Abstract
Meiotic recombination does not occur randomly along a chromosome, but instead tends to be concentrated in small regions, known as “recombination hotspots.” Recombination hotspots are thought to be short-lived in evolutionary time due to their self-destructive nature, as gene conversion favors recombination-suppressing alleles over recombination-promoting alleles during double-strand repair. Consistent with this expectation, hotspots in humans are highly dynamic, with little correspondence in location between humans and chimpanzees. Here, we identify recombination hotspots in two lineages of the yeast Saccharomyces paradoxus, and compare their locations to those found previously in Saccharomyces cerevisiae. Surprisingly, we find considerable overlap between the two species, despite the fact that they are at least 10 times more divergent than humans and chimpanzees. We attribute this unexpected result to the low frequency of sex and outcrossing in these yeasts, acting to reduce the population genetic effect of biased gene conversion. Traces from two other signatures of recombination, namely high mutagenicity and GC-biased gene conversion, are consistent with this interpretation. Thus, recombination hotspots are not inevitably short-lived, but rather their persistence through evolutionary time will be determined by the frequency of outcrossing events in the life cycle.
Keywords: base composition, frequency of outcrossing, hotspot degeneration, Saccharomyces, biased gene conversion
Meiotic recombination is critical for generating diversity and, hence, increasing the efficacy of selection. It also has an important structural role during meiosis, crossovers being necessary for proper chromosome segregation (1). Meiotic recombination does not occur uniformly across the genome and, at least in some species, is predominantly localized in short regions, a few kilobases long, known as recombination hotspots (reviewed in refs. 2 and 3). Recombination hotspots are thought to be evolutionarily unstable because of an inherent self-destructive dynamic. Recombination is initiated by a double-strand break in one chromosome, which is then repaired using the homologous chromosome as a template. Alleles with high recombination-initiation activity are therefore continually being replaced during their repair by the unbroken, low-activity homologs (4–7). Consistent with this expectation, comparisons of hotspot locations in humans and chimpanzees show little or no conservation of hotspot position (8–10).
The brewer's yeast Saccharomyces cerevisiae has long been a model system for studies of recombination, and experimental studies have identified multiple recombination hotspots throughout the genome (3, 11 –14). To study the conservation of recombination hotspots in yeast, we have analyzed recombination rates in the wild yeast Saccharomyces paradoxus. This species has recently emerged as an ideal model for population genomic studies because of its phylogenetic proximity to S. cerevisiae and nondomesticated status (15–17). In this article we identify recombination hotspots from population genomic analyses of an alignment of nearly complete chromosome III sequences from 20 strains of S. paradoxus, 12 from Europe and 8 from Far East Asia (18). The alignment has relatively few gaps and, hence, is ideal for analyzing recombination. The European and Far East lineages of S. paradoxus are phylogenetically independent (i.e., all isolates in one population are more closely related to each other than to any isolate from the other population), with about 1% sequence divergence (approximately the same as between humans and chimpanzees), and they are about 13% divergent from S. cerevisiae (19).
Results and Discussion
Identification and Distribution of Recombination Hotspots Along the Chromosome.
The alignment is 287 kb long and there are 464 nonsingleton SNPs in the European population and 232 in the Far East Asian, giving an average density of one SNP every 0.6 kb in Europe and 1.2 kb in the Far East (Table S1). For each population no more than two alternative nucleotides were found at each site. We estimated the population recombination parameter ρ between neighboring pairs of SNPs using the program rhomap (20) (Fig. 1A). In an idealized population, ρ is equal to 4Ner(1−F)M, where Ne is the effective population size, r the rate of recombination between the two SNPs, F the inbreeding coefficient, and M the frequency of sex (18, 21). Both populations show heterogeneity in the rate of recombination along the chromosome, particularly the European population, which also shows higher rates on average. This difference is not simply because of the smaller sample size in the Far East population, as it persists when the European population is reduced to eight strains (Fig. S1).
Fig. 1.
