Abstract
The upsurge in prevalence of obesity has spawned an epidemic of nonalcoholic fatty liver disease (NAFLD). Previously, we identified a sequence variant (I148M) in patatin-like phospholipase domain-containing protein 3 (PNPLA3) that confers susceptibility to both hepatic triglyceride (TG) deposition and liver injury. To glean insights into the biological role of PNPLA3, we examined the molecular mechanisms by which nutrient status controls hepatic expression of PNPLA3. PNPLA3 mRNA levels, which were low in fasting animals, increased ∼90-fold with carbohydrate feeding. The increase was mimicked by treatment with a liver X receptor (LXR) agonist and required the transcription factor SREBP-1c. The site of SREBP-1c binding was mapped to intron 1 of Pnpla3 using chromatin immunoprecipitation and electrophoretic mobility shift assays. SREBP-1c also promotes fatty acid synthesis by activating several genes encoding enzymes in the biosynthetic pathway. Addition of fatty acids (C16:0, C18:1, and C18:2) to the medium of cultured hepatocytes (HuH-7) increased PNPLA3 protein mass without altering mRNA levels. The posttranslational increase in PNPLA3 levels persisted after blocking TG synthesis with triascin C. Oleate (400 μM) treatment prolonged the half-life of PNPLA3 from 2.4 to 6.7 h. These findings are consistent with nutritional control of PNPLA3 being effected by a feed-forward loop; SREBP-1c promotes accumulation of PNPLA3 directly by activating Pnpla3 transcription and indirectly by inhibiting PNPLA3 degradation through the stimulation of fatty acid synthesis.
Keywords: adiponutrin, SREBP-1c, triglycerides, hepatic steatosis
Nonalcoholic fatty liver disease (NAFLD) is a burgeoning medical problem that affects as much as one-third of the US population (1, 2). The disorder encompasses a spectrum of conditions associated with the accumulation of excess triglyceride (TG) in lipid droplets in hepatoctyes (hepatic steatosis). A subset of individuals with hepatic steatosis develop hepatic inflammation (steatohepatitis), which may progress to cirrhosis (3). Despite the high prevalence of NAFLD, the pathogenesis of the disease is poorly understood.
Previously, we showed that a nonsynonymous sequence polymorphism (I148M) in a member of the patatin-like phospholipase domain-containing protein (PNPLA) family (4), PNPLA3, is associated with increased hepatic TG content and evidence of hepatic injury (5). The PNPLA3-I148M variant is most common in individuals of Hispanic ancestry, the group with the highest prevalence of hepatic steatosis (5, 6) and is associated with increased circulating levels of liver enzymes (7–10), biopsy-proven hepatosteatosis (9), and alcohol-related cirrhosis (11, 12).
PNPLA3, also referred to as adiponutrin, encodes a 481-amino-acid protein that contains a highly conserved patatin-like domain at the N terminus. The progenitor of the PNPLA family, patatin, comprises ∼40% of the dry weight of potato tubers (13) and has nonspecific lipid acyl hydrolase activity (14) mediated by a serine-aspartate dyad (15). PNPLA3 is expressed at the highest levels in adipose tissue of mice (16). The enzyme most closely resembles PNPLA2 (also called ATGL), the major hormone-sensitive lipase of adipose tissue (17, 18), and both PNPLA2 and PNPLA3 have TG hydrolase activity in vitro (19–21). Whereas genetic manipulation of PNPLA2 activity strongly influences TG levels in cultured cells and in mice (19), knockdown (22) or overexpression (16, 21) of PNPLA3 has little impact on cellular TG content (22).
Interestingly, the regulation of PNPLA3 is opposite to that of PNPLA2. PNPLA2 is up-regulated by fasting and suppressed by feeding, whereas PNPLA3 mRNA levels are low in the adipose tissue of fasted mice and increase dramatically in fat and liver with refeeding (4, 16). The increase in PNPLA3 levels upon refeeding appears to be mediated at least in part by insulin: PNPLA3 mRNA levels in the adipose tissue of mice are markedly reduced by streptozotocin administration and can be restored by insulin treatment (22). Moreover, PNPLA3 mRNA levels are elevated in obese, insulin-resistant animals (ob/ob mice and fa/fa rats) (4, 16). Here we sought to define the mechanisms by which nutrient status controls PNPLA3 expression. Our data indicate that nutritional control of PNPLA3 is achieved by a feed-forward loop that coordinates up-regulation of transcription and inhibition of protein degradation.
Results
Tissue Distribution of PNPLA3.
