Abstract
The recent assembly of the silkworm Bombyx mori genome with 432 Mb on 28 holocentric chromosomes has become a reference in the genomic analysis of the very diverse Order of Lepidoptera. We sequenced BACs from two major pests, the noctuid moths Helicoverpa armigera and Spodoptera frugiperda, corresponding to 15 regions distributed on 11 B. mori chromosomes, each BAC/region being anchored by known orthologous gene(s) to analyze syntenic relationships and genome rearrangements among the three species. Nearly 300 genes and numerous transposable elements were identified, with long interspersed nuclear elements and terminal inverted repeats the most abundant transposable element classes. There was a high degree of synteny conservation between B. mori and the two noctuid species. Conserved syntenic blocks of identified genes were very small, however, approximately 1.3 genes per block between B. mori and the two noctuid species and 2.0 genes per block between S. frugiperda and H. armigera. This corresponds to approximately two chromosome breaks per Mb DNA per My. This is a much higher evolution rate than among species of the Drosophila genus and may be related to the holocentric nature of the lepidopteran genomes. We report a large cluster of eight members of the aminopeptidase N gene family that we estimate to have been present since the Jurassic. In contrast, several clusters of cytochrome P450 genes showed multiple lineage-specific duplication events, in particular in the lepidopteran CYP9A subfamily. Our study highlights the value of the silkworm genome as a reference in lepidopteran comparative genomics.
Keywords: comparative genomics, silkworm, Noctuidae, transposable elements, gene clusters
The insect order Lepidoptera is second only to Coleoptera as the most prolific in animal species number, with an estimated total of more than 160,000 species falling into more than 130 families. Assembly of the first lepidopteran genome, that of the domesticated silkworm Bombyx mori, has established a valuable reference for lepidopteran comparative genomics and genetics (1). The radiation of the major clades of Lepidoptera occurred in the late Jurassic less than 150 Mya (2, 3). The superfamily Noctuoidea contains approximately one fourth of all Lepidoptera and includes a very large number of major pest species of agriculture and forestry. It has a fossil record dating back to at least 75 Mya (4), which is close to the time of divergence of the superfamily Bombycoidea to which the silkworm belongs. The diversification and proliferation of lepidopteran species is therefore very recent (5). The insights gained from comparative genomic analyses using a reference genome from a model species such as the silkworm would greatly facilitate research on all Lepidoptera, and in particular on selective targets for innovative pest management at a time when competition for food between humans and insects is becoming a critical challenge for a rapidly growing human population. The 432 Mb genome of B. mori is the first fully sequenced lepidopteran genome (1), and detailed SNP and BAC-based chromosomal maps are available (6, 7). We wished to compare the silkworm genome at a finer scale with that of major lepidopteran pests from the family Noctuidae, Helicoverpa armigera and Spodoptera frugiperda. The Old World cotton bollworm H. armigera is a highly polyphagous pest and ranks as the world's worst pest of agriculture (8, 9). It is closely related to Helicoverpa zea, its New World relative from which it diverged approximately 1.5 Mya (10) and to Heliothis virescens, both major pest species in their own right. Similarly, the fall armyworm S. frugiperda is a major pest of maize and rice in the Americas and represents a genus comprising many pests worldwide. Conservation of synteny would allow a rapid identification of genes in these pest species from the knowledge of the Bombyx genome. The precise definition of conserved segments and of the degree of chromosome rearrangement is more difficult (11–15). Nonetheless, conservation of synteny, when it can be documented, is an extremely useful feature of comparative genomics that validates the use of a model organism and that defines a framework for a finer study of gene and genome evolution.
Overall conservation of synteny in Lepidoptera has been reported in several studies (16–20) suggesting that, over the 100 My separating the Bombyx and butterfly lineages, some degree of gene synteny has thus been maintained. However, a more detailed study was warranted to allow more general conclusions. In particular, we asked whether the holocentric nature of the multiple chromosomes in Lepidoptera (n = 28 in Bombyx) favors scrambling of gene order and masks microsyntenic relationships. The Bombyx genome has among the highest level of repetitive sequences [43.6% (1)] of all insect genomes studied to date [vs. Apis mellifera with just 1% (21)]. We therefore also asked whether the high level of repeated sequences would make it difficult to use Bombyx as a reference genome for lepidopteran pests.
To answer these questions, we studied the gene landscape and patterns of repetitive sequence distribution, as well as synteny and rearrangements at a very fine scale in 15 genomic regions anchored by orthologous genes in the two noctuid species, H. armigera and S. frugiperda, and in the silkworm B. mori. Our study is based on a detailed analysis of high-quality BAC sequences in the two noctuid species, and on the newly assembled complete genome sequence of the silkworm.
Results
Strategy of the Comparative Genome Analysis.