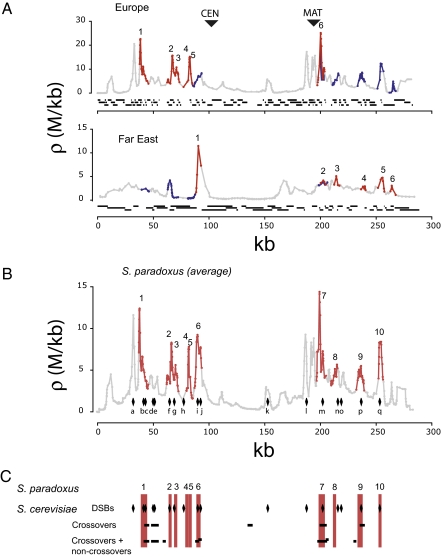
Distribution of recombination on S. paradoxus chromosome III. (A) The population-recombination parameter ρ, calculated between consecutive pairs of SNPs along the chromosome, is shown for the European and Far East populations (the first and last genes in the alignment are VBA3 and HMRA1; SNPs are shown as dots along the lines). The hotspot regions identified in each population are highlighted in red, and the corresponding regions in the other population in blue (for visual clarity, these extend 2 kb on either side). Hotspot numbers correspond to those used in Table 1. Positions of the centromere (CEN) and mating type locus (MAT) are indicated by arrows. The distribution of haplotype blocks, representing regions with no evidence of recombination, is also shown (black lines) (see Methods). Haplotype blocks are longer in the Far East population, consistent with the finding of lower recombination rate. (B) The average ρ for the two populations of S. paradoxus is plotted and hotspots from the combined analysis shown in red. Diamonds indicate the position of DSB hotspots in S. cerevisiae (11). (C) Comparison of hotpsot locations. Vertical red bars show the position of hotspots in S. paradoxus. Diamonds again show the position of DSB hotspots in S. cerevisiae (11) and horizontal lines show the hotspots identified by Mancera et al. (12) in their “crossover” and “total” analyses.
To assess our population-genomic estimates of recombination rates, we tested whether two well-supported experimental results from S. cerevisiae are replicated in our dataset. In S. cerevisiae, rates of recombination are relatively low between the centromere and mating-type locus, and also higher in intergenic regions containing at least one promoter than in those with none (22, 23). We confirm both these results in S. paradoxus. Recombination in the ∼100-kb region between the centromere (CEN) and the mating type (MAT) locus is about half that for the rest of the chromosome (ρ = 1.9 vs. 4.5 Morgans/kb in Europe and 0.9 vs. 2.0 in the Far East). In addition, intergenic regions that are 5′ to at least one of the flanking genes have higher ρ than those that are 5′ to none (P = 0.004 and P = 0.07 for Europe and Far East) (Table S2). This correspondence of results gives us confidence that our dataset is sufficient to detect major features of recombination.
To identify recombination hotspots, we tested for statistically significant increases in ρ compared to flanking regions, using the program sequenceLDhot (24). For each 2-kb window (with a 1-kb offset), the program tests the null hypothesis that ρ in the window is equal to ρ in the flanking 50-kb region (centered on that window). Six recombination hotspots were found in each population (Fig. 1A and Table 1). They are 2 to 5 kb in length, similar to the 3 kb average found in S. cerevisiae chromosome III and the 1 to 2 kb length of human hotspots (3, 12, 25). Exactly the same hotspots are found if each 2-kb window along the chromosome is tested against the flanking 6-kb region. We further evaluated the significance of these hotspots by calculating the number of hotspots expected by chance in chromosomes with constant recombination along their length. Twenty simulated alignments were obtained by evolving chromosomes of the same length and level of polymorphism as the original dataset, using the program ms (26). Only an average of about one such “false” hotspot region was found per simulated alignment (1.2 and 1.4 for Europe and Far East, respectively). All hotspots in Europe have likelihood ratios greater than 10, and in the simulated alignments only 0.2 hotspots per alignment had likelihood ratios at least that high.
Table 1.
