Abstract
The majority (∼70%) of surface buried in protein folding is hydrocarbon, whereas in DNA helix formation, the majority (∼65%) of surface buried is relatively polar nitrogen and oxygen. Our previous quantification of salt exclusion from hydrocarbon (C) accessible surface area (ASA) and accumulation at amide nitrogen (N) and oxygen (O) ASA leads to a prediction of very different Hofmeister effects on processes that bury mostly polar (N, O) surface compared to the range of effects commonly observed for processes that bury mainly nonpolar (C) surface, e.g., micelle formation and protein folding. Here we quantify the effects of salts on folding of the monomeric DNA binding domain (DBD) of lac repressor (lac DBD) and on formation of an oligomeric DNA duplex. In accord with this prediction, no salt investigated has a stabilizing Hofmeister effect on DNA helix formation. Our ASA-based analyses of model compound data and estimates of the surface area buried in protein folding and DNA helix formation allow us to predict Hofmeister effects on these processes. We observe semiquantitative to quantitative agreement between these predictions and the experimental values, obtained from a novel separation of coulombic and Hofmeister effects. Possible explanations of deviations, including salt-dependent unfolded ensembles and interactions with other types of surface, are discussed.
Keywords: Hofmeister salts, m-values, thermodynamics
Salts typically exert both specific (Hofmeister) and nonspecific (coulombic) effects on biomolecular processes (1–6). To manipulate and probe biopolymer processes using salts, it is extremely important to develop quantitative methods to interpret and predict these effects in terms of structure. coulombic, valence-specific effects of salt ions (due to screening of surface charges) are most significant at relatively low salt concentrations (< 0.1 M). At higher concentrations (> 0.1 M), ion-specific effects and relatively nonspecific osmotic effects (due to the lowering of water activity) become increasingly significant. In 1888, Franz Hofmeister discovered that the effectiveness of salts for protein precipitation generally followed a specific order, regardless of the protein being investigated (7). Since then, the so-called Hofmeister series of salt effects has been observed in physical properties of aqueous salt solutions (e.g., surface tension and surface potential) (8, 9), as well as salt effects on a variety of macromolecular processes (e.g., micelle formation, “salting out” nonpolar compounds, and protein folding) (10–13). The general ranking of ions, in decreasing order of effectiveness (best to worst) in driving processes where surface area is buried (e.g., folding and precipitation) or macroscopic surface is lost (transfer of water from the air–water interface to bulk), is as follows (14):
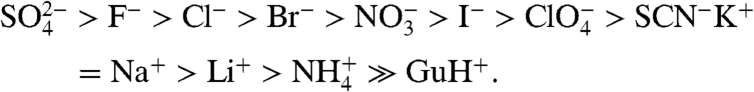 |
Although it is generally accepted that interactions of salts with hydrocarbon surface are unfavorable and salt-specific, following the above order (1, 3, 11, 14–16), less is known about the interactions of Hofmeister salts with polar N and O surfaces, and, consequently, less is known about the origin of their effects on processes that expose or bury mostly polar surface (e.g., DNA melting).
We previously quantified Hofmeister ion exclusion from, or accumulation at, hydrocarbon (C) and amide nitrogen and oxygen (N, O) surfaces and showed how the net exclusion or accumulation of salt ions affects the solubility of model hydrocarbons, peptides, and micelles (13, 17). Our analysis of these studies (13, 17) quantified the conclusion drawn by earlier investigators (15, 18) that the interactions of Hofmeister salts with amide surface are favorable and relatively nonspecific. Thus, the observed Hofmeister ordering of salt effects on solubility of small peptides is primarily due to the ∼75% of the surface that is hydrocarbon. Unfolding of globular proteins exposes a surface similar in composition to the peptides (i.e., 65–75% hydrocarbon and 15–20% amide) (19, 20). By contrast, melting of DNA duplexes exposes a surface that is only ∼35% hydrocarbon, with the remainder being primarily polar N and O (21). These differences in the compositions of the surfaces exposed have striking implications for the prediction of Hofmeister salt effects on DNA melting. Although all salts stabilize DNA against denaturation at low salt concentration for coulombic reasons, we predict from our ASA-based analysis of model compound data that no salt will have a stabilizing noncoulombic effect on DNA duplex stability due to the dominance of polar N, O surface in the ΔASA of melting. Until now, no systematic studies of the effects of the full range of Hofmeister salts on the equilibrium constant of DNA helix formation have been performed.
Effects of neutral and destabilizing Hofmeister salts on the midpoint temperature (Tm) of thermal denaturation of DNA were determined by Hamaguchi and Geiduschek in 1962 (22); the Hofmeister series order is qualitatively observed, but no stabilizing salts were included in the study for direct comparison with our predictions. Gruenwedel and coworkers investigated the effects of “stabilizing” salts (sulfates) on DNA thermal stability (23, 24); curvature in plots of Tm as a function of log[Na2SO4] was observed at high salt concentration (0.3–1.6 M), but the curvature was eliminated when analyzed using Na2SO4 activity. To compare the effects of salts on protein unfolding and DNA melting and to test our qualitative predictions about the differences between the two systems, we have systematically investigated the noncoulombic effects of biochemically relevant salts from the Hofmeister series on equilibrium constants at ∼40 °C for two-state melting of a marginally stable globular protein [the well-characterized DNA binding domain (DBD) of lac repressor, lac DBD (25)] and a marginally stable DNA oligomer duplex.