We sequenced and annotated BACs representing 15 putatively homologous regions of the genome in comparison with the fully assembled genome of B. mori. We sought to cover genomic regions neighboring genes that vary widely in their evolutionary constraints (Table S1). These regions were covered by 18 BACs in H. armigera covering a total of 1.963 Mb of genomic DNA and by 17 BACs in S. frugiperda covering a total of 2.042 Mb of genomic DNA. This represents approximately 0.5% each of the two genomes. We estimated the total sequence overlap between the H. armigera and S. frugiperda sequences to cover 1.22 Mb, with 81.6 ± 8.1 kb of overlap for each pair. The 15 genomic regions were each compared to regions spanning 200 kb of the B. mori genome on 15 scaffolds that were distributed on 11 of the 28 chromosomes. The GC content in the three species was similar (32.7–36.3%). We identified 502, 201, and 274 genes in B. mori, H. armigera, and S. frugiperda, respectively (Table S2). We compared both the complete set of presumed orthologues and a limited set of additional genes of the three species (34–46 genes), which were analyzed in greater detail. These genes were selected because of the high degree of confidence in their annotation, i.e., 18 unique genes and 16 to 28 members of multigene families. Table S2 shows that gene length was greater in B. mori (6.5–8.0 kb) than in S. frugiperda (4.3–4.9 kb) and in H. armigera (3.1–4.9 kb). This was a result of a corresponding variation in intron size that was greatest in B. mori. The intergenic regions represent 41% of the B. mori genome compared with 55% in the noctuids.
Repetitive Sequences and Transposable Elements.
The BACs covering the 15 regions of interest and 20, 20, and 55 additional BACs sequences from H. armigera, S. frugiperda, and B. mori, respectively (that are not included in the synteny study) were used for de novo repeated sequences detection using the REPET pipelines (22). The results (Dataset S1 (XLS)) show that the most abundant classes are long interspersed nuclear elements (LINEs) and terminal inverted repeats (TIRs) in the three lepidopteran subgenomes with a quasi-equal number of consensus for both classes in each species. Class II elements predominate in H. armigera and S. frugiperda with 13 and 11 consensus, respectively, compared with seven and six consensus of class I, whereas elements of class I are the most abundant in the 55 BACs of our B. mori subgenome (76 consensus of class I for 59 of class II).
The total proportions of repeated sequences in H. armigera, S. frugiperda, and B. mori were 16.2%, 8.0%, and 33.8%, respectively. This latter value is close to that found for the whole silkworm genome, i.e., 43.6% for repeated sequences and 35.1% for transposable elements (TEs), respectively (1, 23). Among repeated sequences of the TE type, retrotransposons cover 42.5%, 76.5%, and 68.4% of the total TE sequence in H. armigera, S. frugiperda, and B. mori, respectively, with an expansion of some LINE families in S. frugiperda and B. mori. The TE annotation is available online at the Lepido-DB (http://www.inra.fr/lepidodb), and the description of families and their respective distribution in the three species will be the object of a future study.
Conservation of Synteny.
Quantifying the synteny conservation.
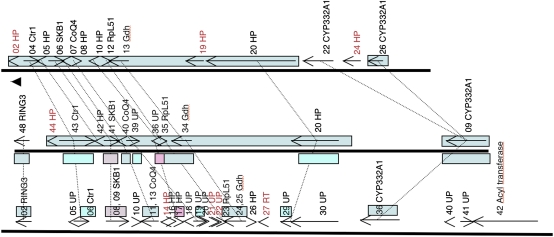
Syntenic genes were detected and annotated (Fig. S1) in the 15 genomic regions as described below in Materials and Methods, and the resulting three-way comparisons are presented in Fig. S2. Syntenic genes can also be visualized online with the Cmap software at the Lepido-DB Web site. Fig. 1 shows an example of such three-way comparisons, with the CYP332A region with a very high degree of synteny conservation.
Fig. 1.
Syntenic relationship at the CYP332A locus. Schemes at scale representing, from top to bottom, H. armigera, S. frugiperda, and B. mori. Arrows represent genes predicted by KAIKOGAAS. Only valid genes are shown (Fig. S1). Synteny links are shown with black dotted lines. In text boxes, gene ID, or for other genes, HP, unknown protein (UP; presence of a match to an EST with a threshold of 10−40 by BlastN). Synteny blocks (as defined in text) are shown as colored boxes spanning genes arrows in the case of H. armigera and S. frugiperda, below and above gene arrows in the case of S. frugiperda and B. mori, respectively. See Fig. S2 for gene name abbreviations.