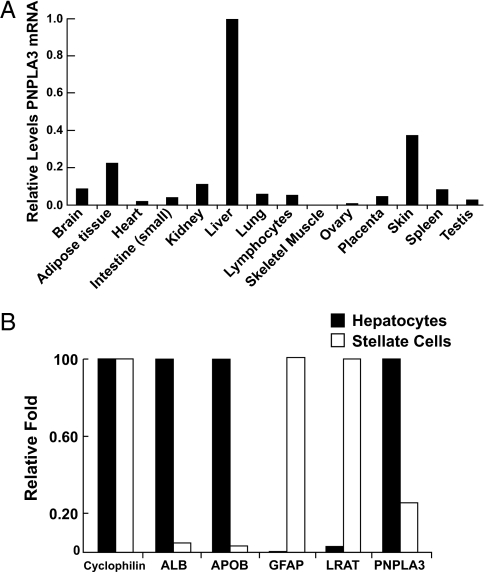
Real-time PCR of cDNA from human tissues indicated that PNPLA3 expression was highest in the liver, followed by skin and adipose tissue (Fig. 1A). This is in contrast to the relative expression levels in mice, where expression is significantly greater in adipose tissue than in liver (12). Next we compared PNPLA3 mRNA levels in freshly isolated mouse hepatocytes and stellate cells, the primary storage site for retinyl esters (Fig. 1B). The level of PNPLA3 mRNA in stellate cells was only 23% of that found in hepatocytes (Fig. 1B). Hoekstra et al. (12) recently reported that PNPLA3 is not expressed in hepatic endothelial cells or Kupffer cells of mice. Taken together, these data indicate that hepatic PNPLA3 is expressed predominantly in hepatocytes in the liver (Fig. 1B).
Fig. 1.
Expression of PNPLA3 in human tissues (A) and mouse liver (B). (A) Relative levels of PNPLA3 and cyclophilin mRNA were determined by quantitative real-time PCR using cDNA from 48 human tissues (Origene). The cDNAs were standardized using cyclophilin as a calibrator. Each bar represents the mean of triplicate measurements expressed as a fraction of the Ct value obtained from liver, which was set to 1. (B) The relative levels of mRNA from genes expressed predominantly in hepatocytes [albumin (ALB) apolipoprotein B (APOB)] and stellate cells [glial fibrillary acidic protein (GFAP), lecithin retinol acyltransferase (LRAT)]. The hepatocytes and stellate cells were fractionated from mouse liver and mRNA levels were quantitated using quantitative real-time PCR as described in Materials and Methods.
Transcriptional Regulation of Expression of PNPLA3 by Fasting and Refeeding Requires LXR and SREBP.
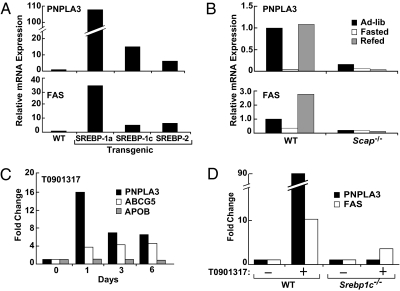
Transcriptional control of many genes in lipid metabolism is orchestrated by the sterol regulatory element binding protein (SREBP) pathway (23). To determine whether PNPLA3 is regulated by SREBPs, we measured hepatic PNPLA3 mRNA levels in mice overexpressing similar levels of the active forms of the three SREBPs: SREBP-1a, SREBP-1c, and SREBP-2 (24). PNPLA3 mRNA levels were increased in all three strains (Fig. 2A). The increase was greatest in the SREBP-1a transgenic mice (>100-fold), followed by the SREBP-1c (12-fold) and SREBP-2 (4-fold) animals. The pattern of expression was similar to that of fatty acid synthase (FAS) (Fig. 2A). As noted previously in adipose tissue (4), PNPLA3 mRNA expression was dramatically reduced with fasting and increased 90-fold with carbohydrate refeeding (Fig. 2B). Unlike other SREBP-1c target genes, PNPLA3 mRNA levels did not show the typical overshoot response with refeeding (23). Whereas FAS was more highly expressed in the livers of carbohydrate refed animals than in livers of mice fed chow diets ad libitum, levels of PNPLA3 mRNA after refeeding were consistently similar to those of mice fed ad libitum.
Fig. 2.
Regulation of PNPAL3 expression by SREBPs (A), fasting and refeeding (B), and LXR (C and D). (A) Relative mRNA levels of PNPLA3 (Upper) and fatty acid synthase (FAS) (Lower) in livers of wild-type (WT) mice and mice expressing SREBP-1a, SREBP-1c, or SREBP-2 transgenes (39, 44–46). Mice were fed a high-protein diet for 2 weeks to induce transgene expression and then fasted for 4 h before killing. Equal aliquots of total RNA from five mice in each group were pooled and mRNA levels were assessed by real-time PCR. (B) WT and SCAP knockout (Scap−/−) mice were fed ad libitum chow diets, fasted for 24 h, or fasted for 24 h and refed a high-carbohydrate diet for 12 h (26). RNA levels were assessed by real-time PCR using pooled samples (five mice/group). (C) Relative levels of mRNA encoding PNPLA3, ABCG5, and APOB in livers of C57BL/6J mice (four mice/group) fed chow diets containing 0.025% of the LXR agonist, T0901317. Pooled hepatic RNA samples were used to probe expression arrays (Affymetrix MG 430 ver. 2, Invitrogen). RNA levels are expressed relative to the level at time 0. (D) WT and SREBP-1c knockout mice were fed chow diets containing 0.025% T0901317 for 12 h and then fasted for 4 h before isolating hepatic mRNA. Equal aliquots of total RNA from five mice in each group were pooled and mRNA levels were assessed by real-time PCR.
