Abstract
Genome-wide association studies (GWAS) facilitate the discovery of genotype–phenotype relations from population-based sequence databases, which is an integral facet of personalized medicine. The increasing adoption of electronic medical records allows large amounts of patients’ standardized clinical features to be combined with the genomic sequences of these patients and shared to support validation of GWAS findings and to enable novel discoveries. However, disseminating these data “as is” may lead to patient reidentification when genomic sequences are linked to resources that contain the corresponding patients’ identity information based on standardized clinical features. This work proposes an approach that provably prevents this type of data linkage and furnishes a result that helps support GWAS. Our approach automatically extracts potentially linkable clinical features and modifies them in a way that they can no longer be used to link a genomic sequence to a small number of patients, while preserving the associations between genomic sequences and specific sets of clinical features corresponding to GWAS-related diseases. Extensive experiments with real patient data derived from the Vanderbilt's University Medical Center verify that our approach generates data that eliminate the threat of individual reidentification, while supporting GWAS validation and clinical case analysis tasks.
The decreasing cost of high-throughput sequencing technologies, in combination with the growing adoption of health information systems, has the potential to facilitate personalized medicine. Such technologies can generate a substantial quantity of detailed data, which can be mined to improve clinical diagnostics, as well as treatments, for a wide range of complex diseases (1). Notably, genome-wide association studies (GWAS) are increasingly applied to identify relationships between specific genomic variations and health-related phenomena; yet the cost associated with studies on populations that are large enough for sufficiently powered claims remains nontrivial (2). Consequentially, it is difficult for investigators to validate published associations. To overcome this problem, various regulations have been developed to encourage organizations to deposit data into repositories for reuse (3). The National Institutes of Health (NIH), for instance, recently defined a policy (4) stating that any GWAS data generated by, or studied with, NIH-sponsorship should be deposited in the Database of Genotypes and Phenotypes (dbGaP) (3) for broad dissemination. At the same time, the NIH acknowledges the need to maintain privacy standards and, thus, requires data to be deidentified. A typical deidentification strategy is based on the Safe Harbor standard of the Health Insurance Portability and Accountability Act (5), whereby records are stripped of a number of potential identifiers, such as personal names and geocodes.
Electronic medical record (EMR) systems are increasingly recognized as an important resource for GWAS (6). They contain detailed patient-level data on large populations, incorporate demographics and standardized clinical terminologies, and can reduce both costs and time of conducting large-scale GWAS. Although EMR data are derived from the primary care setting, the data are often devoid of detailed genomic sequences, which tend to be collected in a research environment. However, when combined and disclosed in a deidentified state, the released data may lead to individual reidentification if genomic sequences are linked to resources that contain patients’ identity (e.g., hospital discharge summaries or the original EMR system) through the standardized clinical features, or “clinical profile,” of a patient. Recently, it was demonstrated (7) that this type of data linkage may lead to compromising the privacy of more than 96% of a cohort of 2,762 patients from the Vanderbilt University Medical Center, involved in an NIH-funded GWAS, because these patients were uniquely identifiable on the basis of the combination of their ICD-9-CM codes (henceforth referred to as ICD codes).† Although such clinical and genomic data have yet to be disseminated, this illustrates the potential for privacy risks. To provide a clearer picture of the reidentification scenario, consider the dataset in Fig. 1A. In this table, each record corresponds to a fictional deidentified patient, comprises a set of ICD codes (derived from an EMR) and a DNA sequence (derived from a research project beyond primary care), and is analyzed in a GWAS on asthma (the ICD codes for asthma are 493.00, 493.01, and 493.02). If a hospital employee knows that Jim was diagnosed with the three ICD codes for asthma during a single hospital visit (e.g., by accessing the first record of the identified EMR data of Fig. 1B), then they would infer Jim’s DNA sequence because there is only one patient in this dataset harboring this specific set of codes. Note that it may also be possible for an attacker to know the ICD codes an individual was diagnosed with during all visits (e.g., when they have access to the entire dataset of Fig. 1B). We will consider this attack later in this article.
Fig. 1.
Biomedical datasets (fictional) and policies used by the proposed anonymization approach. (A) Research data, (B) identified EMR data, (C) utility policy, (D) privacy policy, and (E) a 5-anonymization for the research data.