Taking B. mori scaffolds as a reference, we counted for each region the number of genes maintained in the overlapping portion of the B. mori scaffolds with either of the noctuid BACs (Table 1 and Dataset S2). Among the 270 genes analyzed, 141 (52.2%) were present in either of the noctuids BACs. Most of the analyzed genes (74.8%) were of “known class,” that is, homologous to a gene of known function, having a match to an RNA sequence, or syntenic with a gene of known function; the remaining 25.2% were classified as “HP,” i.e., encoding hypothetical proteins. Among the B. mori identified known genes, 64.4% were found in both of the noctuids BACs, whereas only 16.2% of the HP genes were syntenic. This suggests that known genes, whose sequence is conserved between species, are located in more stable regions of the genome whereas unconserved genes, some of which may be species-specific, lie in more plastic regions. Twenty-six of 141 genes of B. mori (18.3%) that are found in the corresponding BACs from noctuids were in the reverse orientation.
Table 1.
Gene categories with B. mori as reference
| Type | Genes with known function (ID) | Genes with identified mRNA (UP) | Partial sum (ID + UP) | Genes encoding hypothetical protein (HP) | Total genes(ID + UP + HP) |
| Syntenic | 113 | 17 | 130 (64.4%) | 11 (16.2%) | 141 (52.2%) |
| Nonsyntenic | 2 | 70 | 72 (35.6%) | 57 (83.8%) | 129 (47.8%) |
UP, unknown protein.
We counted the number of presumed orthologues found in the corresponding B. mori region for each of the 15 BAC pairs, irrespective of their order or orientation. This is a restrictive view, because in some cases, genes present on the BACs were found outside the 200-kb B. mori region, but within the same scaffold. For example, in the Or83b genomic region of B. mori (Fig. S2), the FBPA genes were located 297 kb away from the Or83b gene on the same scaffold. The data (Dataset S3) show that the number of presumed orthologues was very variable, from a minimum of one (the anchor gene used to select the BAC) to 16 in the TATA binding protein (TBP) region of S. frugiperda and 13 in the CYP4M region of H. armigera. These variations were roughly correlated with gene density that was not evenly distributed. The TBP region of S. frugiperda was characterized by one gene every 4.5 kb, whereas the juvenile hormone acid methyl transferase region had only one gene every 24.6 kb in H. armigera. On average, 69.8% and 50.2% of the genes from H. armigera and S. frugiperda, respectively, were found to be in macrosyntenic conservation, with a median density of eight genes per BAC. In other words, in approximately half the cases, two genes in synteny over 50 kb in B. mori would be found within 34 kb of each other in the two noctuid species. This clearly indicates conservation of synteny at a macroscopic scale. We then analyzed the syntenic relationships at a finer scale.
Fine-scale microsynteny and size of synteny blocks.
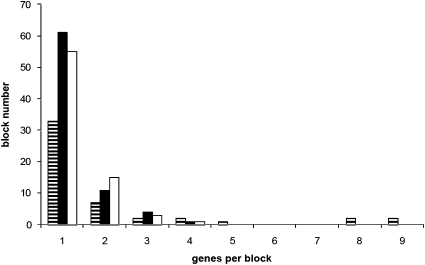
We measured the number and length of synteny blocks, i.e., conserved segment corresponding to any region in which gene content and order are conserved (24). Genes belonging to a TE were excluded. Only validated genes were considered (as detailed earlier). This analysis (Fig. 2) showed that synteny blocks are very small. They spanned 2.01 genes per block on average between the two related noctuids (maximum size of nine genes), and were smaller between B. mori and S. frugiperda or H. armigera (1.29 and 1.32 on average, respectively, with a maximum of four genes). Because of their limited size, synteny blocks were seldom interrupted by lack of sequence extent (i.e., BAC extremities). The average number of genes per BAC or genomic region analyzed was 11 for H. armigera, 16 for S. frugiperda, and 26 for B. mori, far greater than the average block size.
Fig. 2.
Number of genes per synteny block. Black and white striped bars: between H. armigera and S. frugiperda (average size 2.04). Black bars: between B. mori and S. frugiperda (average size 1.29). White bars: between B. mori and H. armigera (average size 1.32).
We then examined the recombination events responsible for the synteny breaks. Apart from inversions, we assumed that the presence of B. mori genes whose orthologues were not found in the two noctuid species has resulted from transpositions to or from unsequenced parts of these genomes (i.e., beyond the boundaries of the BACs). In addition to orthologues we also analyzed paralogues resulting from duplications in one of the species, taking into account the closest paralogue pairs based on phylogenetic analysis of the encoded proteins. We recorded a ratio of 1.0:8.0:3.4 of inversions, transpositions, and duplications, respectively, between the pest genomes (Table 2). This ratio was 1.0:8.7:3.4 between B. mori and H. armigera and 1.0:11.4:3.4 between B. mori and S. frugiperda. We then focused on the overall rate of rearrangements since speciation of the three lepidopteran species. An inversion event results from two double-strand breaks, whereas a transposition requires three (24). The duplications observed within gene families may have resulted from replication slippage or a break-and-join mechanism such as unequal crossing over or transposition and may thus involve zero, two, or three breaks, respectively, and we conservatively counted one break per duplication. The results (Table 3) showed a very high number of breakages. Several genomic regions in our analysis contained variable numbers of duplicated genes from large gene families that may have biased our results by inflating the number of breaks. We therefore corrected for this potential bias by counting only orthologues or best paralogue pairs (Table 3, column 2), by excluding all genes that had undergone duplication in one of the species (Table 3, column 3) or by excluding all BAC regions in which members of large gene families were present (Table 3, column 4). In all three cases, the calculated evolution rate remained very similar. The evolution rate is approximately two breaks per Mb per Mya and has perhaps accelerated within the Noctuidae.
