Abstract
Changes in non–protein-coding regulatory DNA sequences have been proposed to play distinctive roles in adaptive evolution. We analyzed correlations between gene functions and evidence for positive selection in a common statistical framework across several large surveys of coding and noncoding sequences throughout the human genome. Strong correlations with both classifications in gene ontologies and measurements of gene expression indicate that neural development and function have adapted mainly through noncoding changes. In contrast, adaptation via coding changes is dominated by immunity, olfaction, and male reproduction. Genes with highly tissue-specific expression have undergone more adaptive coding changes, suggesting that pleiotropic constraints inhibit such changes in broadly expressed genes. In contrast, adaptive noncoding changes do not exhibit this pattern. Our findings underscore the probable importance of noncoding changes in the evolution of human traits, particularly cognitive traits.
Keywords: adaptive evolution, human origins, protein-coding DNA sequences, noncoding DNA sequences, regulatory DNA sequences
Among the most fundamental unanswered questions about adaptive evolution are whether it proceeds primarily through changes in protein-coding DNA sequences or noncoding regulatory sequences, whether the proportions of coding and noncoding changes vary appreciably among organismal traits, and, if so, why (1, 2). For example, it has been argued that morphological adaptation occurs mainly via noncoding changes, on the grounds that many genes underlying development are active in many contexts, and a noncoding mutation is likely to alter a gene’s activity in only one or a few contexts, avoiding pleiotropic constraints (3–5). These questions have been addressed mainly using case studies of individual genes and traits; however, they now can be addressed on a genomic scale, and doing so is indispensable for assessing whether consistent, intelligible patterns exist.
The human genome is especially suitable for such inquiry, for two reasons. First, there are now many surveys aiming to detect signatures of positive selection on sequences throughout the human genome (6), most focusing on coding sequences but a few focusing on noncoding sequences. Second, the extensive functional annotations of the human genome often make it possible to infer something about the trait through which positive selection on a sequence arose (6). Here we evaluate what several large surveys of the human genome imply about the roles of coding and noncoding changes in adaptive evolution, how these roles vary among gene functions, and what this variation suggests about its causes.
In this work, we analyzed three surveys of coding sequences (7–9) and three surveys of noncoding sequences (10–12) (Methods and Table S1). Each survey aims to detect a signature of positive selection, distinguishes selection on coding versus noncoding sequences, and treats thousands of sequences without a priori bias regarding function. The surveys’ data are diverse, and even when they overlap, different surveys sometimes give different results, because their methods have different sensitivities, not only to signatures of positive selection, but also to confounding factors (13–15). This diversity of data and methods is what makes considering these surveys collectively valuable. Although each survey offers unique insights, we are interested in trends prevailing across all three coding surveys or all three noncoding surveys, which are likely to represent consistent features of adaptation via coding or noncoding changes.
The trends on which we focus are correlations between positive selection and gene functions according to the PANTHER (16) and Gene Ontology (GO) (17) classification systems and the Novartis Gene Expression Atlas (18). Within each survey, certain functional categories and expression domains are enriched with or depauperate of genes scoring high for positive selection, suggesting that changes detected in the survey have played large or small roles, respectively, in the adaptation of these functions. These functional annotations apply directly to proteins and hence coding sequences. In the absence of direct annotations of most noncoding sequences, we associated each noncoding sequence with the nearest coding sequence, because coding sequences often are regulated by nearby noncoding sequences (19). Each survey was published with some analysis of functional enrichment, and some impression of patterns across surveys can be derived from these analyses (6); however, a more detailed and precise understanding can be achieved by analyzing the surveys in a common statistical framework.
Results
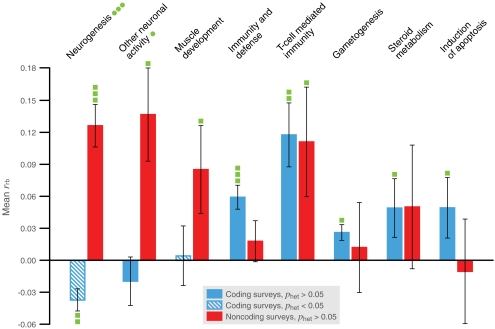
For each PANTHER or GO category, we computed the rank correlation between the score for positive selection and membership in the category (rank-biserial correlation, rrb) and the SE of the correlation within each survey (Methods). We then computed the weighted mean of this correlation and the SEM across coding surveys and across noncoding surveys, weighting so that those surveys estimating the correlation more precisely contribute more heavily to the mean. We are particularly interested in categories for which the mean correlation is significantly positive (Penr < 0.05) or negative (Pdep = 1 − Penr < 0.05) across coding or noncoding surveys, indicating that the category is, respectively, appreciably enriched with or depauperate of positive selection on coding or noncoding changes. We also computed a heterogeneity statistic across coding surveys and across noncoding surveys, and when discussing categories enriched with or depauperate of positive selection across coding or noncoding surveys, we restrict attention to categories for which this statistic is nonsignificant across the same surveys (Phet > 0.05), indicating a lack of appreciable discord among the surveys. Figure 1 plots the results for large PANTHER biological processes, Table 1 lists results for large and middle-sized PANTHER biological processes, and Table S2 is analogous for GO biological processes. Tables S3, S4, S5, S6, and S7 present complete results for large and middle-sized PANTHER and GO biological processes, molecular functions, and cellular components.