Physical locations and parameter estimates for recombination hotspots on chromosome III of S. paradoxus
| Nucleotide diversity |
||||||
| Location* | Length (kb) | ρ (Morgans/kb)† (LR, ρmax) | θπ (×1,000) | θs (×1,000) | Divergence (×1,000)‡ | %GC |
| Europe | ||||||
| 1. MGR1–PDI1–GLK1–GID7 | 5 | 7.3 (20.5, 11.2) | 1.1 | 1.1 | 9.1 | 46 |
| 2. FUS1–HBN1–FRM2 | 2 | 5.8 (11.0, 9.1) | 0.3 | 0.5 | 4.5 | 39 |
| 3. AGP1–KCC4 | 3 | 8.2 (12.8, 10.8) | 1.1 | 1.3 | 6.0 | 44 |
| 4. NFS1–DCC1 | 2 | 4.5 (10.9, 5.4) | 1.2 | 1.3 | 5.0 | 43 |
| 5. BUD3 | 2 | 8.6 (22.1, 15.3) | 1.1 | 1.0 | 2.0 | 41 |
| 6. YCR045C–IMG1–BUD23–ARE1 | 5 | 10.1 (22.1, 25.2) | 1.9 | 1.7 | 3.4 | 47 |
| Hotspot average (SE) | 7.4 (0.82) | 1.1 (0.21) | 1.2 (0.16) | 5.0 (1.0) | 43 (1.2) | |
| Chromosome average (SE)§ | 3.8 (0.38) | 1.2 (0.09) | 1.2 (0.08) | 5.2 (0.30) | 39 (0.4) | |
| P¶ | 0.89 | 0.90 | 0.78 | 0.004 | ||
| Far East | ||||||
| 1. GBP2–SGF29–ILV6 | 3 | 8.9 (6.2, 11.2) | 0.7 | 0.8 | 8.0 | 44 |
| 2. IMG1–BUD23–ARE1 | 2 | 3.8 (6.6, 4.0) | 1.1 | 1.2 | 3.5 | 47 |
| 3. PWP2–YIH1 | 2 | 3.8 (7.2, 4.9) | 0.9 | 1.0 | 4.0 | 47 |
| 4. SSK22–SOL2 | 3 | 2.4 (9.8, 2.7) | 0.7 | 0.8 | 4.4 | 43 |
| 5. TUP1–CSM1–LUG1–ABP1 | 3 | 3.9 (8.3, 4.6) | 0.8 | 0.65 | 2.0 | 40 |
| 6. YCR90C–KIN82 | 2 | 1.8 (5.6, 3.0) | 1.1 | 1.0 | 9.0 | 37 |
| Hotspot average (SE) | 4.1 (1.02) | 0.9 (0.08) | 0.9 (0.08) | 5.2 (1.12) | 43 (1.6) | |
| Chromosome average (SE)§ | 1.7 (0.18) | 0.9 (0.07) | 0.9 (0.07) | 5.3 (0.29) | 39 (0.4) | |
| P¶ | 0.74 | 0.59 | 0.69 | 0.02 | ||
*Genes overlapping each hotspot region are given, with “ – “ indicating intergenes. Numbers correspond to those in Fig. 1A.
†Average ρ across the hotspot estimated using rhomap (likelihood ratio from sequenceLDhot, maximum ρ in the hotspot).
‡Divergence of the composite European or Far East sequence from the common ancestor, calculated using Saccharomyces cariocanus as an outgroup (16).
§Chromosome averages and SEs calculated from 56 nonoverlapping 5-kb windows.
¶P values are for differences between hotspot and chromosomal averages; Wilcoxon rank-sum tests.