Table 2.
Number of rearrangements recorded between species
| Species | Inversions | Transpositions | Duplications |
| Ha-Sf | 5 | 40 | 17 |
| Bm-Ha | 10 | 87 | 34 |
| Bm-Sf | 9 | 103 | 31 |
Table 3.
Number of chromosomal breakages per Mb DNA since divergence of the species
| No. of breakages/Mb (number/Mb/MY) |
|||||
| Species | Divergence time, MYa | Overall values for 15 regions, counting only 1:1 orthologue or best paralogue pairs | Values for 15 regions, excluding gene duplications | Values for seven regions lacking gene families | |
| Ha/Sf | 20–40 | 85 (2.83) | 75 (2.50) | 99 (3.30) | |
| Bm/Ha | 60–100 | 132 (1.65) | 114 (1.43) | 109 (1.37) | |
| Bm/Sf | 60–100 | 150 (1.88) | 136 (1.70) | 165 (2.07) | |
| Mean | — | 122 (2.12) | 108 (1.87) | 125 (2.25) | |
Correlation Between Breakages and TE Density.
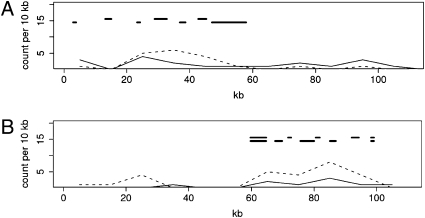
TEs are often a source of genomic rearrangements or can insert at genomic breakages. We recorded the precise coordinates of synteny breaks between the two noctuid genomes, and counted the number of TE copies by a sliding window of 10 kb along the BACs. In many cases, a clear correlation was evidenced. In the case of the CYP4M region (Fig. 3A), a clear association of TE copy density was seen with the synteny breaks corresponding to one gene inversion and to the duplications of the CYP4M genes. Such an association was found also in the H. armigera BAC carrying the gene encoding the ecdysone receptor at the places where duplications of the gene encoding a putative multibinding protein genes have occurred (Fig. 3B).
Fig. 3.
Correlation between synteny breaks and TE copy density. TE copy density is shown as a function of BAC length. 3A, S. frugiperda CYP4M region; 3B, H. armigera EcR region; dotted lines, TE copy density. Thick lines, regions corresponding to synteny breaks; thin lines. gene density.
Evolution of Gene Clusters.
Several gene clusters were covered by our analysis. The aminopeptidase N (APN) of lepidopteran midgut has received wide attention because of its role in the mode of action of Cry toxins of Bacillus thurigiensis (25), and multiple APN cDNAs from a variety of species are available (26). We show here that APN genes are organized in a single, large cluster of nine genes on chromosome 9 of B. mori. Remarkably, this cluster is highly conserved in both order and orientation of the genes in the three species (Fig. S3). In addition to seven previously identified APNs (26), the cluster contained two additional genes: Zn-m1, a member of the more distantly related protease m1 zinc metalloproteases found in insects and vertebrates, and APN-8, related to a fat body–specific transcript previously reported from Spodoptera litura (27).