Fig. 1.
Large PANTHER biological processes enriched with positive selection across coding or noncoding surveys. Plotted categories are all but three of those for which (i) the mean number of genes per survey is at least 50 across all surveys, (ii) the mean rank-biserial correlation (rrb) between score for positive selection and membership in the category is significantly positive (Penr < 0.05) across coding or noncoding surveys, and (iii) heterogeneity is nonsignificant (Phet > 0.05) across the same surveys. The three such categories not plotted are “ectoderm development,” “neuronal activities,” and “mesoderm development,” which are not significantly enriched when their respective subcategories “neurogenesis,” “other neuronal activity,” and “muscle development” are subtracted. Error bars represent SEM- rrb. Green blocks on error bars indicate one-tailed P values for the one-sample z test of mean rrb = 0. One block, 5 × 10−4 < P < 0.05; two blocks, 5 × 10−6 < P < 5 × 10−4; three blocks, P < 5 × 10−6. Similarly, green dots on category names indicate two-tailed P values for two-sample z tests of equal mean, rrb, across coding versus noncoding surveys.
Table 1.
PANTHER biological processes enriched with positive selection
| Category* | Mean n† | Mean rrb | SEM rrb | Penr‡ |
| Across coding surveys | ||||
| Chemosensory perception | 68 | 0.37 | 0.054 | < 10−6 |
| Immunity and defense | 550 | 0.059 | 0.011 | < 10−6 |
| T cell–mediated immunity | 83 | 0.12 | 0.030 | 4.9 × 10−5 |
| IFN-mediated immunity | 30 | 0.16 | 0.061 | 0.0037 |
| Other oncogenesis | 23 | 0.15 | 0.058 | 0.0051 |
| Anterior/posterior patterning | 29 | 0.12 | 0.064 | 0.029 |
| Segment specification | 48 | 0.079 | 0.043 | 0.034 |
| Steroid metabolism | 78 | 0.049 | 0.027 | 0.038 |
| Spermatogenesis and motility | 50 | 0.068 | 0.039 | 0.039 |
| Induction of apoptosis | 73 | 0.049 | 0.028 | 0.042 |
| Across noncoding surveys | ||||
| Neurogenesis | 266 | 0.13 | 0.020 | < 10−6 |
| Other neuronal activity | 54 | 0.14 | 0.044 | 8.9 × 10−4 |
| T cell–mediated immunity | 42 | 0.11 | 0.051 | 0.016 |
| Oncogene | 39 | 0.12 | 0.055 | 0.0017 |
| Muscle development | 59 | 0.085 | 0.041 | 0.020 |
| Blood clotting | 23 | 0.12 | 0.068 | 0.042 |
| Sulfur metabolism | 31 | 0.11 | 0.064 | 0.047 |
*Listed categories satisfy the following: (i) Eeach of at least five surveys treats at least 10 genes, (ii) the mean rank-biserial correlation (rrb) between score for positive selection and membership in the category is significantly positive (Penr < 0.05) across coding or noncoding surveys, and (iii) heterogeneity is nonsignificant (Phet > 0.05) across the same surveys. When one such category contains others, the former is listed only if it still satisfies (i)–(iii) when the latter are subtracted. A total of 163 categories satisfy (i).
†Mean number of genes in the category per survey, rounded to the nearest integer.
‡Upper one-tailed P value for the one-sample z test of mean rrb = 0, rounded upward. Penr is not adjusted for multiple comparisons, which is difficult to do correctly because categories overlap extensively, but Penr < 3.0 × 10−4 would remain significant even under Bonferroni adjustment, which is extremely conservative.