Activation of SREBPs involves transport of the membrane-bound transcription factors from the ER to the Golgi where the proteins undergo cleavage to release a soluble, active transcription factor (23). ER to Golgi transit of SREBP requires its association with a polytopic membrane chaperone protein, SREBP-cleavage activating protein (SCAP) (25). Genetic deletion of SCAP abolishes the fasting-refeeding response of SREBP-responsive genes such as FAS (26). In SCAP knockout mice, mRNA levels of PNPLA3 were very low and were not increased by refeeding (Fig. 2B).
Because up-regulation of SREBP-1c by carbohydrate feeding is dependent upon the liver X receptor (LXR) (27, 28), we examined the effects of an LXR agonist, T0901317 (29), on PNPLA3 mRNA levels in livers of mice. PNPLA3 mRNA levels increased 15-fold within 24 h of initiating treatment with the agonist (Fig. 2C). Levels of Abcg5, an LXR target gene, were also increased by T0901317, whereas those of Apob were not (Fig. 2C). Because SREBP-1c is a direct target gene of the LXR/retinoid X receptor (RXR) heterodimer (30), we administered T0901317 to mice lacking SREBP-1c expression in the liver (Fig. 2D) to determine whether the effect on PNPLA3 is direct or indirect (31). No increase in PNPLA3 was found in these animals. Taken together, these data are consistent with PNPLA3 being a direct target gene of SREBP-1c.
Mapping SREBP-1 Binding Site in Intron 1 of Pnpla3.
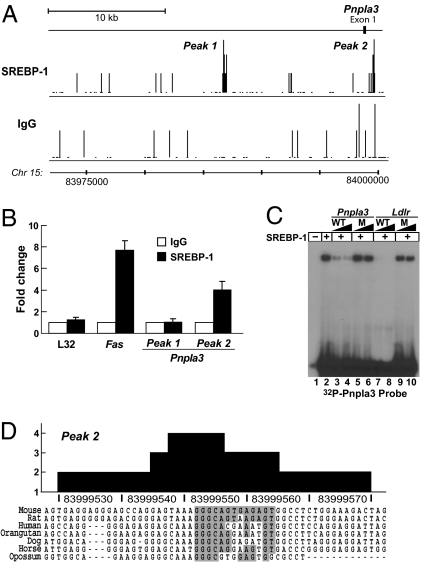
To identify SREBP binding sites that may be involved in transcriptional activation of PNPLA3, we analyzed whole genome chromatin immunoprecipitation and sequencing (ChIP-Seq) experiments performed in mouse liver using an SREBP-1-specific antibody (32). A map of the DNA sequence fragments located near Pnpla3 that were immunoprecipitated with the anti-SREBP-1 antibody versus a control antibody (rabbit IgG) is shown in Fig. 3A. Two sites of apparently specific SREBP-1 binding were identified: one located ∼11 kb 5′ of exon 1 and a second located 1,051 bp 3′ of the first exon (Fig. 3A). To confirm these findings we performed gene-specific ChIP assays (32) using primers designed for the two peaks predicted from the ChIP-Seq analysis. Gene-specific ChIP assays confirmed binding at site 2 but not at site 1 (Fig. 3B). To further examine the specificity of SREBP binding to site 2, electrophoretic mobility shift assays (EMSA) were performed using recombinant SREBP-1 (residues 1–490) and a 32P-labeled oligonucleotide probe corresponding to site 2. The probe was mixed with unlabeled competitor oligonucleotides, either from site 2 or from a known SREBP-1 binding site in the Ldlr promoter (32, 33). The wild-type (WT) but not the mutant oligonucleotides from site 2 in Pnpla3 competed for binding to SREBP-1 (Fig. 3C). The sequence of peak 2 includes a 13-bp motif (5′-GGGCAGTGAGAGT-3′) that is partially conserved between species and shares significant sequence similarity with a recently identified consensus sequence revealed through a genomewide analysis of SREBP-1c binding (32) (z score of 4.421). From these experiments we concluded that the robust transcriptional up-regulation of PNPLA3 expression with carbohydrate feeding is mediated through increased levels of SREBP-1 that result from the action of LXR.
Fig. 3.