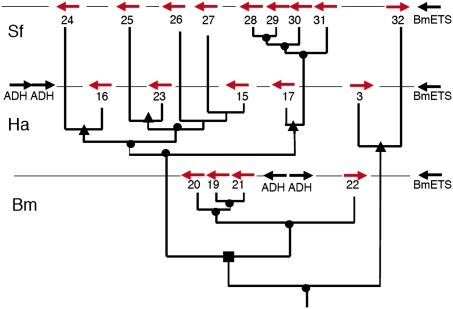
Our analysis also covered five clusters of P450 genes, one of the largest multigene family in insects (28). The CYP genes in the five clusters have evolved at a roughly similar rate, as seen by the similar range in overall protein identity of the encoded enzymes (51–68% between B. mori and the two noctuids). However, this evolution was punctuated by multiple duplication events in the two noctuid lineages, and possible gene loss in the silkworm lineage, so that we found 17 to 22 CYP genes in the noctuids and just nine in B. mori. The CYP9A cluster on chromosome 17 had the most complex evolutionary history (Fig. 4). We found four CYP9A genes in B. mori, and there are no additional CYP9A genes in the genome. In contrast, we found five CYP9A genes in H. armigera and nine in S. frugiperda. We searched the yet unassembled H. armigera genome for other CYP9A genes and found that the four B. mori genes are monophyletic with the only other CYP9A gene that is present in the H. armigera genome, but beyond the boundaries of our BAC coverage. This gene, CYP9A14, is most closely related to CYP9A22 of B. mori, and to date a closest paralogue in S. frugiperda has not been found. Two gene duplications leading to CYP9A19, -20, and -21 therefore occurred in the Bombyx lineage. The CYP9A3 and CYP9A32 genes of the noctuid species are on the opposite strand of the chromosomes respective to the rest of the cluster. They are probable orthologues and their expected third orthologue has probably been lost in the Bombyx lineage. The remainder of the CYP9A genes in the noctuids resulted from seven gene duplications, of which four occurred in the Spodoptera lineage. The rate of evolution varied over time in a lineage-specific manner, and this was probably accompanied by chromosomes rearrangements, as flanking genes have drastically changed in distance and orientation (Fig. 4).
Fig. 4.
Evolution of the CYP9A gene cluster. The CYP9A genes in the three species are shown in their correct orientation and order, but not relative distance, on the BACs or scaffold. From top to bottom: Sf, S. frugiperda; Ha, H. armigera; Bm, B. mori. The phylogenetic tree based on alignments of the CYP proteins is superimposed with its correct topology, but with branch lengths modified for clarity of the figure. Gene duplication events (●), B. mori/noctuid split (i.e., ancient speciation rather than duplication event) (■), and the S. frugiperda/H. armigera split (▲) are indicated. The sequence of events for the CYP9A15, -26, and -27 genes is unresolved. The relative orientation of the genes indicates at least two inversions in addition to the duplication events. Recognized flanking genes [alcohol dehydrogenases (ADH) and BmETS transcription factor] are shown (see Fig. S2 for details).
Large-Scale Chromosomal Inversion.
In the RpL5A genomic region, the rpl5A gene and the three genes immediately downstream of rpl5A are in the same orientation in the three species (Fig. S4). In the flanking regions, three known genes are reversed in B. mori compared with S. frugiperda on one side, and five genes are also reversed in B. mori on the other side. The colinearity all along the BACs from noctuids is shown at the nucleotide level by dot plots, which also show the large inversion between B. mori and the two noctuids (Fig. S4). This inversion dates before the divergence of the two noctuid species. It is possible that other large-scale inversions were missed in the present study because the size of the BACs may be equal or smaller than the size of such inversions.
Discussion
The assembled sequence for Bombyx mori amounts to a 432 Mb genome size, and this size was estimated to 405 Mb for H. virescens (29), a species closely related to H. armigera. Sequenced genomes show enormous variation in TE copy number, which can largely account for differences in genome size (30). Invasion of the silkworm genome by class I TEs may be responsible for its slightly increased size relative to noctuids. The most abundant classes of TE are LINEs and TIRs in the three genomes. In terms of genome coverage, LINEs predominate in B. mori and S. frugiperda and contribute equally with TIRs in H. armigera. The prevalence of non-LTR retrotransposons makes lepidopteran genomes different from that of Drosophila melanogaster, in which LTR retrotransposons are the most abundant (31). Both the rather high repeat coverage and the prevalence of non-LTR retrotransposons make lepidopteran genomes look like mammalian genomes (32, 33).
We chose to measure the evolution rate of chromosomes (12) to further compare the genomic plasticity of Lepidoptera with that of other invertebrates like Diptera and nematodes. Thus, we examined the border of each synteny block to identify the recombination events responsible for the synteny breaks, and deduced a number of breakages per Mb. We calculated 85 breakages per Mb between the pest genomes and (132/150) breakages per Mb between pests and B. mori. These values are higher than the 51 breakages per Mb determined in a comparison of 13% of the Caenorhabditis briggsae genome with the Caenorhabditis elegans genome (12), whose divergence time is estimated at 50 to 120 Mya. These initial values of 0.4 to 1 breakages per Mb per Mya (12) were later refined by the same method to 0.5 to 0.7 breakages per Mb per Mya in a whole-genome comparison (34), suggesting that the values we obtained for our genome sampling are likely to be robust. The evolution rate of lepidopteran genomes (approximately 2 breakages per Mb per Mya) is thus faster than that of nematodes, themselves evolving fourfold faster than Drosophila species (12), whose chromosomes rearrange two orders of magnitude faster than those of mammals and faster than plant chromosomes (35). This very high rate is clearly not correlated to generation time or effective population size (36), because these life history traits can be very similar between Lepidoptera and higher Diptera. Nematodes and Lepidoptera share a common feature, i.e., the holocentric organization of their chromosomes. The scattered organization of centromeric determinants may lead to a greater genomic plasticity as chromosome fragments resulting from double-strand breaks may be maintained and reintegrated elsewhere. A ratio of 1.0:2.3 of inversions to transpositions was described in the comparison of Caenorhabditis species (12). We find ratios of 1.0:8.0 (between noctuids) or 1.0:10.0 (average between noctuids and Bombyx). This higher rate of transposition events may result from a higher proportion of TE in Bombyx than in C. briggsae (45%/22.4%) and from the different nature of these elements (mainly retrotransposons in Bombyx vs. DNA transposons in C. elegans).