Because the EMR-derived data are part of the GWAS study, it should be disseminated to comply with data sharing requirements, but in a manner that mitigates the aforementioned threat. This can be achieved by (i) specifying the sets of diagnosis codes that are linkable to identified resources, and (ii) modifying the linkable codes so that the clinical profiles containing these codes can be linked to a sufficiently large number of individuals on the basis of these codes. Code modification can prevent the linkage of a patient to their DNA sequence, but at the same time it may distort the associations between sets of codes corresponding to GWAS-related diseases and the DNA sequence. Therefore, it is crucial to retain the ability to support clinical association validations when modifying clinical profiles. In this respect, various privacy-preserving data publishing approaches have been proposed by the statistical disclosure control and database communities. Methods developed by the former community are inappropriate for our scenario because they produce data that do not correspond to real-world individuals (8). Thus, despite being able to retain some aggregate statistics, the practical usefulness of the produced data may be low because they do not allow data recipients to examine truthful patient records. At the same time, methods developed by the databases community [e.g., k-anonymization (9) and (h, k, p)-coherence (10)] are unable to guarantee a practically useful result because (i) they tend to assume that an individual is associated with a small set of linkable clinical features of fixed size (9), and (ii) they do not take into account the ability of released data to support clinical association validation when modifying clinical features (9, 10). Further discussion of related work can be found in SI Text.
In this study, we introduce an approach to prevent data linkage attacks via standardized clinical features, while ensuring that the released data are useful for GWAS validation. Our approach extracts a privacy policy in the form of sets of clinical features that require protection and anonymizes each record that contains any of these codes to ensure that it links to no fewer than k individuals with respect to these sets. Anonymization is achieved by replacing clinical features with sets of semantically related codes to satisfy a utility policy, which reflects the distribution of GWAS-related diseases to be preserved. Thus, the proposed approach addresses both limitations of prior work (9, 10), because it can handle records with large sets of clinical features that vary in size and takes into account specific utility requirements. For instance, assuming that an attacker knows sets of clinical features diagnosed during a single visit, our method can be applied to the dataset shown in Fig. 1A, with k = 5 and the utility policy of Fig. 1C, to extract a privacy policy, part of which is shown in Fig. 1D, and to generate the dataset of Fig. 1E. The latter satisfies these specific privacy requirements because each record links to no fewer than five individuals with respect to the sets of ICD codes in the privacy policy, as well as the utility requirements because the associations between diseases and DNA sequences are unaffected; that is, the distribution of the GWAS-related diseases is preserved (e.g., there are six, five, and eight patients diagnosed with asthma, prostate cancer, and pancreatic cancer in either of the datasets shown in Figs. 1A and 1E, respectively). We evaluate our approach with a real dataset involved in an NIH-sponsored GWAS study and demonstrate that anonymized clinical profiles support the validation of GWAS focusing on several diseases and studies focusing on clinical case counts.
Clinical Profile Anonymization Framework.
This section begins with a description of the basic concepts used in our method and then provides algorithms for extracting privacy policies and anonymizing clinical profiles.
Definitions.
In what follows, we define the structure of the considered datasets, the notions of privacy and utility policies, and the anonymization strategy.
Structure of the data.
We consider two types of biomedical datasets, each of which is represented as a database table. The first table D corresponds to the GWAS research dataset. It contains a set of ICD codes and a DNA sequence. We assume that each record in D corresponds to a unique patient. The second table T corresponds to an identified EMR. It contains records of patients’ explicit identifiers (e.g., personal names), service dates, and a set of ICD codes. Examples of D and T are depicted in Fig. 1 A and B, respectively. To protect privacy, our approach constructs a modified version of 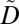 . Each record of
. Each record of 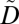 contains a patient's anonymized clinical information and their DNA sequence.
contains a patient's anonymized clinical information and their DNA sequence.
Policies.
In this section, we define the notions of privacy and utility policies. A privacy policy is defined as a set of potentially linkable combinations of ICD codes, referred to as privacy constraints. We define a privacy constraint as supported when all of the ICD codes contained in it appear in 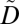 , and as nonsupported otherwise. A privacy policy is satisfied when (i) for each supported privacy constraint, all nonempty subsets constructed by the ICD codes appear at least k times in
, and as nonsupported otherwise. A privacy policy is satisfied when (i) for each supported privacy constraint, all nonempty subsets constructed by the ICD codes appear at least k times in 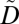 , and (ii) all nonempty subsets constructed by the ICD codes of each nonsupported privacy constraint appear at least k times in
, and (ii) all nonempty subsets constructed by the ICD codes of each nonsupported privacy constraint appear at least k times in 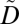 , or do not appear in
, or do not appear in 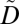 . As an example, consider the privacy policy of Fig. 1D, the dataset of Fig. 1E, and k = 5. A set of ICD codes in brackets denotes that the corresponding patient is diagnosed with any possible combination of these codes. It can be seen that the privacy constraint {157.9} is supported in the dataset of Fig. 1E. This privacy constraint is also satisfied for k = 5 in the same dataset because all of its nonempty subsets, which in this case is only the privacy constraint itself, appear in at least 5 records. Because all privacy constraints of Fig. 1D are satisfied in the dataset of Fig. 1E, the privacy policy is satisfied. The satisfaction of the privacy policy prevents patient reidentification because an attacker cannot use potentially linkable sets of ICD codes to link a DNA sequence to fewer than k patients in the released dataset. In other words, the probability of reidentification remains always at least 1/k.