Detecting statistically significant recombination hotspots from population genomic data are a challenging problem, and even the best algorithms can have low power (20, 24). This low power probably accounts in part for the fact that some peaks in the rhomap estimates are not identified as statistically significant by sequenceLDhot (Fig. 1A). Nevertheless, one region is identified as a hotspot in both populations independently (the IMG1-BUD23-ARE1 hotspot ∼9 kb to the right of MAT). Moreover, it is apparent from Fig. 1A that hotspots in one population correspond to local peaks of recombination in the other population, even if these are not identified as statistically significant by sequenceLDhot. For the 10 hotspots that are statistically significant in only one population, the corresponding regions in the other population have higher ρ than their respective 6-kb flanking regions (paired randomization test, n = 10, P = 0.02). Therefore, to maximize our power to detect hotpots in S. paradoxus, we performed a combined analysis of the two populations. Ten of the 11 hotspots found in the separate analyses of the two populations are also significant in the combined analysis, and no new hotspots were identified (Fig. 1B) (the expected false-positive rate in the combined data is 0.9 hotspots per alignment).
Conservation of Recombination Hotspots.
To test whether recombination hotspots are conserved between S. paradoxus and S. cerevisiae, we compared our hotspots to those identified in two recent experimental studies of S. cerevisiae. The first study is an analysis of double-strand breaks (DSBs) formed during meiosis, in which 17 hotspots were identified on chromosome III that had DSB rates more than 8-fold greater than background (11). Eight of these are in regions homologous to the hotspots we identified in the combined S. paradoxus dataset (Fig. 1 B and C). As our hotspots occupy only 11.2% of the chromosome, the probability of such overlap under the null hypothesis of random placement is (from the binomial distribution) P = 0.00024. In addition, five more of the hotspots identified by Buhler et al. (11) correspond to local peaks in recombination in S. paradoxus that did not reach statistical significance in our analyses (Fig. 1B, hotspots a, j, k, l, and n).
The distribution of meiotic DSBs along a chromosome may not be identical to the distribution of crossovers, as breaks can be repaired without crossing over, for example, using the sister chromatid as a template for repair, or using the homologous chromosome without crossing over (27). Therefore we have also compared our data to the results of an analysis of 51 meioses in S. cerevisiae that identified five crossover hotspots on chromosome III (12). Three of these hotspots overlap hotspots in S. paradoxus (i.e., the middle of the smaller region is contained within the larger region) (Fig. 1C). To test if this much overlap would be expected by chance, we randomized hotspot locations separately in each of the two species, and for each randomization recorded the number of overlaps. In only 2% of randomizations were three or more overlaps observed. Mancera et al. (12) also identified five additional hotspots when combining both crossover and noncrossover data, and two of these overlap with another S. paradoxus hotspot (Fig. 1C). Overall, 4 of the 10 hotspots found in S. paradoxus overlap hotspots in S. cerevisiae, and 5 of the 10 hotspots in S. cerevisiae overlap those in S. paradoxus.
All three studies identify the same region as the hottest hotspot, corresponding to the well-known ARE1 (YCR048R) hotspot region (23, 28, 29) (hotspot 7 in Fig. 1B), suggesting there may be similarities between species in both location and intensity of recombination hotspots. These similarities are found despite the fact that completely different methodologies were used in the two species (population genomic vs. experimental). For those hotspots found in only one of the two species, it is not clear if this is because of real differences in the location of hotspots or to low power in the analyses.
Thus, the distribution of hotspots in S. cerevisiae and S. paradoxus appears to be much more similar than that between humans and chimpanzees, despite the fact that the yeasts are more than 10-times more divergent at the sequence level. These similarities between yeast species are most parsimoniously explained by conservation from the common ancestor, although we cannot formally exclude the possibility of recurrent hotspot evolution at the same sites because of limited availability of alternative sites for hotspots in a smaller genome. This relatively high conservation presumably reflects a slower rate of hotspot loss by gene conversion. We attribute this difference to the low frequency of sex and outcrossing in yeast. At the population level, the change in gene frequencies due to biased gene conversion, and hence the rate of deterioration of a particular hotspot, will be proportional to the frequency of sex and the level of heterozygosity (5). Previous work has indicated that S. paradoxus in nature goes through meiosis only once every 1,000 generations, and only 1% of matings are outcrossed (15, 18). The effect of gene conversion will therefore be reduced by a factor of 105 relative to that in an otherwise comparable obligately outcrossed species. Hotspots may also be maintained if the hotspot sequence is functionally relevant for a reason other than recombination, and it is likely that a smaller fraction of the yeast genome is selectively neutral than in the human genome [upper bounds of ∼35% and ∼88%, respectively (18, 30)].