The synteny block size we measured is very small for species having diverged approximately 20 Mya (2.0 genes per block between the two noctuid species) or between 60 and 100 Mya (3, 5) (1.3 genes per block in our study for noctuids/B. mori). This is to be compared to synteny block size observed between Drosophila pseudoobscura with D. melanogaster, in which the average number of genes in syntenic blocks is 10 (37) and the divergence time approximately 55 Mya. Similarly, the gene-based microsynteny blocks as calculated with single copy orthologues in mosquitoes was 3.9 genes per block between Aedes aegypti and Anopheles gambiae, species that diverged 150 Mya (38). This figure decreases to 2.4 genes per block between Aedes aegypti and D. melanogaster, for which the divergence time is approximately 250 Mya.
The high genome fragmentation evidenced in our study is paradoxical, however, because the small size of the synteny blocks masks a higher order of synteny conservation, i.e., the “noise” of multiple insertions, deletions and inversions masks the “signal” that is apparent from many of our three-pair comparisons: In 11 of the 15 regions, there are six or more (as many as 16) genes found on the corresponding BAC/region of the other two species. Clearly there are constraints that prevent the total scrambling of gene order in these genomes. When estimating the relationship between protein sequence identity and gene order of orthologues in insect genomes, it was suggested that gene order would be lost below an average of 50% identity between orthologue pairs (13), but here synteny conservation was observed for many genes below that threshold.
The higher order of gene conservation was our initial expectation, based on previous studies in Lepidoptera (16). In a study of 72 orthologous loci between B. mori and Heliconius melpomene, a very high degree of conserved synteny was observed, with 21 linkage groups (LGs) of the butterfly mapped with one to seven genes in common with the silkworm. However, only one chromosomal inversion was noted, and it is probable that the density of markers per LG did not allow the detection of more rearrangements (17). A study on Bicyclus anynana with 462 markers documented a high degree of macrosyntenic relationships between the 28 chromosomes of B. mori and the 23 LGs of B. anynana. LG10 had 15 genes and LG 21 four genes in conserved order (19). The remainder of the chromosomes showed one or more rearrangements or inversions. Similarly, 124 of 131 orthologues retained the same order on the chromosomes of B. mori and Manduca sexta (20), and these authors noted the paradox of conserved synteny on holocentric chromosomes, in which increased chromosome rearrangements are more likely. With the higher resolution of BAC sequencing, high colinearity of genes was observed between two related species of Heliconius, H. melpomene, and H. erato, in two regions covering 180 kb and 280 kb of sequence (18). Just one gene was present in an indel block, although nine such indel blocks characterized by repetitive sequences were noted on a 180-kb span. A comparison of H. erato and B. mori over approximately 190 kb of sequence revealed 11 conserved gene, with one translocation and two inversions (18), thus foreshadowing our results. On 15 regions we observed both a high degree of gene conservation or macrosynteny and a high amount of local rearrangements.
Our observation of small synteny blocks in a background of high macrosynteny in Lepidoptera contrasts with Drosophila, in which paracentric inversions (i.e., within one chromosome arm or Muller element) are common and result in a lack of synteny (37, 39). This was evident in a direct comparison of two Drosophila genomes (37) and shown to explain most gene order shuffling in a study of 12 species (39). Furthermore, it was pointed out (37) that the fitness costs associated with such chromosomal rearrangements are reduced because there is no crossing over in the male, avoiding the generation of aneuploidy with dicentric/acentric chromosomes, whereas in the female such chromosomes are directed to polar bodies and not to gametes. The holocentric chromosomes of Lepidoptera appear to allow inversions only on much more limited chromosomal regions, possibly those between the multiple equivalents of centromeres. Perhaps holocentric chromosomes are then more resistant to large-scale rearrangements, thus explaining our paradoxical high degree of conserved synteny over the noise of the small chromosomal rearrangements. As it is the female that has no crossing over in Lepidoptera, this hypothesis suggests how negative fitness costs associated with chromosomal inversions might be reduced. The hypothesis needs to be tested on defined, larger, replication units. When the H. armigera genome sequence and those of other lepidopteran species become available, a larger dataset will become available for analysis by specific algorithms (e.g., ref .40) to answer these questions. Sequences functioning as centromeres would also need to be identified and mapped physically.