. As an example, consider the privacy policy of Fig. 1D, the dataset of Fig. 1E, and k = 5. A set of ICD codes in brackets denotes that the corresponding patient is diagnosed with any possible combination of these codes. It can be seen that the privacy constraint {157.9} is supported in the dataset of Fig. 1E. This privacy constraint is also satisfied for k = 5 in the same dataset because all of its nonempty subsets, which in this case is only the privacy constraint itself, appear in at least 5 records. Because all privacy constraints of Fig. 1D are satisfied in the dataset of Fig. 1E, the privacy policy is satisfied. The satisfaction of the privacy policy prevents patient reidentification because an attacker cannot use potentially linkable sets of ICD codes to link a DNA sequence to fewer than k patients in the released dataset. In other words, the probability of reidentification remains always at least 1/k.
A utility policy is defined as a set of diseases in which each disease (henceforth referred to as utility constraint) is a set of ICD codes derived from D. A utility constraint is said to be satisfied when the number of records associated with this disease in 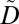 is equal to the number of records harboring at least one of the ICD codes contained in the utility constraint in D. A utility policy is satisfied if all of its utility constraints are satisfied. Consider, for example, the utility policy of Fig. 1C and the utility constraint asthma corresponding to the set of ICD codes {493.00, 493.01, 493.02}. The latter constraint is satisfied, because the number of patients suffering from asthma in the dataset of Fig. 1E [i.e., the records harboring (493.00, 493.01, 493.02)] is equal to the number of patients harboring at least one of the ICD codes related to asthma in the dataset of Fig. 1A. Similarly, it can be verified that the utility constraints for prostate cancer and pancreatic cancer are satisfied as well; therefore, the utility policy of Fig. 1C is satisfied. It is important to note that satisfying a utility policy ensures that the distribution of diseases contained in this policy will not be affected by anonymization. This allows all associations between diseases and DNA sequences present in the original dataset to be preserved in the anonymized dataset.
is equal to the number of records harboring at least one of the ICD codes contained in the utility constraint in D. A utility policy is satisfied if all of its utility constraints are satisfied. Consider, for example, the utility policy of Fig. 1C and the utility constraint asthma corresponding to the set of ICD codes {493.00, 493.01, 493.02}. The latter constraint is satisfied, because the number of patients suffering from asthma in the dataset of Fig. 1E [i.e., the records harboring (493.00, 493.01, 493.02)] is equal to the number of patients harboring at least one of the ICD codes related to asthma in the dataset of Fig. 1A. Similarly, it can be verified that the utility constraints for prostate cancer and pancreatic cancer are satisfied as well; therefore, the utility policy of Fig. 1C is satisfied. It is important to note that satisfying a utility policy ensures that the distribution of diseases contained in this policy will not be affected by anonymization. This allows all associations between diseases and DNA sequences present in the original dataset to be preserved in the anonymized dataset.
Anonymization strategy.
Our approach creates 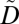 from D by replacing every ICD code of D with a unique anonymized item, which is represented as a set of ICD codes. Disparate anonymized codes are mutually exclusive, such that there are no ICD codes in multiple anonymized items. For instance, the ICD code 493.00 in the first record of Fig. 1A is replaced by the anonymized item (493.00, 493.01, 493.02) when these data are modified to that of Fig. 1E. The process by which ICD codes are replaced with anonymized items is formally presented in SI Text.
from D by replacing every ICD code of D with a unique anonymized item, which is represented as a set of ICD codes. Disparate anonymized codes are mutually exclusive, such that there are no ICD codes in multiple anonymized items. For instance, the ICD code 493.00 in the first record of Fig. 1A is replaced by the anonymized item (493.00, 493.01, 493.02) when these data are modified to that of Fig. 1E. The process by which ICD codes are replaced with anonymized items is formally presented in SI Text.
Our anonymization strategy may replace ICD codes with semantically consistent but more general terms, typically specified by user-defined taxonomies (9) or, by default, the ICD coding hierarchy (http://www.cdc.gov/nchs/icd/icd9cm.htm). This process is referred to as generalization (9). Alternatively, ICD codes may also be suppressed (i.e., removed from the anonymous result) (10). As we explain later in this article, this occurs when generalization does not suffice to satisfy the specified privacy policy. Both generalization and suppression effectively increase the number of patients to which a privacy constraint is associated, thereby reducing the risk of reidentification.