Identification of SREBP-1c binding sites in Pnpla3. (A) Sequence reads from ChIP-Seq experiments were mapped onto the mouse genome in the University of California Santa Cruz (UCSC) Genome Browser. The numbers of sequence reads corresponding to each position at the Pnpla3 locus are shown for the IgG control and SREBP-1 antibodies. (B) Gene-specific ChIP analysis using liver chromatin enriched by precipitation with an antibody to SREBP-1(solid bars) or control IgG (open bars). (C) 32P-labeled probe corresponding to the SREBP-1 binding motif from peak 2 (5′-ACTCTCACTGCCC-3′) was incubated with recombinant SREBP-1 (200 ng) and analyzed by electrophoretic mobility shift assay. Increasing concentrations of unlabeled probe (100-, or 300-fold molar excess) for wild-type Pnpla3 (lanes 3 and 4) or for the Ldlr SRE (lanes 7 and 8) were included in the binding reactions as indicated. In lanes 5 and 6, a 100- or 300-fold molar excess of a mutant version of the Pnpla3 site 2 probe was added. In lanes 9 and 10, a mutant oligonucleotide that decreases SREBP-1 binding to Ldlr was added to the reaction. (D) The evolutionary conservation of site 2 sequence for SREBP-1 binding. The boxed sequence matches the recently described SREBP-1 binding motif (32) with a z score of 4.421 (Materials and Methods).
Posttranslational Regulation of PNPLA3 by Fatty Acids.
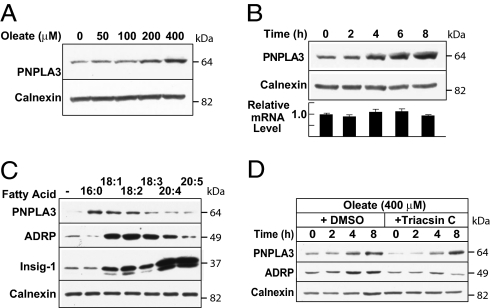
In cultured hepatocytes (HuH7 cells) stably expressing recombinant PNPLA3 under control of a CMV promoter, treatment with oleate to increase lipid droplet formation markedly increased the immunofluorescent signal associated with PNPLA3 (21). This finding suggested that PNPLA3 may undergo posttranslational regulation by fatty acids, the major end products of the SREBP-1c pathway. To test this hypothesis, we examined the effects of fatty acids on the levels of PNPLA3 in these cells. Immunoblot analysis consistently showed that levels of PNPLA3 increased in a dose-dependent manner in cells treated with oleate (Fig. 4A). The increase in PNPLA3 protein mass was apparent within 4 h and was not associated with any significant increase in PNPLA3 mRNA (Fig. 4B).
Fig. 4.
Fatty acid-stimulated posttranslational regulation of PNPLA3. (A) Dose–response, (B) time course, and (C) effect of fatty acid saturation and chain length on PNPLA3 protein mass. (A) HuH-7 cells stably expressing PNPLA3-V5 were treated with oleate for 8 h. Immunoblotting of the cell lysates was performed using anti-V5 and calnexin antibodies. (B) Cells were treated with 400 μM oleate for the indicated times before immunoblotting. RNA was extracted from duplicate sets of cells and the abundance of PNPLA3 mRNA was assayed using real-time PCR. (C) Cells transfected with 0.5 μg of pTK-Insig1-myc (34) were treated for 6 h with vehicle or 400 μM palmitate (C16:0), oleate (C18:1), linoleic acid (C18:2), linolenic acid (C18:3), arachidonic acid (C20:4), and eicosapentaenoic acid (C20:5), respectively. Cell lysates were subjected to immunoblotting. (D) Cultured cells were treated with 400 μM oleate plus DMSO or 5 μM triacsin C for the indicated times before immunoblotting.
To determine the specificity of fatty acid stabilization of PNPLA3, we examined the effect of various fatty acids on cellular levels of the protein. Addition of selected saturated fatty acids (palmitate, C16:0), monounsaturated fatty acids (oleate, C18:1), or polyunsaturated fatty acids (linoleic acid, C18:2) was associated with increased PNPLA3 protein expression. In contrast to these results, the very long chain polyunsaturated fatty acids arachidonic acid (C20:4) and eicosapentaenoic acid (C20:5) increased levels of INSIG 1 protein, as previously reported (34), but did not affect the amount of PNPLA3 (Fig. 4C). We also examined the effect of the various fatty acids on expression levels of adipose differentiation-related protein (ADRP), a lipid droplet-associated protein. Although ADRP levels also increased in cells treated with some of the fatty acids, the pattern did not completely mirror that of PNPLA3.
Treatment of cells with fatty acids leads to the formation and accumulation of TGs, which are stored as lipid droplets. To determine whether the increase in PNPLA3 protein mass was a direct effect of the fatty acids or an indirect consequence of TG accumulation, we treated cells with 400 μM oleic acid in the presence of triacsin C, a fungal metabolite that inhibits long fatty acyl CoA synthetase (35). Inhibition of TG synthesis by triacsin C did not attenuate the increase in PNPLA3 associated with oleate treatment (Fig. 4D). In contrast, the oleate-induced increase in ADRP was inhibited by triacsin C, confirming its activity in limiting TG accumulation.