In conclusion, our microsynteny study provides insight into the extent of genome conservation that underlies macrosynteny and emphasizes the significance of the B. mori genome as a reference for Lepidoptera. It is logical to assume that the broad coverage of multiple chromosomes, and of genomic regions of varying degrees of gene richness, is sufficient justification to allow the extension of our conclusions to the whole genome. Despite a higher average size of genes in B. mori, and a different complement of repetitive sequences/TEs, the organization of the genomes shows a high degree of macrosynteny conservation. Bombyx can therefore be used as a first reference for genomic/genetic studies in Lepidoptera. Our study also highlights the very rapid evolution of the three genomes. Gene clusters from rapidly evolving multigene families such as the P450s show evidence of recent and multiple gene duplication events that are specific for both noctuid genomes; however, we report a high number of synteny breaks or small size of synteny blocks, evidence for more frequent rearrangements than in the Drosophila lineage for instance. Therefore, the view that emerges from our comparative genome analysis depends on the focus. From up close, the fragmentation is obvious, but on a larger scale conservation is maintained despite, or perhaps because of, the holocentric nature of the lepidopteran genomes. This higher order level of chromosome organization and the evolutionary constraints that have led to it in Lepidoptera remain a matter of conjecture.
Materials and Methods
BAC library construction, probe selection, and screening, BAC sequencing and annotation by Kaiko (Silkworm) Genome Automated Annotation System (KAIKOGAAS) followed by manual curation are described in detail in SI Materials and Methods. The fully annotated BACs and B. mori genome regions are available at the Lepido-DB Web site. To detect syntenic genes, each BAC sequence was compared by BlastX (threshold <10−5) to the sequences of the peptides predicted by KAIKOGAAS on the corresponding BAC in the other noctuid species or on a 200-kb region of the B. mori genome. We determined reciprocal best hits that resulted from a comparison by BlastP between two sets of peptides at a threshold of 10−6 over 75% of the peptide length. Only a partial genomic sequence (i.e., our BAC collection) is available for the two noctuid species that were compared in the present study; therefore, the reciprocal best hits may not always represent strictly orthologous genes. In the case of unique genes in the B . mori genome, this assumption can reasonably be made, however. When a gene has duplicated in one species, the closest paralogue and thus most likely orthologue was defined using phylogenetic trees built with all paralogous proteins.
To measure microsynteny block length (24), all segments within which gene order and orientation was conserved between two genomic regions were identified and the number of genes they contain was counted. The pipeline for definition of the gene categories used in our analysis of synteny conservation is outlined and explained in SI Materials and Methods and Fig. S1.
Supplementary Material
Acknowledgments
This work was supported by Genoscope project 2004/43 Génomique comparée des Lépidoptères; the Integrated Research Project for Plant, Insect and Animal Using Genome Technology (Insect Genome) by MAFF, Japan; Institut National de la Recherche Agronomique/Plant Health and Environment (H.S. and C.G.); and Agence Nationale de la Recherche Grants 06 BLAN 0346 (to R.F.), 07 GPLA 004, and 07 BLAN 0057 (to E.P.) including a postdoctoral fellowship. T.F. is the recipient of an Institut National de la Recherche Agronomique PhD fellowship. We thank Drs. D. G. Heckel, E. Jacquin-Joly, H. H. Rees, Y. Park, and T. Shinoda for kindly providing us with some hybridization probes, and reviewers for thoughtful comments.
Footnotes
The authors declare no conflict of interest.
Data deposition: The BAC sequences reported in this paper have been deposited in the GenBank database (accession nos. FP340404–FP340438; see Table S1 for a list of probes and accession numbers).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0910413107/-/DCSupplemental.