Notably, our generalization strategy eliminates the need of taxonomies, which may be deficient or nonexistent (11), and allows the production of more fine-grained anonymizations with higher utility than those constructed by alternative generalization strategies (9). As an example, consider diabetes mellitus type 2, a disease that is associated with a set of ICD codes of the form 250.xy, where x is an integer in [0, 9] and y ∈ {0, 2}. Current generalization strategies (e.g., ref. 9) would replace any set of this type of code with 250 (denoting diabetes mellitus) when a taxonomy that associates five-digit ICD codes to their three-digit counterparts is applied. By doing so, the generalization process makes it impossible to distinguish between diabetes mellitus type 1 and type 2 and yields anonymized data that are meaningless for validating GWAS on diabetes mellitus type 2 (12). In contrast, our strategy allows a utility constraint corresponding to diabetes mellitus type 2 to be specified and guarantees that the number of patients diagnosed with diabetes mellitus type 2 will be equal in the original and anonymized clinical profiles, when this utility constraint is satisfied. This effectively preserves all associations between this disease and DNA regions and thus allows for validation of GWAS studies.
Privacy policy extraction.
To help data owners formulate a privacy policy, we present the privacy policy extraction (PPE) algorithm. We assume that an attacker knows (i) a patient's explicit identifiers, (ii) a certain set of ICD codes for each patient, and (iii) whether a patient's record is contained in D. We discuss how such knowledge can be obtained in Discussion.
Given k and a filtering condition (i.e., a set of ICD codes deemed as potentially identifying), PPE derives a privacy policy in two steps. First, it iteratively populates the privacy policy with sets of ICD codes that satisfy the filtering condition. Subsequently, PPE retains the minimal number of privacy constraints required to satisfy the derived policy. This is performed by discarding privacy constraints that do not require protection (e.g., all nonempty subsets induced by the ICD codes of these constraints that correspond to at least k patients) or receive protection when their supersets in the privacy policy are already protected as explained above (e.g., satisfying the privacy constraint {493.00, 493.01, 493.02} implies that the privacy constraint {493.00} will be satisfied as well).
We consider two concrete filtering conditions. Alternative filtering conditions are also possible and can be formulated by data owners according to their expectations about attackers’ knowledge. The first one corresponds to a single-visit case, in which data owners treat the set of ICD codes for each patient's visit (corresponding to a distinct service date) as potentially identifying. In this case, it is assumed that an attacker cannot use sets of ICD codes that span two or more visits. This is difficult when publicly available hospital discharge summaries are used, because it requires associating deidentified records with different service dates to the same patient (9). The second filtering condition corresponds to the all-visits case, in which all ICD codes of a patient are treated as potentially identifying. Clearly, this is the strictest privacy policy one can adopt.
As an example, consider the application of PPE to Mary’s records in Fig. 1B using k = 5 and the single-visit filtering condition. First, the privacy constraints {157.0, 185} and {157.1, 157.2, 157.3}, which correspond to the service dates 01/02/08 and 07/09/09, respectively, are added to the privacy policy. However, because the first privacy constraint is a subset of five records in D, it is removed from the privacy policy. In contrast, {157.1, 157.2, 157.3} is not a subset of any other privacy constraint, and thus the extracted privacy policy is {157.1, 157.2, 157.3}.
Utility-guided anonymization of clinical profiles.
Given a dataset D, a utility policy, a privacy policy, and a parameter k, we sketch Utility-Guided Anonymization of CLInical Profiles (UGACLIP), an algorithm to construct an anonymized dataset 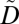 that is protected from reidentification and useful in validating GWAS. UGACLIP aims to satisfy the privacy policy by iteratively satisfying each of its privacy constraints in a series of steps. First, it selects the privacy constraint that is currently associated with the most patients. Second, it satisfies this constraint by iteratively generalizing the most infrequent ICD code (or anonymized item) with another ICD code (or anonymized item) in a way that satisfies the corresponding utility constraint while minimizing an information loss measure. If a privacy constraint cannot be satisfied by generalization, the ICD code (or anonymized item) is suppressed. Third, if the privacy constraint remains unprotected, it is satisfied by suppression. At this point, the algorithm proceeds to the next iteration. The process terminates when the privacy policy is satisfied. The pseudocode of UGACLIP can be found in SI Text.
that is protected from reidentification and useful in validating GWAS. UGACLIP aims to satisfy the privacy policy by iteratively satisfying each of its privacy constraints in a series of steps. First, it selects the privacy constraint that is currently associated with the most patients. Second, it satisfies this constraint by iteratively generalizing the most infrequent ICD code (or anonymized item) with another ICD code (or anonymized item) in a way that satisfies the corresponding utility constraint while minimizing an information loss measure. If a privacy constraint cannot be satisfied by generalization, the ICD code (or anonymized item) is suppressed. Third, if the privacy constraint remains unprotected, it is satisfied by suppression. At this point, the algorithm proceeds to the next iteration. The process terminates when the privacy policy is satisfied. The pseudocode of UGACLIP can be found in SI Text.