Background on Solute m-Values and Chemical Potential Derivatives μ23
Noncoulombic Hofmeister effects of salts on biopolymer processes such as folding and binding are quantified by m-values, defined as derivatives with respect to salt concentration (m3) of the observed standard free energy change of the process 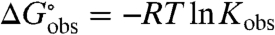 , where Kobs is the observed equilibrium concentration quotient (expressed in terms of concentrations and not thermodynamic activities):
, where Kobs is the observed equilibrium concentration quotient (expressed in terms of concentrations and not thermodynamic activities):
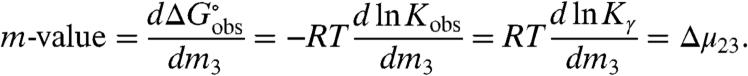 |
[1] |
In Eq. 1, Kγ is the quotient of biopolymer activity coefficients corresponding to the concentration quotient Kobs and μ23 = RTd ln γ2/dm3†; μ23 is closely related to the preferential interaction coefficient Γμ3 (4, 5), which can be predicted if the ion distributions near the biopolymer are known (26, 27).
Classically, solubility measurements have been used to determine quantities analogous to μ23. The observable is the derivative 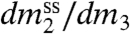 , the change in solubility (
, the change in solubility (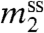 , the concentration in the saturated solution) of a model compound or biopolymer as the concentration of the perturbing solute (m3) is changed. This derivative is related to by the following approximate equation (valid at low to moderate
, the concentration in the saturated solution) of a model compound or biopolymer as the concentration of the perturbing solute (m3) is changed. This derivative is related to by the following approximate equation (valid at low to moderate 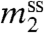 ):
):
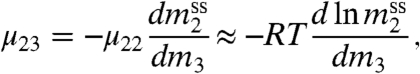 |
[2] |
where μ22 = (RT/m2)(1 + d ln γ2/d ln m2) can be approximated as RT/m2 for sparingly soluble model compounds.
As a first level of interpretation of an experimental value of Δμ23/RT or μ23/RT (see Eqs. 1 and 2), we dissect it into additive contributions from chemically distinct, coarse-grained surface types (17, 28–31). This is analogous to the Tanford-Bolen approach, which assumes that a solute m-value for protein unfolding can be decomposed into additive contributions from the 20 side chains and the peptide backbone units exposed in unfolding. Our use of chemically distinct surface types (e.g., aliphatic and aromatic hydrocarbon, amide oxygen, and nitrogen) allows one to use fewer terms in the dissection and to obtain a more straightforward molecular interpretation of the values obtained. We propose that the contribution of each type of surface (i) to μ23/RT is the product of a solute interaction potential [contribution per unit of ASA; (μ23/RTASA)i] and the ASA of that surface. The experimental value of the chemical potential derivative μ23/RT is therefore represented as the sum of terms:
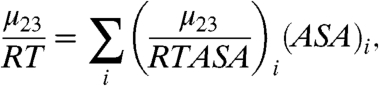 |
[3] |
where the interaction potential (μ23/RTASA)i quantifies the interaction of the salt of interest with one Å2 of surface of type i on any compound or biopolymer, and (ASA)i is the water-accessible area in Å2 of surface type i on the model compound or biopolymer being analyzed. (The relevant surface for a biopolymer process with a corresponding m-value, or Δμ23, is the ASA exposed or buried, or ΔASA.) The observed chemical potential derivatives μ23/RT are model-independent thermodynamic quantities; the solute potentials (μ23/RTASA)i therefore provide a model-independent description of the effect of the solute per unit area of a particular type of water-accessible surface on the biomolecule or model compound.
If salt interaction potentials are known for various biopolymer surface types and ΔASA composition is available from structural data, then Eq. 3 can be used to predict the effect of any salt on any biopolymer process. We previously analyzed salt concentration and identity dependences for hydrocarbon and peptide solubility (Eq. 2) and reported salt (and salt ion) partition coefficients [Kp, where μ23/RTASA ∝ (Kp - 1) quantifying exclusion or accumulation near (aliphatic and aromatic) hydrocarbon and amide surfaces] (17). Here, the solubility data analyzed in ref. 17 were globally fit for each salt to determine values of (μ23/RTASA)i (as in ref. 31), presented in Table 1.
Table 1.
Salt-surface interaction potentials from ASA-based analysis of solubility/distribution studies with model compounds
| μ23/(RTASA) × 103, (m-1 Å-2)* | |||
| Salt | Aliphatic C | Aromatic C | Amide (O,N) |
| Na2SO4 | 6.0 ± 0.1 | 5.9 ± 0.2 | −6.8 ± 0.5 |
| (NH4)2SO4 | 3.8 ± 0.1 | −3.3 ± 0.3 | |
| KF | 3.4 ± 0.1 | 2.4 ± 0.1 | −4.6 ± 0.3 |
| GuH2SO4† | 2.2 ± 0.3 | 0.3 ± 0.5 | −5.8 ± 1.0 |
| NaCl | 2.2 ± 0.1 | 2.0 ± 0.1 | −4.2 ± 0.2 |
| KCl | 2.0 ± 0.1 | 1.7 ± 0.1 | −4.2 ± 0.2 |
| KBr | 1.7 ± 0.1 | 1.2 ± 0.2 | −4.1 ± 0.4 |
| NaClO4 | 0.9 ± 0.1 | 1.1 ± 0.1 | −7.0 ± 0.3 |
| NH4Cl | 0.8 ± 0.3 | −1.2 ± 0.8 | |
| GuHCl | 0.3 ± 0.1 | −0.8 ± 0.2 | −3.7 ± 0.4 |
*Model compound dataset is identical to that in ref. 17. Ammonium salts were not included in ref. 17 due to scarcity of data; values here are estimates obtained from analysis of two [(NH4)2SO4] or three (NH4Cl) solubility μ23/RT values and one vapor pressure osmometry μ23/RT value (see Fig. S1). Salts are assumed to be neither accumulated at, nor excluded from, small amounts of ester oxygen surface (6–13% of total ASA) on model peptides.
†Obtained from Na2SO4, NaCl, and GuHCl values assuming ion additivity.