References
- 1.The International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Labandeira CC, Dilcher DL, Davis DR, Wagner DL. Ninety-seven million years of angiosperm-insect association: Paleobiological insights into the meaning of coevolution. Proc Natl Acad Sci USA. 1994;91:12278–12282. doi: 10.1073/pnas.91.25.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 4.Gall LF, Tiffney BH. A Fossil Noctuid Moth Egg from the Late Cretaceous of Eastern North America. Science. 1983;219:507–509. doi: 10.1126/science.219.4584.507. [DOI] [PubMed] [Google Scholar]
- 5.Grimaldi D, Engel MS. Evolution of the Insects. Cambridge: Cambridge University Press; 2005. p. 755. [Google Scholar]
- 6.Yamamoto K, et al. Construction of a single nucleotide polymorphism linkage map for the silkworm, Bombyx mori, based on bacterial artificial chromosome end sequences. Genetics. 2006;173:151–161. doi: 10.1534/genetics.105.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto K, et al. A BAC-based integrated linkage map of the silkworm Bombyx mori. Genome Biol. 2008;9:R21. doi: 10.1186/gb-2008-9-1-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitt GP. The ecology of Heliothis species in relation to agroecosystems. Annu Rev Entomol. 1989;34:17–52. [Google Scholar]
- 9.Sharma HC. Heliothis/Helicoverpa Management: Emerging Trends and Strategies for Future Research. New Delhi: Oxford & IBH Publishing; 2005. [Google Scholar]
- 10.Behere GT, et al. Mitochondrial DNA analysis of field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) and of its relationship to H. zea. BMC Evol Biol. 2007;7:117. doi: 10.1186/1471-2148-7-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadeau JH, Sankoff D. Counting on comparative maps. Trends Genet. 1998;14:495–501. doi: 10.1016/s0168-9525(98)01607-2. [DOI] [PubMed] [Google Scholar]
- 12.Coghlan A, Wolfe KH. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 2002;12:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zdobnov EM, Bork P. Quantification of insect genome divergence. Trends Genet. 2007;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Zdobnov EM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 15.Bourque G, Pevzner PA, Tesler G. Reconstructing the genomic architecture of ancestral mammals: Lessons from human, mouse, and rat genomes. Genome Res. 2004;14:507–516. doi: 10.1101/gr.1975204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahara K, et al. Conserved synteny of genes between chromosome 15 of Bombyx mori and a chromosome of Manduca sexta shown by five-color BAC-FISH. Genome. 2007;50:1061–1065. doi: 10.1139/g07-082. [DOI] [PubMed] [Google Scholar]
- 17.Pringle EG, et al. Synteny and chromosome evolution in the lepidoptera: Evidence from mapping in Heliconius melpomene. Genetics. 2007;177:417–426. doi: 10.1534/genetics.107.073122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papa R, et al. Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. BMC Genomics. 2008;9:345. doi: 10.1186/1471-2164-9-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beldade P, Saenko SV, Pul N, Long AD. A gene-based linkage map for Bicyclus anynana butterflies allows for a comprehensive analysis of synteny with the lepidopteran reference genome. PLoS Genet. 2009;5:e1000366. doi: 10.1371/journal.pgen.1000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasukochi Y, et al. Extensive conserved synteny of genes between the karyotypes of Manduca sexta and Bombyx mori revealed by BAC-FISH mapping. PLoS One. 2009;4:e7465. doi: 10.1371/journal.pone.0007465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quesneville H, et al. Combined evidence annotation of transposable elements in genome sequences. PLOS Comput Biol. 2005;1:166–175. doi: 10.1371/journal.pcbi.0010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38:1046–1057. doi: 10.1016/j.ibmb.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Sankoff D. Comparative mapping and genome rearrangement. Ames, IA: Iowa State Univ; 1999. pp. 124–134. [Google Scholar]
- 25.Soberon M, Gill SS, Bravo A. Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci. 2009;66:1337–1349. doi: 10.1007/s00018-008-8330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelucci C, et al. Diversity of aminopeptidases, derived from four lepidopteran gene duplications, and polycalins expressed in the midgut of Helicoverpa armigera: Identification of proteins binding the delta-endotoxin, Cry1Ac of Bacillus thuringiensis. Insect Biochem Mol Biol. 2008;38:685–696. doi: 10.1016/j.ibmb.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budatha M, Meur G, Kirti PB, Dutta-Gupta A. Characterization of Bacillus thuringiensis Cry toxin binding novel GPI anchored aminopeptidase from fat body of the moth Spodoptera litura. Biotechnol Lett. 2007;29:1651–1657. doi: 10.1007/s10529-007-9453-z. [DOI] [PubMed] [Google Scholar]
- 28.Feyereisen R. Evolution of insect P450. Biochem Soc Trans. 2006;34:1252–1255. doi: 10.1042/BST0341252. [DOI] [PubMed] [Google Scholar]
- 29.Taylor M, Zawadzki J, Black B, Kreitman M. Genome size and endopolyploidy in pyrethroid-resistant and susceptible strains of Heliothis virescens (Lepidoptera: Noctuidae) J Econ Entomol. 1993;86:1030–1034. [Google Scholar]
- 30.Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- 31.Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 32.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 33.Cordaux R. The human genome in the LINE of fire. Proc Natl Acad Sci USA. 2008;105:19033–19034. doi: 10.1073/pnas.0810202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein LD, et al. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranz JM, Casals F, Ruiz A. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res. 2001;11:230–239. doi: 10.1101/gr.162901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L. Chromosome evolution in eukaryotes: A multi-kingdom perspective. Trends Genet. 2005;21:673–682. doi: 10.1016/j.tig.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhutkar AS, et al. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao H, Bourque G. Recovering genome rearrangements in the mammalian phylogeny. Genome Res. 2009;19:934–942. doi: 10.1101/gr.086009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.