As an example, consider applying UGACLIP to the data shown in Fig. 1A using the utility and privacy policies of Figs. 1 C and D, respectively, and k = 5. Because all privacy constraints correspond to one patient, UGACLIP arbitrarily selects the privacy constraint {157.1, 157.2, 157.3} and attempts to satisfy it by replacing 157.3 (the most infrequent ICD code in this constraint) and 157.2 with the anonymized item (157.2, 157.3), which corresponds to four patients. This generalization minimizes information loss and satisfies the utility constraint corresponding to pancreatic cancer (i.e., the number of patients with this disease in the data of Fig. 1A is unaffected by generalization). However, the privacy constraint remains unprotected and thus (157.2, 157.3) is further generalized to the anonymized item (157.1, 157.2, 157.3). The latter generalization satisfies the utility constraint corresponding to pancreatic cancer as well as the privacy constraint {157.1, 157.2, 157.3}, because (157.1, 157.2, 157.3) corresponds to at least five patients. At this point the first iteration of UGACLIP has completed, and the algorithm proceeds by considering the next privacy constraint {157.9}. After all iterations, UGACLIP produces the anonymized clinical profiles depicted in Fig. 1E.
In UGACLIP, we use the aforementioned generalization strategy, which replaces a number of ICD codes with an anonymized item corresponding to their set according to the specified utility policy. In addition, because there are many possible ways to generalize data that satisfy the specified utility and privacy policies, our method incorporates an information loss measure to select a generalization that less distorts data (see SI Text for details). However, it may be impossible to protect a privacy constraint using generalization only. This occurs when a constraint contains subsets of ICD codes that appear fewer than k times in 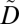 after applying all generalizations that satisfy the utility policy. In this case, we apply suppression to remove ICD codes from the privacy constraint until the constraint is satisfied. We note that applying suppression may result in violating the utility policy because it may reduce the number of patients associated with certain diseases, resulting in potentially invalid GWAS findings. However, such an effect can be mitigated by limiting the number of allowable suppressed ICD codes via a user-specified threshold. Fortunately, GWAS tend to focus on statistically significant associations, which involve frequent ICD codes that are unlikely to be suppressed in practice.
after applying all generalizations that satisfy the utility policy. In this case, we apply suppression to remove ICD codes from the privacy constraint until the constraint is satisfied. We note that applying suppression may result in violating the utility policy because it may reduce the number of patients associated with certain diseases, resulting in potentially invalid GWAS findings. However, such an effect can be mitigated by limiting the number of allowable suppressed ICD codes via a user-specified threshold. Fortunately, GWAS tend to focus on statistically significant associations, which involve frequent ICD codes that are unlikely to be suppressed in practice.
Results
Datasets and Experimental Setup.
We evaluated our approach with two sets of patient records derived from a deidentified version of the Vanderbilt University Medical Center EMR system (13). The first dataset contains the diagnosis codes of 2,762 patients and was constructed for the purposes of an NIH-sponsored GWAS. This is referred to as the Vanderbilt Native Electrical Conduction dataset (VNEC), and it was also used by Loukides et al. (7). The genomic sequences and primary clinical phenotype are expected to be deposited in dbGaP. However, it is our goal to determine which components of the profile can be shared in addition. The second dataset is a version of VNEC with a reduced set of ICD codes and comprises 1,335 patient records. It models a scenario in which data owners know which diagnoses can be applied as cases for other studies (1) and is referred to as the Vanderbilt Native Electrical Conduction Known Controls dataset (VNECKC).
We instantiated UGACLIP with privacy policies derived by PPE using both the single-visit and all-visits filtering conditions and values of k between 2 and 25. We manually constructed utility policies that include ICD codes associated with diseases that are relevant to existing GWAS studies. Specifically, we considered GWAS for several diseases, which are summarized elsewhere (14) and appear at least k times in the datasets. Diseases that correspond to fewer than k patients in a dataset must be suppressed by UGACLIP to satisfy the privacy policy. The diseases contained in the utility policy for both datasets and for k = 5 are illustrated in Table 1. The additional utility policies we applied, as well as the associations between diseases and ICD codes in these policies, can be found in SI Text.
Table 1.