Fatty Acids Inhibit Degradation of PNPLA3.
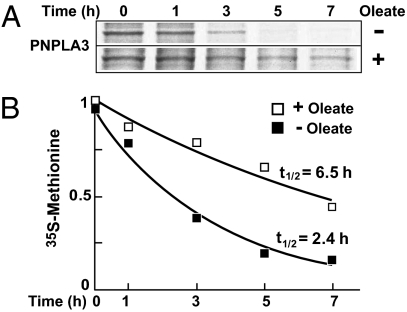
The fatty acid-induced increase in PNPLA3 may be due to increased translational efficiency or to decreased protein degradation. To distinguish between these possibilities, we used pulse-chase studies to measure the rate of degradation of PNPLA3 in the presence and absence of oleate. In cells incubated in FCS, the decrease in labeled PNPLA3 during the 7-h chase followed a monoexponential decay curve with a half-life of 2.4 h (Fig. 5). Treatment of the cells with 400 μM oleate extended the half-life of the protein to 6.7 h. Although we found evidence that PNPLA3 undergoes ubiquitination (Fig. S1A), we also found that addition of proteosome inhibitors (MG-132) resulted in increased PNPLA3 mRNA as well as protein levels (Fig. S1B). A similar increase in mRNA levels in response to proteosome inhibitors has been observed for other stably expressed recombinant proteins under the control of a CMV promoter (36).
Fig. 5.
Fatty acids increase half-life of PNPLA3. (A) Cells were treated with BSA or 400 μM oleate for 16 h and then grown in methionine- and cysteine-depleted DMEM. After 1 h, 0.2 mCi/mL 35S-labeled methionine/cysteine was added. One hour later the medium was changed to complete medium and the cells were grown for the indicated times. PNPLA3 was purified from cell lysates using cobalt beads and resolved by SDS/PAGE gels. The gels were dried before autoradiography. (B) The levels of radiolabeled PNPLA3 were measured by densitometry using ImageJ software. The measured values for both treatments were fitted into monoexponential decay model using Microsoft Excel.
These experiments indicate that PNPLA3 is nutritionally regulated at the transcriptional level through insulin-stimulated up-regulation via LXR and SREBP-1 and at the posttranscriptional level by fatty acids, an end-product of SREBP-1c action.
Discussion
The major finding of this study is that PNPLA3 undergoes coordinated transcriptional and posttranslational regulation that ensures greatly increased protein levels following the transition from the fasted to the fed state. Transcriptional regulation of PNPLA3 is effected by SREBP-1c through carbohydrate-mediated activation of LXR/RXR. The carbohydrate-induced increase in PNPLA3 mRNA levels was recapitulated by hepatic overexpression of SREBP-1a and SREBP-1c and abolished by genetic deletion of SCAP, a protein required for activation of the SREBPs (23). An additional level of nutritional regulation of PNPLA3 was revealed by the observation that PNPLA3 protein levels were consistently increased by fatty acid treatment in cultured liver cells stably expressing PNPLA3 under the control of a heterologous promoter. The fatty-acid-induced increase in PNPLA3 protein levels was not blocked by triacsin C, a potent inhibitor of acyl Co-A synthetase (35) and is therefore likely to be a direct effect of fatty acids rather than a secondary consequence of TG accumulation. Taken together, these findings reveal that nutritional control of PNPLA3 is effected by a feed-forward loop that is initiated by transcriptional up-regulation and amplified by coordinate inhibition of protein degradation.
The responses of PNPLA3 to pharmacological and genetic manipulation of LXR and SREBP-1c are consistent with a model proposed by Brown and Goldstein in which SREBP-1c coordinates the responses of many genes involved in fatty acid and TG metabolism to fasting and refeeding (23). In this model, insulin released in response to carbohydrate feeding increases activity of LXR/RXR heterodimers, which induce transcription of SREBP-1c via two LXR binding sites in the SREBP-1c promoter (27, 30, 37). In the present study, treatment with the LXR agonist T0901317 (29) increased PNPLA3 expression in wild-type, but not in SREBP-1c knockout mice (31), indicating that LXR promotes PNPLA3 transcription by activating SREBP-1c, rather than by direct activation of the PNPLA3 gene.
Chromatin imunoprecipitation assays and EMSA experiments indicated that SREBP-1 interacts directly with Pnpla3 via a response element in the first intron. Most SREBP-1 binding sites identified by genomewide ChIP are located in the proximal promoters of their target genes, but ∼10% are in introns (32). The sequence of the response element in PNPLA3 resembles a recently identified class of SREs found in 76% of SREBP-1 binding sites identified in a genomewide analysis using chromatin from mouse liver (5′-GGGANNTGTAGT-3′) (32). Whereas the antibody used in this experiment does not distinguish between SREBP-1a and -1c, SREBP-1c is the major SREBP-1 isoform in liver (38) and is regulated in response to fasting and refeeding whereas SREBP-1a is not (39). These data are consistent with PNPLA3 being a direct target of SREBP-1c.