Results and Analysis
Salt Effects on lac DBD and DNA Melting.
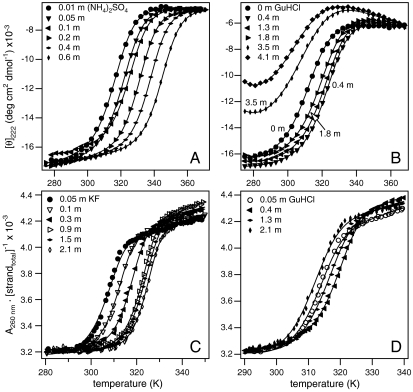
Fig. 1 shows typical unfolding/melting curves for lac DBD and a 12-bp DNA duplex; for each process the effects of salts from both “stabilizing” and “destabilizing” ends [(NH4)2SO4 and KF; GuHCl] of the Hofmeister series are shown. For both processes, at low salt concentrations (≲0.5 m), increases in concentrations of all salts increase the stability of the ordered, higher charge density state (i.e., the midpoint temperature for the melting transition increases with salt concentration). Higher concentrations of GuHCl reverse this initial trend and destabilize both lac DBD and DNA (Fig. 1B and D). The effects of the “stabilizers” at high concentrations are starkly different for the two processes. As shown in Fig. 1A, over the whole range of (NH4)2SO4 concentrations for which the entire transition can be observed (10–600 mmol/kg), the midpoint temperature increases monotonically with concentration. (This is also observed qualitatively at higher concentrations, where melting does not reach completion by 98 °C.) In contrast, the stabilizing effect of KF on DNA duplex formation reaches a plateau at high concentrations (Fig. 1C).
Fig. 1.
Thermal melting transitions of lac DBD and DNA oligomer at different salt concentrations. Protein unfolding data for Na2SO4 (A) and GuHCl (B) and 12 bp DNA melting data for KF (C) and GuHCl (D). Mean residue molar ellipticities [θ]222 and absorbances (A260) are plotted vs. temperature. Curves indicate global fits to the two-state melting transitions.
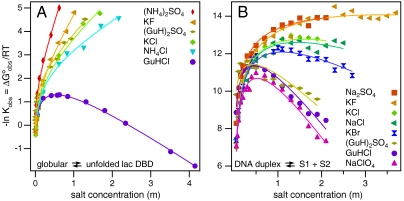
Melting equilibrium constants and standard free energy changes (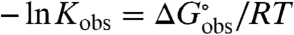 ) at ∼40 °C, obtained from fits of the transition curves (see Materials and Methods), are shown as a function of salt concentration in Fig. 2. Qualitatively, the protein unfolding data are consistent with a large body of Tm data as a function of salt concentration (32–36); our study is the only systematic investigation of effects of salts from the full range of the Hofmeister series on protein unfolding and DNA duplex dissociation in which isothermal equilibrium constants and
) at ∼40 °C, obtained from fits of the transition curves (see Materials and Methods), are shown as a function of salt concentration in Fig. 2. Qualitatively, the protein unfolding data are consistent with a large body of Tm data as a function of salt concentration (32–36); our study is the only systematic investigation of effects of salts from the full range of the Hofmeister series on protein unfolding and DNA duplex dissociation in which isothermal equilibrium constants and 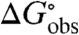 as a function of concentration are obtained. For both processes, the curves exhibit two qualitatively different regions of effects of salts on thermal stability. As mentioned above, at low concentrations, all salts, whether typically considered stabilizers or destabilizers, increase the stability of the native state to similar extents as salt concentration is increased. At higher salt concentrations, Fig. 2 shows very different effects of salts from the “stabilizing” end of the Hofmeister series on protein unfolding and DNA melting. For lac DBD unfolding in all salts except GuHCl,‡ native state stability continues to increase with salt concentration up to the highest concentrations investigated; the effectiveness of the salts as stabilizing agents differs though, manifesting in different high-concentration values of Δμ23/RT (equivalent to m-value/RT). This is not observed for DNA melting in high concentrations of salts; on the contrary, the range of effects observed here includes only varying degrees of effectiveness at destabilizing the duplex. No salt is observed to stabilize the duplex at concentrations above ∼1.5 m; increasing concentrations of KF and Na2SO4, salts containing anions from the “stabilizing” end of the Hofmeister series, appear to have no effect on thermal stability of the duplex at these high salt concentrations.
as a function of concentration are obtained. For both processes, the curves exhibit two qualitatively different regions of effects of salts on thermal stability. As mentioned above, at low concentrations, all salts, whether typically considered stabilizers or destabilizers, increase the stability of the native state to similar extents as salt concentration is increased. At higher salt concentrations, Fig. 2 shows very different effects of salts from the “stabilizing” end of the Hofmeister series on protein unfolding and DNA melting. For lac DBD unfolding in all salts except GuHCl,‡ native state stability continues to increase with salt concentration up to the highest concentrations investigated; the effectiveness of the salts as stabilizing agents differs though, manifesting in different high-concentration values of Δμ23/RT (equivalent to m-value/RT). This is not observed for DNA melting in high concentrations of salts; on the contrary, the range of effects observed here includes only varying degrees of effectiveness at destabilizing the duplex. No salt is observed to stabilize the duplex at concentrations above ∼1.5 m; increasing concentrations of KF and Na2SO4, salts containing anions from the “stabilizing” end of the Hofmeister series, appear to have no effect on thermal stability of the duplex at these high salt concentrations.
Fig. 2.
Equilibrium constants (ln Kobs) for lac DBD unfolding and DNA oligomeric duplex melting are plotted as a function of molal salt concentration (mol/kg). Curves are fit to a three- or four-parameter equation for the separation of Hofmeister from coulombic salt effects (see Materials and Methods).