The low frequency of sex and outcrossing in natural populations means that features of recombination that are observed in the laboratory (e.g., a conversion bias in favor of recombination-suppressing alleles) may have relatively small evolutionary effects. We now analyze two other laboratory-observed features of recombination, and estimate the magnitude of their effects on yeast genome evolution.
Recombination and Mutation Rate.
Recombinational repair of DSBs in mitotic cells is associated with a 100-fold increase in the mutation rate (31). Assuming that meiotic recombination shows an equivalent increase in mutation rate—for which there is some evidence (32)—and if sex is common, one might expect increased sequence divergence at recombination hotspots. However, S. paradoxus hotspots are neither more diverse nor more divergent than the chromosome average (Table 1). In addition, there is no correlation between rates of recombination and either levels of polymorphism or rates of divergence across 5-kb segments of the chromosome (Fig. S2). The lack of positive association between recombination rates and divergence (also reported by ref. 32) is fully consistent with recombination being rare, as the mutational effect of recombination will be swamped by the mutations occurring in the intervening asexual generations. For example, in S. cerevisiae the hottest hotspot recombines in about 25% of meioses (12). If sex occurs only once every 1,000 generations, then its average mutation rate over the whole life cycle is only 100 × 0.25 × 1/1,000 = 2.5% higher than a region that never recombines, too small a difference to be detected. Again, if hotspots tend to be in more functionally constrained regions of the genome, this could also contribute to the lack of association with increased divergence.
Recombination and Base Composition.
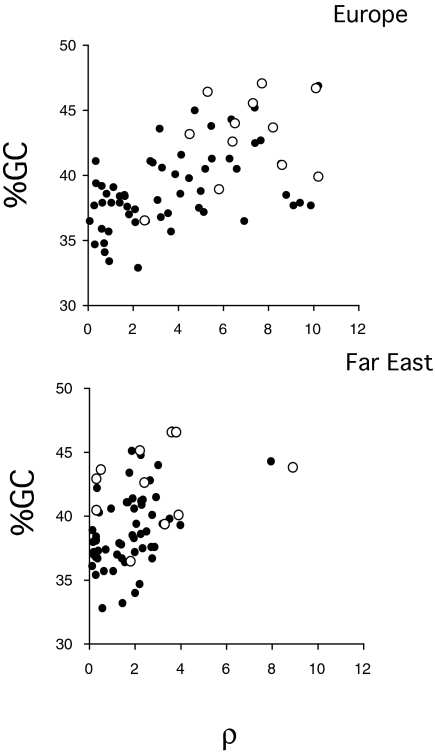
Experiments have shown that biased gene conversion in S. cerevisiae not only favors recombination-suppressing alleles, but also, independently, G and C nucleotides over A and T (12, 33). This bias occurs as a result of the mismatch-repair system acting on heteroduplex DNA formed during meiosis converting AT nucleotides into GC nucleotides. This repair bias may in turn have evolved to counteract the AT-bias in mutation (34). Recombination hotspots in S. cerevisiae have higher GC content than the rest of the genome (23), a result also found in S. paradoxus (43 vs. 39%, for both Europe and Far East, P = 0.004 and 0.02, respectively) (Table 1), and there is a highly significant positive correlation between ρ and GC content across nonoverlapping 5-kb segments of the genome (n = 56 windows; Kendall's τ: 0.30, P = 0.001, in both populations) (Fig. 2). Correlations between recombination rate and GC content can be explained by biased gene conversion in favor of Gs and Cs, or by high GC content promoting recombination (35). Similar correlations have previously been found in humans (36). However, if we consider only the substitutions that have occurred since the common ancestor of the European and Far East lineages, and are therefore recent, hotspots show a pronounced bias in the opposite direction, toward increased AT content (Table 2). The absolute number of AT to GC changes in hotspots is about 40% lower than the number of GC to AT changes. By contrast, the numbers of changes in the nonhot regions are about equal, indicating that GC content is at equilibrium.