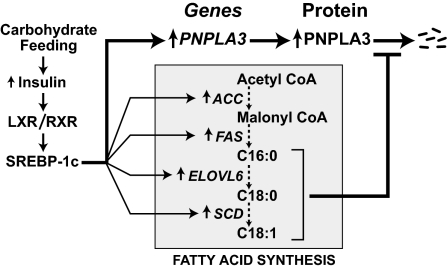
The coordinate regulation of PNPLA3 by SREBP and fatty acids forms a coherent feed-forward loop (40) that couples transcriptional and posttranslational controls to amplify the effects of nutritional stimuli on expression of the enzyme (Fig. 6). Under fasting conditions, low transcription of PNPLA3 mRNA and efficient degradation of PNPLA3 protein ensure very low levels of PNPLA3 expression in the liver. Rapid protein degradation is a common feature of regulatory circuits because it allows for rapid homeostatic response (40). The cost of rapid protein turnover is reduced efficiency: greater protein production is required to achieve a given steady state level. In coherent feed-forward loops, suppression of protein degradation allows for an efficient rebound of protein levels when transcription is activated. During carbohydrate refeeding, SREBP-1c stimulates PNPLA3 transcription as it up-regulates fatty acid biosynthesis. Fatty acid accumulation retards degradation of PNPLA3 protein, leading to a marked postprandial rebound in PNPLA3 levels. The specific fatty acids that most effectively stabilize PNPLA3 correspond to those that accumulate when SREBP-1c is activated: oleate and palmitate (which stabilize PNPLA3) are abundant in livers of SREBP-1c transgenic mice, whereas arachidonate and docosahexaenoic acid (which do not affect PNPLA3 degradation) are relatively depleted (41).
Fig. 6.
A feed forward loop for nutritional regulation of PNPLA3. Carbohydrate feeding activates SREBP-1c through the heterodimer LXR/RXR. SREBP-1c transcriptionally activates Pnpla3 as well as several genes encoding enzymes in the fatty acid biosynthetic pathway. Accumulation of fatty acids, specifically C16:0, C18:0, and C18:1, inhibit degradation of PNPLA3.
Previous studies have shown that fatty acids also induce a posttranslational increase in the expression of another SREBP-1c target, the polytopic ER membrane protein INSIG1 (34). Thus coordinate transcriptional and posttranslational regulation may be a common feature of SREBP-1c target genes. Fatty acid treatment of cells promotes the formation of lipid droplets, which may serve to stabilize the protein. The finding that triacsin C, which inhibits TG formation, did not interfere with fatty acid-induced PNPLA3 accumulation in hepatocytes (Fig. 4D) argues against this scenario. Triacsin C also failed to block the effects of arachidonate (20:4) on INSIG1 (34). Interestingly, different fatty acids delay degradation of INSIG1 and PNPLA3. Up-regulation of INSIG1 is specific to unsaturated long-chain fatty acids, including C18:1, C18:2, C18:3, C20:4, and C22:4; the saturated fatty acids C16:0 and C18:0 do not affect INSIG1 levels (34). In contrast, PNPLA3 levels are increased by palmitate as well as oleate and linoleate, and were not increased by C20:4 and C22:4 fatty acids. Further studies will be required to determine the mechanisms by which certain fatty acids increase cellular levels of PNPLA3.
We have tested the hypothesis that fatty acids retard PNPLA3 protein degradation by inhibiting proteasomal degradation of the enzyme, as has been reported for INSIG1 (34). Although we have evidence that PNPLA3 undergoes ubiqitination (Fig. S1), we are uncertain as to the significance of this pathway. Treatment of cells with agents that inhibit proteasomal degradation (MG-132 or ALLN) caused a significant increase in PNPLA3 mRNA, as well as PNPLA3 protein (Fig. S1). Thus the effect of these agents on PNPLA3 mRNA levels is likely an artifact of proteosome inhibitors on the expression of recombinant genes under the control of a CMV promoter (36).
The regulation of PNPLA3 may provide a clue to the physiological role of the protein, which remains obscure. Other genes that are specifically activated by SREBP-1c in response to refeeding are primarily involved in anabolic processes such as lipogenesis (31). In contrast, major mediators of lipolysis, such as adipocyte TG lipase (ATGL, PNPLA2) and hormone sensitive lipase, are inhibited rather than activated by refeeding (42, 43). Thus although partially purified recombinant PNPLA3 can catalyze the hydrolysis of TGs in vitro (20, 21), the marked up-regulation of PNPLA3 in response to refeeding suggests that this enzyme is involved in lipid remodeling, rather than catabolism.
Materials and Methods
Materials.
Materials used in the experiments are listed in SI Materials and Methods.