In order to quantify the Hofmeister m-values, coulombic effects must be extracted. We have fit the curves in Fig. 2 to equations containing nonlinear Poisson Boltzmann (NLPB)-predicted functional forms expected for coulombic effects of salts on protein unfolding and oligomeric duplex dissociation, respectively (see Materials and Methods for more details) along with a term that is linear in salt concentration [the slope of which is m-value/RT, or h (Eq. 7), quantifying the Hofmeister effect]. Hofmeister coefficients obtained from these fits are shown in Table 2; the functional forms used appear justified by the quality of the fits for both transitions at all concentrations of salts studied and the reasonable coulombic parameters obtained (see Materials and Methods). This dimension of the work will be discussed in a forthcoming paper.
Table 2.
Comparison of experimental Hofmeister salt m-values for protein (lac DBD) unfolding and 12-bp DNA melting (Eq. 7) with predictions from an ASA analysis of model compound data
| lac DBD unfolding m-value/RT, (m-1) | 12-bp DNA duplex melting m-value/RT, (m-1) | |||
| Salt | Experimental | Predicted* | Experimental | Predicted† |
| Na2SO4 | — | — | −0.1 ± 0.2 | −0.2 ± 0.5 |
| (NH4)2SO4 | 4.6 ± 0.2 | 5.4 ± 0.2 | — | — |
| KF | 2.3 ± 0.1 | 3.9 ± 0.5 | −0.4 ± 0.1 | −1.3 ± 0.3 |
| (GuH)2SO4 | 1.5 ± 0.1 | 1.0 ± 0.5 | −2.6 ± 0.3 | −3.1 ± 0.3 |
| KCl | 1.3 ± 0.1 | 1.6 ± 0.2 | −1.0 ± 0.1 | −1.8 ± 0.2 |
| NaCl | — | — | −1.1 ± 0.1 | −1.7 ± 0.2 |
| NH4Cl | 0.8 ± 0.1 | 0.9 ± 0.6 | — | — |
| KBr | — | — | −1.5 ± 0.1 | −2.1 ± 0.3 |
| GuHCl | −1.2 ± 0.1 | −1.3 ± 0.3 | −3.6 ± 0.2 | −3.5 ± 0.4 |
| NaClO4 | — | — | −3.6 ± 0.2 | −4.3 ± 0.3 |
*Based on 92% of ΔASA; remaining 8% assigned zero contribution.
†Based on 64% of ΔASA; remaining 36% assigned zero contribution.
Comparison Between Observed m-Values for lac DBD Unfolding and Predictions from Model Compound Analysis.
For lac DBD unfolding, which exposes mainly hydrocarbon surface, Hofmeister effects of salts range from very stabilizing [4.6 m-1 for (NH4)2SO4] to moderately destabilizing (-1.2 m-1 for GuHCl). Hydrocarbon and amide N and O comprise ∼90% of the ΔASA exposed in this process, so the interaction potentials in Table 1, along with the composition of this ΔASA, can be applied to predict the high-concentration Hofmeister slope (m-value/RT = Δμ23/RT = h; Eqs. 1, 3, and 7). To obtain the ΔASA for this process, a model for the unfolded state is required. In previous analyses (20, 25), we successfully used a fully extended model for the unfolded state; initial calculations using this maximum ΔASA suggested that this state was not appropriate for solutes that interact strongly (here unfavorably) with hydrocarbon surface. The recent web application ProtSA generates sequence-dependent atomic models of unfolded proteins (39, 40); tests for selected proteins indicate consistency with experimental residual dipolar couplings and small-angle x-ray scattering data (41). ProtSA was therefore used to generate an unfolded ensemble for the 51 residue lac DBD, consisting of 1,919 acceptable conformations. The average ASA is ∼6400 Å (see Materials and Methods), approximately 85% of the accessible area calculated for a completely extended chain. The 40 conformers of Protein Data Bank (42) PDB ID code 1OSL [first 51 residues of both monomers of the nonspecifically bound lac headpiece dimer (43)] were used to model the folded state. The resulting average ΔASA composition is presented in Table S1.
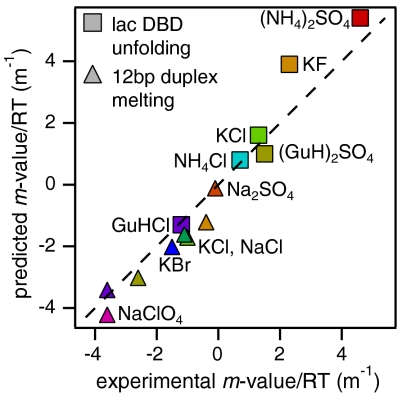
A quantitative comparison of predicted values of Δμ23/RT (using the interaction coefficients in Table 1 and the ΔASA composition in Table S1) with experimental h coefficients (Eq. 7) is shown in Fig. 3 and Table 2. Over the entire range of effects (h values from -1.2 to +4.6 m-1), good to excellent agreement is attained. For the set of six salts studied with lac DBD, a root mean square deviation of 0.8 is observed for predicted and observed values of h. The predictions for strongly perturbing salts [e.g., KF, (NH4)2SO4], are the farthest from the experimentally observed values. For example, the predicted Hofmeister m-value/RT for KF (3.9 m-1) is 70% larger than the observed value of 2.3 m-1. Deviations at the highly excluded end of the spectrum of salts could be explained by an effect of the salt identity on the unfolded ensemble, possibly analogous to what we previously observed for the model process of micelle formation, where better agreement was observed between calculated and observed salt effects when more headgroups were assumed to be buried in excluded salt solutions (13). Here, compaction of the unfolded state in excluded salt solutions could result in the larger discrepancies observed for those salts.
Fig. 3.