Satisfied utility constraints for k = 5 and the single-visit case (✓ denotes that a utility constraint is satisfied)
|
VNEC |
VNECKC |
||||
| Disease | UGACLIP | ACLIP | UGACLIP | ACLIP | |
| Asthma | ✓ | ✓ | |||
| Attention deficit with hyperactivity | |||||
| Bipolar I disorder | ✓ | ✓ | |||
| Bladder cancer | |||||
| Breast cancer | ✓ | ✓ | |||
| Coronary disease | ✓ | ✓ | ✓ | ||
| Dental caries | ✓ | ✓ | |||
| Diabetes mellitus type 1 | ✓ | ✓ | |||
| Diabetes mellitus type 2 | ✓ | ✓ | ✓ | ||
| Lung cancer | ✓ | ✓ | |||
| Pancreatic cancer | ✓ | ✓ | |||
| Platelet phenotypes | |||||
| Preterm birth | ✓ | ✓ | |||
| Prostate cancer | ✓ | ✓ | |||
| Psoriasis | ✓ | ||||
| Renal cancer | ✓ | ||||
| Schizophrenia | ✓ | ||||
| Sickle-cell disease | ✓ | ||||
Because there are no directly comparable anonymization approaches to UGACLIP, we compare it against a variant of this algorithm, referred to as ACLIP (Anonymization of CLInical Profiles). ACLIP uses the same privacy policy as UGACLIP, but it does not take the utility policy into account. Rather, ACLIP follows the approach of Xu et al. (10) in that it attempts to maximize data utility by minimizing the amount of information loss incurred to anonymize clinical profiles.
In the following experiments, we evaluate the reidentification risk, as well as the effectiveness of our method, in terms of its ability to (i) construct useful anonymizations for validating GWAS and (ii) perform studies focusing on clinical case counts.
Reidentification Risk Evaluation.
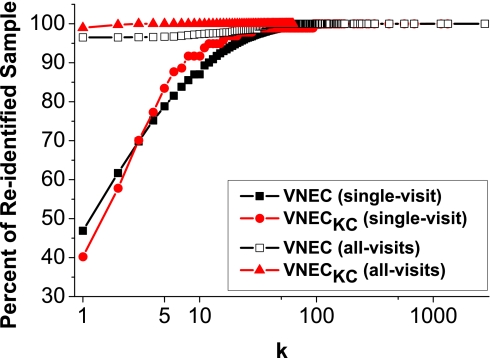
The risk of reidentification was quantified by measuring the number of records that share a set of potentially linkable ICD codes extracted by our algorithm. This number is referred to as distinguishability and is equal to the inverse of the probability of associating a patient to his DNA sequence with respect to the extracted sets of potentially linkable codes after data release. Records with a distinguishability score of 1 are regarded as uniquely identifiable and correspond to patients whose identity can be revealed after data linkage, whereas those with a score of <5 are defined as unpublishable. Fig. 2 reports the result for both datasets and filtering conditions. Notice that, for the single-visit case, at least 40% of the records of each dataset are uniquely identifiable, whereas 75% are unpublishable. Furthermore, because of the stricter privacy policy used, the corresponding statistics for the all-visits case increased to 96% and 99%, respectively. This validates the feasibility of the considered attack and the need for a formal approach to preserve privacy when releasing clinical profiles.
Fig. 2.
Reidentification risk (shown as a cumulative distribution function).
Utility Constraint Satisfaction.
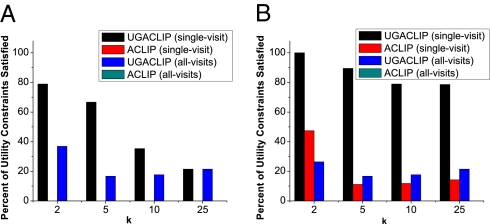
We next evaluated the effectiveness of UGACLIP at generating anonymizations that assist in GWAS validation. First we measured the number of utility constraints the anonymized clinical profiles satisfy. The result for the VNEC dataset is shown in Fig. 3A. It can be seen that the anonymizations generated by UGACLIP with k = 5, as is often in applications satisfied more than 66% of the specified utility constraints for the single-visit case. As expected, the number of satisfied utility constraints dropped for the all-visits case. However, UGACLIP still satisfied at least 16.7% of the utility constraints for all k values. This is in contrast to ACLIP, which failed to construct a useful result for GWAS (no utility constraints were satisfied in any case). The result of applying UGACLIP and ACLIP to the VNECKC dataset reported in Fig. 3B is quantitatively similar to that of Fig. 3A. These results suggest that our approach is effective in anonymizing data guided by a utility policy. Interestingly, applying both methods to VNECKC resulted in an increased number of satisfied utility constraints. This is because restricting clinical profiles to diseases related to GWAS makes VNECKC less sparse than VNEC, which increases the chance of grouping ICD codes in a way that satisfies utility constraints.
Fig. 3.
Utility constraint satisfaction at various levels of protection for (A) VNEC and (B) VNECKC.