Fig. 2.
Correlation between GC content and the population recombination parameter ρ (Morgans/kb) for Europe and Far East; points are values measured in nonoverlapping 5-kb windows. The correlations are significant in both populations (Kendall's τ: 0.30, P = 0.001 for both Europe and Far East). Open points give the values for the 11 hotspot regions in the two populations.
Table 2.
Rates of nucleotide substitution in ancestral GC or AT sites during the differentiation of European or Far East sequences from their common ancestor
| P§ | P¶ | |||||
| Ancestral site | No. Sites | Change I* | Change II† | u/v‡ | GCtoAT = ATtoGC | (u/v)H=(u/v)NH |
| Europe | ||||||
| Whole chromosome | ||||||
| GC | 109,040 | 613 | 79 | 1.53 | 0.67 | |
| AT | 171,193 | 629 | 98 | |||
| Hotspot regions | ||||||
| GC | 8,345 | 58 | 8 | 2.15 | 0.016 | <0.001 |
| AT | 10,537 | 34 | 3 | |||
| Nonhot regions | ||||||
| GC | 100,695 | 555 | 71 | 1.49 | 0.25 | |
| AT | 160,656 | 595 | 95 | |||
| Far East | ||||||
| Whole chromosome | ||||||
| GC | 109,040 | 652 | 70 | 1.64 | 0.47 | |
| AT | 171,193 | 625 | 93 | |||
| Hotspot regions | ||||||
| GC | 6,353 | 45 | 4 | 2.42 | 0.023 | <0.001 |
| AT | 8,535 | 25 | 2 | |||
| Nonhot regions | ||||||
| GC | 102,687 | 607 | 66 | 1.6 | 0.86 | |
| AT | 162,658 | 600 | 91 |
*Total number of GC to AT or AT to GC changes.
†Total number of GC to CG or AT to TA changes.
‡u = (number of GC to AT changes)/(total ancestral GC sites) and v = (number of AT to GC changes)/(total ancestral AT sites); all ratios are significantly different from 1, P < 0.001.
§Probability that absolute numbers of GC to AT changes are equal to numbers of AT to GC changes (from Change I column); binomial sign tests.
¶Probability that u/v in hotspots is equal to u/v in nonhot regions.
These results are consistent with biased gene conversion having had a significant effect some time in the evolutionary past, allowing recombination hotspots to build up a high GC content, and then, more recently, a reduction in the force of biased gene conversion, such as would be expected if the frequency of sex and outcrossing had subsequently decreased in these lineages. The reduced sex and outcrossing would have relaxed the GC pressure due to gene conversion, resulting in a rebound toward higher AT content. Nonhot regions, which did not have an elevated GC content due to biased gene conversion, would not be affected by the change in mating system, and not show a rebound. Hotspots show a higher rate of GC to AT changes (relative to AT to GC changes) than nonhot regions (Table 2), as expected when only some nucleotide sites are free to change and others are functionally constrained. The correlation between recombination and GC content is then a holdover from an ancestral condition in which outcrossing was more frequent than it is now.
Population genetic calculations confirm that biased gene conversion for GC nucleotides should currently be a weak force. In S. cerevisiae, when a site that is heterozygous for GC vs. AT is involved in a recombination event, on average, the GC nucleotide is transmitted to 50.6% of the spores, compared with the Mendelian 50% (12). If a hotspot recombines in 25% of meioses, and the frequency of outcrossing is 10−5 per cell division, then the selection coefficient in favor of GC nucleotides will be s = (2(0.506)−1)(0.25)(10−5) = 3 × 10−8 (s = 0 for unbiased transmission) (5). This value is less than the reciprocal of the effective population size of S. paradoxus (Ne∼107) (18), and therefore biased gene conversion is not expected to significantly affect GC composition.