Plot of predicted vs. observed Hofmeister salt Δμ23/RT values for lac DBD unfolding and DNA oligomeric duplex melting. Predicted values are determined from the ΔASA compositions for the two processes (Table S1) and the interaction potentials in Table 1. Observed values of Δμ23/RT are the h coefficients (Eq. 7) obtained from the global fits shown in Fig. 2. The dashed line represents equality of predicted and observed values.
Comparison Between Observed m-Values for 12 bp DNA Melting and Predictions from Model Compound Analysis.
Hofmeister coefficients for DNA duplex melting, obtained from the fits in Fig. 2, are shown in Table 2. As might be expected from the accumulation of all salts at (amide) nitrogen and oxygen surface, the values of h (Eq. 7) for DNA melting are all negative, indicating that the Hofmeister effects of all salts studied on duplex DNA are destabilizing. For example, Hofmeister effects of salts range from completely neutral (h = -0.1 m-1 for Na2SO4) to modestly destabilizing (h = -1.5 m-1 for KBr) to very destabilizing (h = -3.6 m-1 for GuHCl and NaClO4).
In order to use model compound data to predict effects of Hofmeister salts on DNA melting (h coefficients), a model for the partially unstacked DNA strands is necessary. No program to do this (i.e., comparable to ProtSA) is currently available. As an approximation, we assume that (i) the only surface area that is newly exposed upon melting is on the DNA bases and (ii) the amount of surface area exposed on each base is proportional to the overall extent of base unstacking in each denatured strand (see Table S1). An additional assumption arises in classification of surface area types. Unlike lac DBD unfolding, melting of the DNA duplex exposes functional groups not included in the model compound analysis (e.g., amino nitrogens and heteroatomic aromatic rings). As a first approximation, carbonyl oxygens and adjacent nitrogens are classified as “amide” (N, O), and all types of surface not in Table 1 are assigned null interaction (i.e., salts are neither accumulated nor excluded).
Shown in Fig. 3 (and in Table 2) is the comparison between the predicted values obtained as described above and the observed Hofmeister effects obtained from the fits. The predictions here are only for 64% of the surface, yet the root mean square deviations for the predicted and observed salt m-values for duplex melting (0.7) and lac DBD unfolding (0.8) are similar. Remarkably, the predictions correctly capture the difference in rank order of (GuH)2SO4 for DNA melting vs. protein unfolding. Whereas (GuH)2SO4 has long been known to be a relatively inert protein cosolute [i.e., neither a strong stabilizer nor a strong denaturant (3)], the increased amount of N and O surface exposed in DNA helix dissociation and unstacking shifts this salt toward much greater accumulation (see Table 1) and hence destabilization. Except for Na2SO4 and GuHCl, predicted values of Δμ23/RT are consistently more negative (by 0.5–0.9 m-1) than observed values. This suggests that the salts interact favorably (μ23/RTASA = -0.7 × 10-3) with the composite of the surface types not included in the calculation; we are testing this prediction in model compound studies.
Concluding Discussion.
Salts of ions from the extremes of the Hofmeister series are widely used at high concentrations in biochemistry (e.g., ammonium sulfate for precipitation and crystallization; guanidinium chloride or thiocyanate for denaturation), but the molecular basis of their ion-specific effects remains controversial. Hofmeister ions have often been discussed as either water structure breakers (“chaotropes”) or structure makers (“kosmotropes”), and Hofmeister effects have been attributed to the “ordering” or “disordering” of bulk water structure effected by a particular ion; however, proposals of long-range ordering (or disordering) of water in concentrated salt solutions are not supported by water activity data (44) nor by spectroscopic results (45, 46). Pioneering investigations of Nandi and Robinson and others [salt effects on solubility of model compounds (10, 11, 15, 47)], von Hippel and others [recycling chromatography on polyacrylamide columns (18, 48)], and Arakawa and Timasheff [preferential interactions of salts with native proteins (16)] have emphasized the importance of the competition between salt ions and water for interactions with molecular and macromolecular surfaces. The various effects of different ions on water structure (bulk and/or local), although potentially of interest, are not the direct origins of thermodynamic effects on biomolecular processes. Instead, nonuniform ion distributions (accumulation and exclusion) near surfaces are responsible for the thermodynamic effects of concentration of Hofmeister salts on a very wide range of aqueous processes (4, 5, 28).
The idea that direct interactions of ions (in competition with water) with nonpolar surface (e.g., air–water and molecular hydrocarbon) are the source of the Hofmeister effect has gained traction in recent years (13, 17, 49–51). Our work quantifying the accumulation or exclusion of Hofmeister ions near various biochemically relevant surfaces (13, 17, 52, 53) has allowed us to make qualitative predictions about the difference between Hofmeister effects on protein folding and DNA melting. Additionally, we have obtained isothermal m-values (i.e., h coefficients) for the effects of a wide range of Hofmeister salts on globular protein unfolding and DNA duplex melting. Results for both processes are in semiquantitative to quantitative agreement with our predictions based on the model compound analyses. The deviations observed for lac DBD could be explained by an effect of the salt identity on the unfolded ensemble, similar to what we previously observed for the model process of micelle formation (13), whereas those for DNA could indicate the existence of favorable, relatively nonspecific interactions of the salts studied with the 35% of DNA ΔASA for which we have no model compound data.
Materials and Methods
Calculation of Kobs and 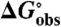 from CD and UV Data.
from CD and UV Data.