Real-time PCR Assays of mRNA Abundance.
The relative expression levels of PNPLA3 in human tissues were assayed in cDNA samples prepared from 48 human tissues (Human Normal cDNA Panel, OriGene) and standardized to 2 ng using cyclophilin as a calibrator. For details, see SI Materials and Methods. All research protocols involving mice were reviewed and approved by the Institutional Animal Care and Use Committee (University of Texas Southwestern). Hepatocytes and stellate cells were isolated from livers of mice as described (26). All mice were housed in colony cages in rooms maintained on 12 h light/12 h dark cycle and fed a regular chow diet (Harlan Teklad). Mice expressing a SREBP-1a, SREBP-1c, or SREBP-2 transgene (all under control of a phosphoenolpyruvate carboxykinase promoter) in the liver, mice lacking SREBP cleavage-activating protein (Scap−/−), and control mice were provided by Jay Horton (26, 44–46). A total of five 12- to 16-week-old male TgSREBP-1a, -1c, and -2 mice and an equal number of male littermate controls were fed a high protein/low carbohydrate diet for 2 weeks before being killed. Five 10- to 12-week-old liver-specific Scap−/− mice and wild-type littermate controls were either fed ad libitum, fasted for 24 h, or fasted for 24 h and then refed for 12 h with a fat-free, high-carbohydrate diet (MP Biochemicals catalog no. 901683) before being killed (26). RNA samples from five mice were pooled and first-strand cDNAs were synthesized from 2 μg RNA using a reverse-transcription kit (Applied Biosystems) and mRNA levels were determined by real-time PCR.
ChIP-Seq.
Deeply sequenced ChIP-Seq data sets for SREBP-1 antibody versus rabbit IgG in mouse liver tissues were generated as described (32). Gene-specific ChIP assays were performed (32) using primers designed for the two sites predicted from the read mapping profile in the ChIP-Seq analysis. The qPCR oligonucleotide pairs are provided in SI Materials and Methods.
EMSA were performed with recombinant SREBP-1 protein (amino acids 1–490) (32). DNA sequences for the SREBP-1 peaks were used to search for motifs using MEME (47). MEME represents motifs as position-dependent letter-probability matrices (PWM). The PWM for the SREBP-1 motif was used to find a “z score” for any 13-bp sequence; each letter in the sequence has a likelihood given in the PWM, and these are summed to determine the score, with a higher score meaning it is more likely to be the motif in question. Scores for every position along a chromosome (except coding and repeat regions) were determine using the PWM. The z scores for the putative SREBP-1 sites at the PNPLA3 locus were calculated by determining the raw score from the PWM, subtracting the average score for the chromosome, and dividing the result by the standard deviation.
Expression of PNPLA3 in Cultured Cells.
Human hepatoma cells (HuH-7) stably expressing PNPLA3 were generated as described (21). Cells were seeded at 4 × 105 cells per 60-mm dish and cultured in medium A [high glucose (4.5 mg/mL) DMEM, 10% FCS, G418 (1 mg/mL), pencillin (100 IU/mL), streptomycin (100 μg/mL)]. The next day, the medium was switched to medium B [DMEM; 10% delipidated FCS (DFCS), 50 μM compactin, 50 μM mevalonate, penicillin/streptomycin; G418 (0.4 μg/mL)]. After 24 h, 400 μM of BSA-bound fatty acids was added to the medium. For details of preparation of fatty acids, see SI Materials and Methods. At the indicated times, cells were collected, resuspended in 100 μL lysis buffer (PBS plus 1% Triton X-100, 0.1% SDS and protease inhibitors), briefly vortexed, and placed on ice for 15 min. After removal of insoluble material, the supernatants were subjected to immunoblotting (21) or used to immunoprecipitate PNPLA3.
Pulse-Chase Labeling of PNPLA3.
HuH-7 cells stably expressing PNPLA3-V5-His were plated at a density of 106/100 mm dish. The next day, the cells were treated with BSA or 400 μM oleate for 16 h. Cells were grown in DMEM (minus methionine and cysteine) for 1 h before addition of 0.2 mCi/mL of 35S labeled-methionine/cysteine. After 1.5 h, cells were returned to complete medium containing BSA or oleate, cultured for the indicated times, and lysed in 1.9 mL of buffer C (PBS, 1% Triton X-100 and protease inhibitors). Lysates were incubated with 40 μL of HisPur Cobalt resin (50%) for 4 h at 4 °C. PNPLA3 was eluted from the beads in 100 μL of buffer C containing 200 mM imidazole. Loading buffer (5×) was added, the samples were boiled for 5 min, and then the proteins were size-fractionated on 8% SDS/PAGE gels. The gels were dried and subjected to autoradiography.