We further examined which of the utility constraints the anonymized clinical profiles satisfied. Table 1 depicts the result for k = 5 and the single-visit case. Notice that the anonymized version of VNEC produced by UGACLIP validates GWAS-related associations for the majority of the diseases. This, however, was not possible for some diseases, such as Attention deficit with hyperactivity, for which all corresponding ICD codes had to be suppressed to satisfy the privacy policy. Specifically, UGACLIP performed 167 suppressions, in which 0.19% of the ICD codes were removed from VNEC. In contrast, ACLIP generated clinical profiles that fail to satisfy any of the utility constraints and thus do not validate GWAS for the selected diagnoses. Regardless, both anonymization methods performed relatively better for VNECKC. Again, this was because the clinical profiles in this dataset are less sparse. In particular, UGACLIP was able to satisfy all but three of the utility constraints, significantly outperforming ACLIP. Experimental results for the range of tested k values and for the all-visits case are provided in SI Text.
Support of Clinical Case Counts.
We investigated the effectiveness of our method in generating anonymizations that assist studies focusing on clinical case counts, which are beyond GWAS-specific validation. This is important to investigate because it may be difficult for scientists to a priori predict all possible uses of GWAS data. In this set of experiments, we assumed that data users issue queries to learn the number of patient records harboring a combination of ICD codes that appears in at least 10% of the records. This task is crucial in data-mining applications (10).
However, anonymized clinical profiles may not allow such queries to be answered accurately because an anonymized item can be interpreted as any of the nonempty subsets of ICD codes it contains. For instance, a data recipient will be uncertain about the number of records harboring the ICD code 493.00 when using the anonymized data shown in Fig. 1E. Thus, to evaluate utility, we adopted a metric called relative error (RE) (15), which reflects the number of records that are incorrectly retrieved in the answer to a query when issued against anonymized clinical profiles. In our experiments, we measured the mean and SD of the RE scores for a workload of queries. Details of the process are provided in SI Text.
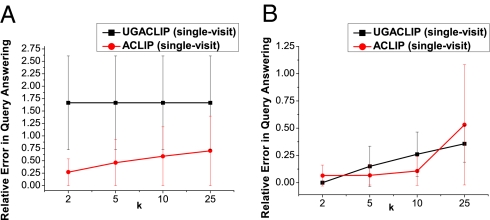
The results for the single-visit case and for the VNEC dataset are reported in Fig. 4A. Observe that ACLIP resulted in a relatively small error, <0.75 on average, for all tested values of k, outperforming UGACLIP. This is because the majority of the combination of ICD codes contained in the queries did not correspond to diseases used as control variables in GWAS, whose distribution UGACLIP was configured to preserve. Thus, as expected, UGACLIP generalized other ICD codes slightly more to preserve the distribution of these diseases. Yet it should be noted that the RE scores for ACLIP and UGACLIP were approximately within 1 SD of each other for all k values. This suggests that UGACLIP is capable of producing anonymizations that support both GWAS and studies focusing on clinical case counts in general. The RE statistics for the single-visit case and for the VNECKC dataset are illustrated in Fig. 4B. As can be seen, the RE scores for both methods were small, and the performance of UGACLIP was comparable to, and in some cases better than, that of ACLIP. This is because the queried combinations of ICD codes were contained in the utility policy used in UGACLIP and thus were not substantially distorted. The results for the all-visits case were quantitatively similar to those of Fig. 4 A and B and can be found in SI Text.
Fig. 4.
Relative error in query answering for the single-visit case and for (A) VNEC and (B) VNECKC. Points correspond to the mean RE, and error bars are of 1 SD.
Discussion
In this section, we discuss the feasibility of data linkage attacks in practice and the limitations of our methodology.
Feasibility of Data Linkage.
It should be acknowledged that to reidentify individuals using disseminated patient-level ICD codes in combination with genomic information, an attacker must harbor knowledge of a patient's identity, as well as some portion of a patient's clinical profile. In this study, we assumed that an attacker possesses three types of knowledge: (i) a patient's identity, (ii) selected ICD codes, and (iii) whether a patient is included in the released research sample. We note that knowledge of the first two types can come in the form of background knowledge (16) or may be solicited by exploiting external data sources, such as publicly available voter lists combined with hospital discharge summaries (9) or the identified EMR system available to the primary care environment (7). In this respect, we note that data access policies that limit the availability of ICD codes often exist in practice. For instance, EMR users at Vanderbilt University Medical Center (13) are required to sign a confidentiality agreement that explicitly prohibits reidentification attempts, and record-level data in dbGaP are available only to researchers approved by a data access committee (4). Nonetheless, such policies provide no formal privacy protection guarantees, and medical record breaches have occurred (9). Regarding the knowledge of the third type, we recognize that it can be inferred by applying the procedure used to create the research sample from a larger patient population, which is often described in the literature.