Circular dichroism data at 222 nm (see SI Text for experimental details), characterizing the helicity of the lac DBD, were converted to mean residue ellipticity ([θ]222, in units of deg cm2 dmol-1), and the unfolded fraction (fU) of the population was calculated as a function of temperature:
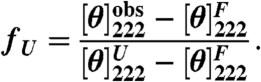 |
[4] |
Completely analogously, the DNA absorbance data at 260 nm can be used to obtain the single-stranded fraction of the population (fss) as a function of temperature. The use of Eq. 4 and the analogous equation containing the absorbances of the double- and single-stranded components requires the establishment of baselines denoting the native and denatured states. For lac DBD unfolding in all salts except GuHCl, linear baselines with a single linked slope and floated intercepts (due to variations in instrumental baseline) were determined by globally fitting datasets for a particular salt. A previous study of thermal unfolding of lac DBD in urea found that the upper baseline was a quadratic function of temperature (25); this behavior is not observed for the salts in the current dataset. For GuHCl, the upper baseline slope was allowed to float, due to clear differences between the different salt concentrations; i.e., as GuHCl concentration increases, the upper baseline gets steeper (cf. Fig. 1B). For DNA melting, baseline slopes and intercepts were floated.
For two-state protein unfolding (F↔U), the observed equilibrium constant for the process is simply the equilibrium concentration ratio of the unfolded and folded populations (determined by the upper and lower baselines):
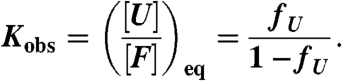 |
[5] |
For two-state DNA oligomeric duplex melting (duplex↔S1 + S2), the observed equilibrium constant for the process depends on the total strand concentration ([str]total = [S1] + [S2] + [duplex]) but is equally easily determined using the fraction of the population in the melted (single-stranded) state:
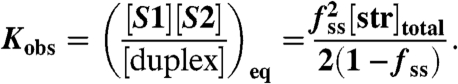 |
[6] |
To determine various thermodynamic parameters for different temperatures and solution conditions, nonlinear regression was performed using IgorPro 5.04B to fit Kobs to the constant ΔCp van’t Hoff equation (25).
Extraction of Coulombic and Hofmeister Contributions to Observed Salt Effects on Protein Unfolding and DNA Duplex Melting.
In order to make comparisons to the predictions of Hofmeister salt effects based on the model compound analysis, long-range coulombic salt effects for both processes must be extracted. NLPB calculations at low salt concentrations indicate a linear dependence of ln Kobs on the logarithm of the salt concentration (ln m3). Although this appears sufficient for modeling the coulombic contribution to lac DBD stability at all salt concentrations, NLPB calculations indicate that an additional quadratic dependence on ln m3 is required for the DNA oligomer. Hofmeister effects on ln Kobs are typically observed to be linear in salt concentration (13). Assuming additivity of free energy contributions for coulombic and Hofmeister effects yields a simple equation to describe the salt dependences of ln Kobs over the full range of concentrations:
| [7] |
In Eq. 7, the parameter h is the Hofmeister slope (linear in m3) at high concentrations. All other parameters describe the coulombic effects (where x = 0 for lac DBD unfolding), where the term b is an offset, equal to the fit value of ln Kobs at 1-m salt and h = 0.
For the fit to lac DBD unfolding data as a function of salt concentration (Fig. 2), the “coulombic” parameters in Eq. 7 (b and c) were linked for salts of the same valence. Uni-divalent and uni-univalent salts have similar values of c (∼0.5 and ∼0.7, similar to the NLPB results of 0.52 and 0.62) and b (∼2.4). For the four-parameter fits to the DNA melting data, the limiting slope in ln(m3), c, was linked for all salts of the same valence. The value obtained for the uni-univalent salts (c = 1.66) is within 5% of the NLPB calculated value (1.74). For KF, KCl, NaCl, and KBr, b and x were also linked. For GuHCl and NaClO4, these parameters were not linked to the other 1∶1 salts, and in the case of GuHCl (also GuH2SO4), x had to be constrained to be greater than zero. For the two uni-divalent salts, c was constrained to be within 10% of the NLPB calculated value (1.23) based on the results for the uni-univalent salts. The coefficient x varies widely depending on the structural model used for the NLPB calculations; the values obtained are reasonably consistent with those observed in the NLPB calculations (see Table S2).
Surface Area Calculations.
Water-accessible surface areas (ASA) are calculated using Surface Racer (54) with the Richards’ set of van der Waals radii (55) and a 1.4 Å probe radius for water. A unified atom model is used, wherein hydrogens are treated as part of the atom to which they are covalently bonded. Table S1 contains the results and additional details of the lac DBD and oligomeric duplex ΔASA calculations.
Supplementary Material
Acknowledgments.
The authors gratefully acknowledge Demian Riccardi, Ruth Saecker, and Kirk Vander Meulen for many helpful discussions, Darrell McCaslin for technical assistance with the CD spectrometer (Biophysics Instrumentation Facility), Melissa Witmer Anderson for assistance with lac DBD purification, Wayne Kontur for initial experimental guidance, Mike Capp for quantifying the interactions of ammonium salts with acetyl alanine methylamide, and Jorge Estrada for providing the PDB files for the lac DBD unfolded ensemble. The authors also thank the reviewers for their thoughtful comments and suggestions. This work was supported by National Institutes of Health Grant GM47022 (to M.T.R.)
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913376107/DCSupplemental.
†Throughout, subscripts 1, 2, and 3 refer to water, biopolymer or model compound, and salt, respectively.