Conclusions
Our analyses demonstrate that multiple recombination hotspots along the third chromosome are conserved between S. paradoxus and S. cerevisiae, despite the considerable divergence of the DNA sequences involved. The finding of hotspot conservation in yeast contradicts the expectation of hotspot self-destruction because of biased gene conversion. It is also unexpected in view of the transient, highly dynamic hotspots observed in humans (9, 10, 37). This conservation indicates a relatively slow rate of hotspot loss in S. paradoxus compared with overall sequence divergence, which we attribute to the rarity of outcrossed sexual events in the life cycle of yeast, perhaps combined with the genome being under greater functional constraint. A low frequency of sex in natural populations can also explain the absence of a correlation between hotspots and either divergence or diversity, as the mutagenic effect of recombination is swamped by the mutations that occur in the intervening asexual generations. The analysis of recombination and base composition is consistent with low frequencies of sex and outcrossing in recent times, but also points to an ancient ancestor that was more sexual, in which a correlation developed between hotspots and GC content that is currently disappearing.
Thus, genome sequences may contain imprints of the mating system as it existed millions of years ago. If our interpretation is correct, then the current locations of recombination hotspots trace back to this more sexual ancestor: perhaps, we speculate, to an ancestor that existed before the evolution of mating-type switching allowed increased rates of inbreeding (38). Surveys of recombination rates and base composition in more divergent yeasts could test this prediction.
Methods
Rates of Recombination and Hotspot Identification.
The population recombination rate ρ was calculated for consecutive pairs of SNPs using a coalescent-based method implemented in the program rhomap of the package LDhat [ver 2.1 (20)]. We excluded singleton SNPs (on the grounds that they are uninformative in estimating rates of recombination), and SNPs for which any strain had missing data. We ran rhomap with 1,100,000 iterations, the first 10% of which were discarded as burn-in; after the burn-in, samples were taken every 100th iteration. Recombination hotspots were identified using sequenceLDhot (24); regions with a likelihood ratio >5.41 were considered significant (P < 0.01), as has been used previously for humans (39). Log-likelihoods from the two populations were summed for the combined analysis and compared to a critical value of 7.29 (P < 0.01). The program PHASE v2.1 was used to estimate background recombination rates (40), with settings ×10 and −k999, as recommended in the manual and accompanying documentation.
Haplotype Blocks.
In addition to the analysis of rates of recombination, we also searched for regions along the chromosome with no evidence of recombination (haplotype blocks), using custom Perl scripts. For this search, we used a modification of the four gametes test: given a pair of biallelic SNPs (e.g., two A/C polymorphisms), a recombination event would have had to have taken place if all four possible combinations (e.g., AA, AC, CA, CC) of alleles at the two sites are observed among the different strains in the population. Haplotype blocks are regions with consecutive polymorphic sites where only a maximum of three of the above combinations are observed for all pairwise combinations of these sites; a haplotype block becomes interrupted when the next polymorphic site (in either direction) is incompatible (i.e., contains all four combinations of alleles) with any of the sites already included in that block. Note that haplotype blocks defined in this way can overlap. There are 110 such haplotype blocks in the European population, averaging 2.2 kb in length, with 5.4 SNPs per block (Fig. 1 and Table S1), the longest block being 9.6 kb. In the Far East population, we found fewer and longer blocks, implying less recombination.
Nucleotide Diversity and Base Composition.
Nucleotide diversity (θπ and θs) was calculated using Variscan (41), including only sites that have data for at least four strains in each population (i.e., option numnuc = 4). To analyze divergence or GC content, a composite sequence for each of the two populations was constructed by choosing a random valid nucleotide (A, T, C, or G) from all strains within each population and for every site in the alignment. To determine whether changes occurred in the European or Far East lineages, we used Saccharomyces cariocanus as an outgroup (18).
Correlates of Recombination.
To test for correlates of recombination along the chromosome, the alignment was divided into nonoverlapping 5-kb windows. Conventional P values from analyses using data from consecutive windows can be misleadingly low if both variables are autocorrelated (i.e., neighboring windows are more similar than distant windows) (42, 43). We calculated the first order autocorrelation coefficient (r) for each of our variables, using the acf function of R (Table S3) (www.r-project.org). There is significant autocorrelation among the 5-kb windows in recombination rates, but not in any of the other variables.