Supplementary Material
Acknowledgments
We thank Christine Zhao, Zifen Wang, Norma Anderson, and Michele Alkalay for excellent technical assistance and Jay Horton, Guosheng Liang, and David Russell for helpful discussions. The work was supported by National Institutes of Health HL92550 and HL20948 to J.C.C. and H.H.H. and HL48044 to T.F.O.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1003585107/DCSupplemental.
References
- 1.Browning JD, et al. Prevalence of hepatic steatosis in an urban population in the United States: Impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 2.de Alwis NM, Day CP. Non-alcoholic fatty liver disease: The mist gradually clears. J Hepatol. 2008;48(Suppl 1):S104–S112. doi: 10.1016/j.jhep.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Day CP, James OF. Steatohepatitis: A tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 4.Baulande S, Lasnier F, Lucas M, Pairault J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem. 2001;276:33336–33344. doi: 10.1074/jbc.M105193200. [DOI] [PubMed] [Google Scholar]
- 5.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browning JD, Kumar KS, Saboorian MH, Thiele DL. Ethnic differences in the prevalence of cryptogenic cirrhosis. Am J Gastroenterol. 2004;99:292–298. doi: 10.1111/j.1572-0241.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 7.Yuan X, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotronen A, et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52:1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- 9.Sookoian S, et al. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kollerits B, et al. A common variant in the adiponutrin gene influences liver enzyme values. J Med Genet. 2010;47:116–119. doi: 10.1136/jmg.2009.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42:21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 12.Hoekstra M, et al. The expression level of non-alcoholic fatty liver disease-related gene PNPLA3 in hepatocytes is highly influenced by hepatic lipid status. J Hepatol. 2010;52:244–251. doi: 10.1016/j.jhep.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Racusen D, Foote M. A major soluble glycoprotein of potato tubers. J Food Biochem. 1980;4:43–52. [Google Scholar]
- 14.Galliard T. The enzymic deacylation of phospholipids and galactolipids in plants. Purification and properties of a lipolytic acyl-hydrolase from potato tubers. Biochem J. 1971;121:379–390. doi: 10.1042/bj1210379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rydel TJ, et al. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a ser-asp catalytic dyad. Biochemistry. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 16.Lake AC, et al. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J Lipid Res. 2005;46:2477–2487. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Kienesberger PC, Oberer M, Lass A, Zechner R. Mammalian patatin domain containing proteins: A family with diverse lipolytic activities involved in multiple biological functions. J Lipid Res. 2009;50(Suppl):S63–S68. doi: 10.1194/jlr.R800082-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ. Characterization of the human patatin-like phospholipase family. J Lipid Res. 2006;47:1940–1949. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann R, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins CM, et al. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 21.He S, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kershaw EE, et al. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55:148–157. [PMC free article] [PubMed] [Google Scholar]
- 23.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimano H, et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda M, et al. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci USA. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denechaud PD, et al. ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J Clin Invest. 2008;118:956–964. doi: 10.1172/JCI34314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz JR, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang G, et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 32.Seo YK, et al. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc Natl Acad Sci USA. 2009;106:13765–13769. doi: 10.1073/pnas.0904246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Südhof TC, Russell DW, Brown MS, Goldstein JL. 42 bp element from LDL receptor gene confers end-product repression by sterols when inserted into viral TK promoter. Cell. 1987;48:1061–1069. doi: 10.1016/0092-8674(87)90713-6. [DOI] [PubMed] [Google Scholar]
- 34.Lee JN, Zhang X, Feramisco JD, Gong Y, Ye J. Unsaturated fatty acids inhibit proteasomal degradation of Insig-1 at a postubiquitination step. J Biol Chem. 2008;283:33772–33783. doi: 10.1074/jbc.M806108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omura S, Tomoda H, Xu QM, Takahashi Y, Iwai Y. Triacsins, new inhibitors of acyl-CoA synthetase produced by Streptomyces sp. J Antibiot (Tokyo) 1986;39:1211–1218. doi: 10.7164/antibiotics.39.1211. [DOI] [PubMed] [Google Scholar]
- 36.Biasini E, et al. Proteasome inhibition and aggregation in Parkinson's disease: A comparative study in untransfected and transfected cells. J Neurochem. 2004;88:545–553. doi: 10.1046/j.1471-4159.2003.02152.x. [DOI] [PubMed] [Google Scholar]
- 37.Yoshikawa T, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimomura I, Shimano H, Korn BS, Bashmakov Y, Horton JD. Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J Biol Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
- 42.Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: Ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem. 2004;279:47066–47075. doi: 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- 43.Sztalryd C, Kraemer FB. Regulation of hormone-sensitive lipase during fasting. Am J Physiol. 1994;266:E179–E185. doi: 10.1152/ajpendo.1994.266.2.E179. [DOI] [PubMed] [Google Scholar]
- 44.Horton JD, et al. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimano H, et al. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 47.Bailey TL. Curr Protoc Bioinformatics. Brisbane, Australia: University of Queensland; 2002. Discovering novel sequence motifs with MEME. Chap 2, Unit 2.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.