References
- 1.von Hippel PH, Schleich T. Ion effects on the solution structure of biological macromolecules. Accounts Chem Res. 1969;2:257–265. [Google Scholar]
- 2.Barkley MD, Lewis PA, Sullivan GE. Ion effects on the lac repressor—operator equilibrium. Biochemistry. 1981;20:3842–3851. doi: 10.1021/bi00516a027. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin RL. How Hofmeister ion interactions affect protein stability. Biophys J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timasheff SN. Control of protein stability and reactions by weakly interacting cosolvents: The simplicity of the complicated. Adv Protein Chem. 1998;51:355–432. doi: 10.1016/s0065-3233(08)60656-7. [DOI] [PubMed] [Google Scholar]
- 5.Record MT, Zhang W, Anderson CF. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: A practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv Protein Chem. 1998;51:281–353. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 6.Kozlov AG, Lohman TM. Effects of monovalent anions on a temperature-dependent heat capacity change for Escherichia coli SSB tetramer binding to single-stranded DNA. Biochemistry. 2006;45:5190–5205. doi: 10.1021/bi052543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmeister F. Zur lehre von der wirkung der salze. Arch Exp Pathol Pharmakol. 1888;24:247–260. [Google Scholar]
- 8.Frumkin A. Phase-boundary forces and adsorption at the interface air: Solutions of inorganic salts. Z Phys Chem. 1924;109:34–48. [Google Scholar]
- 9.Jarvis NL, Scheiman MA. Surface potentials of aqueous electrolyte solutions. J Phys Chem. 1968;72:74–78. [Google Scholar]
- 10.Long FA, McDevit WF. The activity coefficient of benzene in aqueous salt solutions. J Am Chem Soc. 1952;74:1773–1777. [Google Scholar]
- 11.Nandi PK, Robinson DR. The effects of salts on the free energy of the peptide group. J Am Chem Soc. 1972;94:1299–1308. doi: 10.1021/ja00759a042. [DOI] [PubMed] [Google Scholar]
- 12.Ray A, Némethy G. Effects of ionic protein denaturants on micelle formation by nonionic detergents. J Am Chem Soc. 1971;93:6787–6793. doi: 10.1021/ja00754a014. [DOI] [PubMed] [Google Scholar]
- 13.Pegram LM, Record MT. Quantifying accumulation or exclusion of H+, HO-, and Hofmeister salt ions near interfaces. Chem Phys Lett. 2008;467:1–8. doi: 10.1016/j.cplett.2008.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Hippel PH, Wong KY. Neutral salts. The generality of their effects on the stability of macromolecular conformations. Science. 1964;145:577–580. doi: 10.1126/science.145.3632.577. [DOI] [PubMed] [Google Scholar]
- 15.Schrier EE, Schrier EB. The salting-out behavior of amides and its relation to the denaturation of proteins by salts. J Phys Chem. 1967;71:1851–1860. doi: 10.1021/j100865a049. [DOI] [PubMed] [Google Scholar]
- 16.Arakawa T, Timasheff SN. Preferential interactions of proteins with salts in concentrated solutions. Biochemistry. 1982;21:6545–6552. doi: 10.1021/bi00268a034. [DOI] [PubMed] [Google Scholar]
- 17.Pegram LM, Record MT. Thermodynamic origin of Hofmeister ion effects. J Phys Chem B. 2008;112:9428–9436. doi: 10.1021/jp800816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamabata A, von Hippel PH. Model studies on the effects of neutral salts on the conformational stability of biological macromolecules. II. Effects of vicinal hydrophobic groups on the specificity of binding of ions to amide groups. Biochemistry. 1973;12:1264–1271. doi: 10.1021/bi00731a004. [DOI] [PubMed] [Google Scholar]
- 19.Courtenay ES, Capp MW, Record MT. Thermodynamics of interactions of urea and guanidinium salts with protein surface: Relationship between solute effects on protein processes and changes in water-accessible surface area. Protein Sci. 2001;10:2485–2497. doi: 10.1110/ps.ps.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong J, Capp MW, Saecker RM, Record MT. Use of urea and glycine betaine to quantify coupled folding and probe the burial of DNA phosphates in lac repressor–lac operator binding. Biochemistry. 2005;44:16896–16911. doi: 10.1021/bi0515218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holbrook JA, Capp MW, Saecker RM, Record MT. Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: Interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry. 1999;38:8409–8422. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 22.Hamaguchi K, Geiduschek EP. The effect of electrolytes on the stability of the deoxyribonucleate helix. J Am Chem Soc. 1962;84:1329–1338. [Google Scholar]
- 23.Gruenwedel DW, Hsu C-H. Salt effects on the denaturation of DNA. Biopolymers. 1969;7:557–570. doi: 10.1002/bip.1969.360070412. [DOI] [PubMed] [Google Scholar]
- 24.Gruenwedel DW, Hsu CH, Lu DS. The effects of aqueous neutral-salt solutions on the melting temperatures of deoxyribonucleic acids. Biopolymers. 1971;10:47–68. doi: 10.1002/bip.360100106. [DOI] [PubMed] [Google Scholar]
- 25.Felitsky DJ, Record MT. Thermal and urea-induced unfolding of the marginally stable lac repressor DNA-binding domain: A model system for analysis of solute effects on protein processes. Biochemistry. 2003;42:2202–2217. doi: 10.1021/bi0270992. [DOI] [PubMed] [Google Scholar]
- 26.Pierce V, Kang M, Aburi M, Weerasinghe S, Smith PE. Recent applications of Kirkwood-Buff theory to biological systems. Cell Biochem Biophys. 2008;50:1–22. doi: 10.1007/s12013-007-9005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni HH, Anderson CF, Record MT. Quantifying the thermodynamic consequences of cation (M2+, M+) accumulation and anion (X-) exclusion in mixed salt solutions of polyanionic DNA using Monte Carlo and Poisson-Boltzmann calculations of ion-polyion preferential interaction coefficients. J Phys Chem B. 1999;103:3489–3504. [Google Scholar]