Supplementary Material
Acknowledgments
We thank Gilean McVean and Paul Fearnhead for help with programs analyzing recombination, Richard Bourgon and Eugenio Mancera for calculating the experimental GC bias in transmission during meiosis, and Tim Barraclough for commenting on a previous draft. This work was supported by the Biotechnology and Biological Sciences Research Council and the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908774107/DCSupplemental.
References
- 1.Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: The molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 2.Buard J, de Massy B. Playing hide and seek with mammalian meiotic crossover hotspots. Trends Genet. 2007;23:301–309. doi: 10.1016/j.tig.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 4.Boulton A, Myers RS, Redfield RJ. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci USA. 1997;94:8058–8063. doi: 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Boston, MA: Harvard University Press; 2006. [Google Scholar]
- 6.Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 7.Pineda-Krch M, Redfield RJ. Persistence and loss of meiotic recombination hotspots. Genetics. 2005;169:2319–2333. doi: 10.1534/genetics.104.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 9.Ptak SE, et al. Fine-scale recombination patterns differ between chimpanzees and humans. Nat Genet. 2005;37:429–434. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- 10.Winckler W, et al. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- 11.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:2797–2806. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultes NP, Szostak JW. A poly (dAdT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White MA, Wierdl M, Detloff P, Petes TD. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc Natl Acad Sci USA. 1991;88:9755–9759. doi: 10.1073/pnas.88.21.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson LJ, et al. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43–52. doi: 10.1534/genetics.166.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liti G, Louis EJ. Yeast evolution and comparative genomics. Annu Rev Microbiol. 2005;59:135–153. doi: 10.1146/annurev.micro.59.030804.121400. [DOI] [PubMed] [Google Scholar]
- 17.Replansky T, Koufopanou V, Greig D, Bell G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 2008;23:494–501. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc Natl Acad Sci USA. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 20.Auton A, McVean G. Recombination rate estimation in the presence of hotspots. Genome Res. 2007;17:1219–1227. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutter AD, Baird AC, Charlesworth D. High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics. 2006;174:901–913. doi: 10.1534/genetics.106.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerton JL, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fearnhead P. SequenceLDhot: detecting recombination hotspots. Bioinformatics. 2006;22:3061–3066. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 25.Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 26.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 27.Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-strand DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 28.Borde V, et al. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 29.Nag DK, White MA, Petes TD. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 30.Parker SCJ, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noor MA. Mutagenesis from meiotic recombination is not a primary driver of sequence divergence between Saccharomyces species. Mol Biol Evol. 2008;25:2439–2444. doi: 10.1093/molbev/msn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- 34.Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petes D, Merker JD. Context dependence of meiotic recombination hotspots in yeast: the relationship between recombination activity of a reporter construct and base composition. Genetics. 2002;162:2049–2052. doi: 10.1093/genetics/162.4.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marais G. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 2003;19:330–338. doi: 10.1016/S0168-9525(03)00116-1. [DOI] [PubMed] [Google Scholar]
- 37.Jeffreys AJ, Neumann R. The rise and fall of a human recombination hot spot. Nat Genet. 2009;41:625–629. doi: 10.1038/ng.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Jeffreys AJ, Neumann R, Panayi M, Myers S, Donnelly P. Human recombination hot spots hidden in regions of strong marker association. Nat Genet. 2005;37:601–606. doi: 10.1038/ng1565. [DOI] [PubMed] [Google Scholar]
- 40.Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J. VariScan: Analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics. 2005;21:2791–2793. doi: 10.1093/bioinformatics/bti403. [DOI] [PubMed] [Google Scholar]
- 42.Chatfield C. The Analysis of Time Series: An Introduction. London: Chapman & Hall/CRC; 2003. [Google Scholar]
- 43.Hahn MW. Accurate inference and estimation in population genomics. Mol Biol Evol. 2006;23:911–918. doi: 10.1093/molbev/msj094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.