It is also important to mention that there is another type of data linkage that can be exploited to associate individuals to their released genomic information (17). To perform this linkage, an attacker needs access to an individual's identity and DNA, as well as to a reference pool of DNA containing individuals from the same genetic population as the identified individual. We emphasize that data owners should consider both types of data linkage before releasing their data and devise their policies according to their expectations about the capabilities of attackers.
Limitations.
The proposed approach is limited in certain aspects, which we highlight to suggest opportunities for further research. First, as is true of all data anonymization methods, our approach leaves the decision of selecting a suitable privacy protection level (i.e., k and the filtering condition of PPE) to data owners or policy officials. This may be a difficult task, because it requires prediction of adversarial knowledge. Yet, if such knowledge is underestimated, it may lead to the compromise of patients’ privacy. On the other hand, if an attacker's knowledge is overestimated, it may result in excessively distorted data (16).
A second limitation of this work is that the UGACLIP algorithm does not guarantee that the anonymized clinical profiles will incur the least amount of information loss possible to satisfy the specified utility policy. The design of approximation algorithms that offer such guarantees is important to address the growing size of GWAS-related datasets, but is also challenging due to the computational complexity of the problem (10).
Conclusions
In this article we proposed a method to anonymize patient-specific clinical profiles, which should be disseminated to support biomedical studies. Our method (i) offers provable protection from individual reidentification based on clinical features, (ii) allows sensitive patterns of ICD codes to be automatically extracted, and (iii) generates anonymizations that help validate GWAS and perform clinical case analysis tasks. These features were experimentally verified using patients’ data from clinical profiles derived from a real GWAS cohort.
Supplementary Material
Acknowledgments
This research was funded by National Human Genome Research Institute Grant U01HG004603 and National Library of Medicine Grant 1R01LM009989.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911686107/DCSupplemental.
*This Direct Submission article had a prearranged editor.
†The International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9-CM) is the official system of assigning codes to diagnoses associated with inpatient, outpatient, and physician office utilization in the United States.
References
- 1.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 2.Gurwitz D, Lunshof JE, Altman RB. A call for the creation of personalized medicine databases. Nat Rev Drug Discov. 2006;5:23–26. doi: 10.1038/nrd1931. [DOI] [PubMed] [Google Scholar]
- 3.Mailman MD, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Institutes of Health Policy for sharing of data obtained in NIH supported or conducted genome-wide association studies. 2007 NOT-OD-07-088. Available at: http://grants.nih.gov/grants/guide/notice-files/NOT-OD-07-088.html. Accessed February 16, 2010. [Google Scholar]
- 5.US Department of Health and Human Services Standards for privacy of individually identifiable health information; final rule. Federal Register 45 (2002) 2002 parts 160 and 164. [PubMed] [Google Scholar]
- 6.Department of Biomedical Informatics, Vanderbilt University School of Medicine. Electronic Medical Records and Genomics (eMERGE) Network Available at. http://www.gwas.net. Accessed February 16, 2010.
- 7.Loukides G, Denny JC, Malin B. The disclosure of diagnosis codes can breach research participants’ privacy. J Am Med Inform Assoc. 2009 doi: 10.1136/jamia.2009.002725. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yancey WE, Winkler WE, Creecy RH. In: Inference Control in Statistical Databases. Domingo-Ferrer J, editor. Heidelberg: Springer; 2002. pp. 49–60. [Google Scholar]
- 9.Sweeney L. k-anonymity: A model for protecting privacy. Int J Uncertainty, Fuzziness Knowledge-Based Systems. 2002;10:557–570. [Google Scholar]
- 10.Xu Y, Wang K, Fu AWC, Yu PS. Anonymizing transaction databases for publication. In: Li Y, Liu B, Sarawagi S, editors. Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York: ACM; 2008. pp. 767–775. [Google Scholar]
- 11.Rogers JE. Quality assurance of medical ontologies. Methods Inf Med. 2006;45:267–274. [PubMed] [Google Scholar]
- 12.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roden DM, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeFevre K, DeWitt DJ, Ramakrishnan R. Mondrian multidimensional k-anonymity. In: Liu L, Reuter R, Whang KY, Zhang J, editors. Proceedings of the International Conference on Data Engineering. Los Alamitos: IEEE Computer Society; 2006. p. 25. [Google Scholar]
- 16.Machanavajjhala A, Gehrke J, Kifer D, Venkitasubramaniam M. l-Diversity: Privacy beyond k-anonymity. In: Liu L, Reuter R, Whang KY, Zhang J, editors. Proceedings of the International Conference on Data Engineering. Los Alamitos: IEEE Computer Society; 2006. p. 24. [Google Scholar]
- 17.Homer N, et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.