- 28.Courtenay ES, Capp MW, Saecker RM, Record MT. Thermodynamic analysis of interactions between denaturants and protein surface exposed on unfolding: Interpretation of urea and guanidinium chloride m-values and their correlation with changes in accessible surface area (ASA) using preferential interaction coefficients and the local-bulk domain model. Proteins: Struct, Funct, Genet. 2000;41:72–85. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Felitsky DJ, et al. The exclusion of glycine betaine from anionic biopolymer surface: Why glycine betaine is an effective osmoprotectant but also a compatible solute. Biochemistry. 2004;43:14732–14743. doi: 10.1021/bi049115w. [DOI] [PubMed] [Google Scholar]
- 30.Cannon JG, Anderson CF, Record MT. Urea-amide preferential interactions in water: Quantitative comparison of model compound data with biopolymer results using water accessible surface areas. J Phys Chem B. 2007;111:9675–9685. doi: 10.1021/jp072037c. [DOI] [PubMed] [Google Scholar]
- 31.Capp MW, et al. Quantifying interactions of the osmolyte glycine betaine with molecular surfaces in water: Thermodynamics, structural interpretation, and prediction of m-values. Biochemistry. 2009;48:10372–10379. doi: 10.1021/bi901273r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Von Hippel PH, Wong KY. The effect of ions on the kinetics of formation and the stability of the collagenfold. Biochemistry. 1962;1:664–674. doi: 10.1021/bi00910a020. [DOI] [PubMed] [Google Scholar]
- 33.KomsaPenkova R, Koynova R, Kostov G, Tenchov B. Thermal stability of calf skin collagen type I in salt solutions. BBA-Protein Struct M. 1996;1297(2):171–181. doi: 10.1016/s0167-4838(96)00092-1. [DOI] [PubMed] [Google Scholar]
- 34.Richard AJ, et al. Thermal stability landscape for Klenow DNA polymerase as a function of pH and salt concentration. BBA-Proteins Proteom. 2006;1764:1546–1552. doi: 10.1016/j.bbapap.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 35.Sedlak E, Stagg L, Wittung-Stafshede P. Effect of Hofmeister ions on protein thermal stability: Roles of ion hydration and peptide groups. Arch Biochem Biophys. 2008;479:69–73. doi: 10.1016/j.abb.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Tadeo X, Lopez-Mendez B, Castano D, Trigueros T, Millet O. Protein stabilization and the Hofmeister effect: The role of hydrophobic solvation. Biophys J. 2009;97:2595–2603. doi: 10.1016/j.bpj.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scholtz JM, Baldwin RL. Perchlorate-induced denaturation of ribonuclease A: Investigation of possible folding intermediates. Biochemistry. 1993;32:4604–4608. doi: 10.1021/bi00068a017. [DOI] [PubMed] [Google Scholar]
- 38.Maison W, Kennedy R, Kemp D. Chaotropic anions strongly stabilize short, n-capped uncharged peptide helicies: A new look at the perchlorate effect. Angew Chem Int Ed Engl. 2001;40:3819–3821. doi: 10.1002/1521-3773(20011015)40:20<3819::AID-ANIE3819>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 39.Bernado P, Blackledge M, Sancho J. Sequence-specific solvent accessibilities of protein residues in unfolded protein ensembles. Biophys J. 2006;91:4536–4543. doi: 10.1529/biophysj.106.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estrada J, Bernadó P, Blackledge M, Sancho J. ProtSA: A web application for calculating sequence specific protein solvent accessibilities in the unfolded ensemble. BMC Bioinformatics. 2009;10:104–111. doi: 10.1186/1471-2105-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernado P, et al. A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc Natl Acad Sci USA. 2005;102:17002–17007. doi: 10.1073/pnas.0506202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalodimos CG, et al. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 44.Robinson RA, Stokes RH. Electrolyte Solutions. London: Butterworths; 1959. pp. 483–490. [Google Scholar]
- 45.Omta AW, Kropman MF, Woutersen S, Bakker HJ. Negligible effect of ions on the hydrogen-bond structure in liquid water. Science. 2003;301:347–349. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 46.Omta AW, Kropman MF, Woutersen S, Bakker HJ. Influence of ions on the hydrogen-bond structure in liquid water. J Chem Phys. 2003;119:12457–12461. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 47.Nandi PK, Robinson DR. The effects of salts on the free energies of nonpolar groups in model peptides. J Am Chem Soc. 1972;94:1308–1315. doi: 10.1021/ja00759a043. [DOI] [PubMed] [Google Scholar]
- 48.von Hippel PH, Peticolas V, Schack L, Karlson L. Model studies on the effects of neutral salts on the conformational stability of biological macromolecules. I. Ion binding to polyacrylamide and polystyrene columns. Biochemistry. 1973;12:1256–1264. doi: 10.1021/bi00731a003. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu S, McLaren WM, Matubayasi N. The Hofmeister series and protein-salt interactions. J Chem Phys. 2006;124:234905. doi: 10.1063/1.2206174. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Cremer PS. Interactions between macromolecules and ions: The Hofmeister series. Curr Opin Chem Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 51.Lund M, Vrbka L, Jungwirth P. Specific ion binding to nonpolar surface patches of proteins. J Am Chem Soc. 2008;130:11582–11583. doi: 10.1021/ja803274p. [DOI] [PubMed] [Google Scholar]
- 52.Pegram LM, Record MT. Partitioning of atmospherically relevant ions between bulk water and the water/vapor interface. Proc Natl Acad Sci USA. 2006;103:14278–14281. doi: 10.1073/pnas.0606256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pegram LM, Record MT. Hofmeister salt effects on surface tension arise from partitioning of anions and cations between bulk water and the air-water interface. J Phys Chem B. 2007;111:5411–5417. doi: 10.1021/jp070245z. [DOI] [PubMed] [Google Scholar]
- 54.Tsodikov OV, Record MT, Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J Comput Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 55.Livingstone JR, Spolar RS, Record MT. Contribution to the thermodynamics of protein folding from the reduction in water-accessible nonpolar surface area. Biochemistry. 1991;30:4237–4244. doi: 10.1021/bi00231a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.