patterns in the Neolithic Yangzi river valley, China. Antiquity. 2008;82:351–366. [Google Scholar]
- 22.Chang KC. The Archaeology of Ancient China. New Haven, CT: Yale Univ Press; 1986. [Google Scholar]
- 23.Chi Z, Hung H. The emergence of agriculture in southern China. Antiquity. 2010;84:11–25. [Google Scholar]
- 24.Higham CFW. Early Culture of Mainland Southeast Asia. Bangkok: River Books; 2002. [Google Scholar]
- 25.Cucchi T, Fujita M, Dobney K. New insights into pig taxonomy, domestication and human dispersal in Islands South East Asia: Molar shape analysis of Sus remains from Niah caves (Sarawak) International Journal of Osteoarchaeology. 2009;19:508–530. [Google Scholar]
- 26.Cucchi T, Yuan J, Hulme-Beaman K, Dobney K. New evidence for early Neolithic pig domestication at Jiahu, Henan Province, China: Clues from tooth shape analyses using geometric morphometrics. J Archaeol Sci. 2010 in press. [Google Scholar]
- 27.Hofreiter M, et al. Lack of phylogeography in European mammals before the last glaciation. Proc Natl Acad Sci USA. 2004;101:12963–12968. doi: 10.1073/pnas.0403618101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valdiosera CE, et al. Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos) Proc Natl Acad Sci USA. 2008;105:5123–5128. doi: 10.1073/pnas.0712223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svensson E, Gotherstrom A. Temporal fluctuations of Y-chromosomal variation in Bos taurus. Biol Lett. 2008;4:752–754. doi: 10.1098/rsbl.2008.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gongora J, et al. Indo-European and Asian origins for Chilean and Pacific chickens revealed by mtDNA. Proc Natl Acad Sci USA. 2008;105:10308–10313. doi: 10.1073/pnas.0801991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu CY, et al. The type I Lanyu pig has a maternal genetic lineage distinct from Asian and European pigs. Anim Genet. 2007;38:499–505. doi: 10.1111/j.1365-2052.2007.01646.x. [DOI] [PubMed] [Google Scholar]
- 32.Mittermeier RA, et al. Hotspots Revisited: Earth's Biologically Richest and Most Endangered Terrestrial Ecoregions. Chicago: Univ of Chicago Press; 2005. [Google Scholar]
- 33.Dobson M. Patterns of distribution in Japanese land mammals. Mammal Rev. 1994;24:91–111. [Google Scholar]
- 34.Yang YM, et al. Biodiversity and biodiversity conservation in Yunnan, China. Biodivers Conserv. 2004;13:813–826. [Google Scholar]
- 35.Barthlott W, et al. Geographic patterns of vascular plant diversity at continental to global scales. Erdkunde. 2007;61:305–315. [Google Scholar]
- 36.Pelkey J. The Phula languages in synchronic and diachronic perspective. PhD thesis. Department of Linguistics. Melbourne: La Trobe University; 2008. [Google Scholar]
- 37.Liu YP, et al. Multiple maternal origins of chickens: Out of the Asian jungles. Mol Phylogenet Evol. 2006;38:12–19. doi: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Jones GF. In: Genetics of the Pig. Rothchild MF, Ruvinksy A, editors. Oxford: CAB Intl; 1998. pp. 17–50. [Google Scholar]
- 39.Giuffra E, et al. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chattopadyaya UC. In: Indian archaeology in Retrospect. II. Protohistory. Settar S, Korisettar R, editors. New Delhi: Manohar; 2002. pp. 365–422. [Google Scholar]
- 41.Meadow RH. In: South Asian Archaeology 1985. Frifelt K, Sørensen P, editors. London: Curzon Press; 1989. pp. 167–175. [Google Scholar]
- 42.Meadow R, Patel AK. In: Indian Archaeology in Retrospect. II. Protohistory. Settar S, Korisettar R, editors. New Delhi: Manohar; 2002. pp. 391–408. [Google Scholar]
- 43.Fuller DQ. Agricultural origins and frontiers in South Asia: A working synthesis. J World Prehist. 2006;20:1–86. [Google Scholar]
- 44.Higham CFW. Aspects of economy and ritual in prehistoric northeast Thailand. J Archaeol Sci. 1975;2:245–288. [Google Scholar]
- 45.Hongo H, et al. Variation in mitochondrial DNA of Vietnamese pigs: Relationships with Asian domestic pigs and Ryukyu wild boars. Zoolog Sci. 2002;19:1329–1335. doi: 10.2108/zsj.19.1329. [DOI] [PubMed] [Google Scholar]
- 46.Higham C. In: Examining the Farming/Language Dispersal Hypothesis. Bellwood P, Renfrew C, editors. Cambridge: McDonald Institute for Archaeological Research; 2003. pp. 223–232. [Google Scholar]
- 47.Pawley A. In: Examining the Farming/Language Dispersal Hypothesis. Renfrew C, Bellwood P, editors. Cambridge: McDonald Institute for Archaeological Research; 2003. pp. 251–273. [Google Scholar]
- 48.Van Driem G. In: Archaeology and Language II: Archaeological Data and Linguistic Hypotheses. Blench R, Spriggs M, editors. London: Routledge; 1998. pp. 67–102. [Google Scholar]
- 49.Blench RM. In: Perspectives on the Phylogeny of East Asian Languages. Blench RM, Sagart L, Sanchez-Mazas A, editors. London: Curzon; 2005. pp. 31–50. [Google Scholar]
- 50.Luetkemeier E, Sodhi M, Schook LB, Malhi RS. Multiple Asian pig origins revealed through genomic analyses. Mol Phylogenet Evol. 2010;54:680–686. doi: 10.1016/j.ympev.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 51.Guo SC, et al. Origin of mitochondrial DNA diversity of domestic yaks. BMC Evol Biol. 2006;6:73. doi: 10.1186/1471-2148-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 53.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 54.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.