The strongest pattern evident in these results is that neural development appears to have adapted primarily through noncoding changes. Across noncoding surveys, the PANTHER biological process “neurogenesis” is highly enriched with positive selection (Penr < 10−6). “Neurogenesis” has no subcategories in PANTHER, but finer resolution is available in GO biological processes, where “regulation of neuron differentiation,” “axon guidance,” “regulation of axonogenesis,” “brain development,” “neuron migration,” “positive regulation of neurogenesis,” and “negative regulation of neurogenesis” are enriched across noncoding surveys (Penr = 4.5 × 10−5, 5.2 × 10−4, 0.0024, 0.0026, 0.0072, 0.0079, and 0.014, respectively), whereas “axon guidance” and “negative regulation of neurogenesis” are depauperate across coding surveys (Pdep = 0.021 and < 10−6, respectively). This pattern arises largely from different genes in different surveys. Of the 86 “neurogenesis” genes scoring high (P < 0.05) for positive selection in at least one noncoding survey, only 8 score high in two surveys, and none do so in all three surveys. (Note that these surveys generally treat different sequences associated with a given gene.)
Likewise, neural function appears to have adapted primarily through noncoding changes, although this pattern is weaker. The PANTHER biological process “other neuronal activity” is enriched with positive selection across noncoding surveys (Penr = 8.9 × 10−4) and remains so when genes also in “neurogenesis” are excluded (Penr = 0.018). The GO biological process “regulation of synaptic transmission” is enriched across noncoding surveys (Penr = 0.0094), the GO molecular functions “GABA receptor activity” and “ionotropic glutamate receptor activity” are enriched across noncoding surveys (Penr = 0.027 and 0.045, respectively) and highly depauperate across coding surveys (Pdep < 10−6 for both), and the GO cellular component “synapse” is marginally enriched across noncoding surveys (Penr = 0.075) and depauperate across coding surveys (Pdep = 1.6 × 10−4). No such category is enriched across coding surveys, apart from the olfaction-related categories mentioned below.
Other trends prevailing across the noncoding surveys include enrichment with positive selection of several aspects of development in addition to neural development. Most conspicuously, the PANTHER biological process “muscle development” is enriched (Penr = 0.020). However, some developmental categories also are enriched across coding surveys, including the PANTHER biological processes “anterior/posterior patterning” and “segment specification” (Penr = 0.029 and 0.034, respectively).
Consistent with earlier reports (6–9), the leading themes of adaptation through coding changes appear to be immunity and olfaction, represented by the PANTHER biological processes “immunity and defense” and “chemosensory perception” (Penr < 10−6 for both), the GO biological processes “defense response” and “sensory perception of smell” (Penr = 0.0011 and < 10−6, respectively), and many other categories. Also conspicuous is sperm function, represented by the PANTHER biological process “spermatogenesis and motility” (Penr = 0.039) and the GO cellular component “acrosome” (Penr = 0.0087), among other categories. Few such categories are depauperate of positive selection across noncoding surveys, however. Indeed, the PANTHER biological process “T cell–mediated immunity” is enriched with positive selection across both coding and noncoding surveys (Penr = 4.9 × 10−5 and 0.016, respectively). In conjunction with the distinctive role of noncoding changes in neural adaptation, these results raise the possibility that a wider range of traits have been amenable to adaptation via noncoding changes than via coding changes.
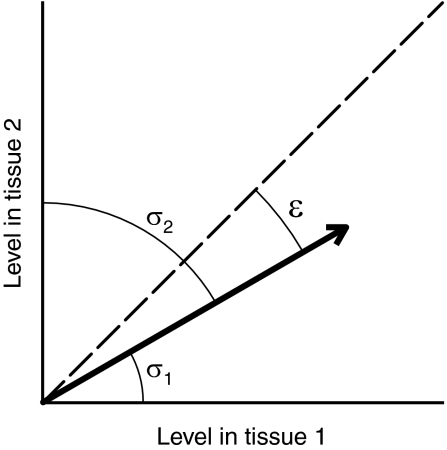
To examine how adaptive coding and noncoding changes relate to gene expression, we turned to the Novartis Gene Expression Atlas. For each gene and each of the 73 noncancerous tissues in the atlas, we computed a specificity score between 0 and 1 representing the specificity of the gene’s expression to the tissue (9, 12). The score is high if the expression is very specific to the tissue and is low even for the gene’s tissue of maximal expression if the gene is nearly as highly expressed in other tissues. For each gene, we also computed an evenness score between 0 and 1, representing the evenness of the gene’s expression across all 73 tissues. The score is high if the expression is not very specific to any tissue. Figure 2 illustrates these scores in the simplest context of two tissues.
Fig. 2.
Specificity and evenness scores. The vector represents expression of a given gene in two tissues. The dashed line corresponds to equal expression in these tissues. The specificity scores of the gene are s1 = cos2σ1 and s2 = cos2σ2. The gene’s expression is higher in tissue 1, so s1 > s2. The evenness score of the gene is e = cos2ε. If the gene’s expression were lower in tissue 1 or higher in tissue 2, then e would be greater.
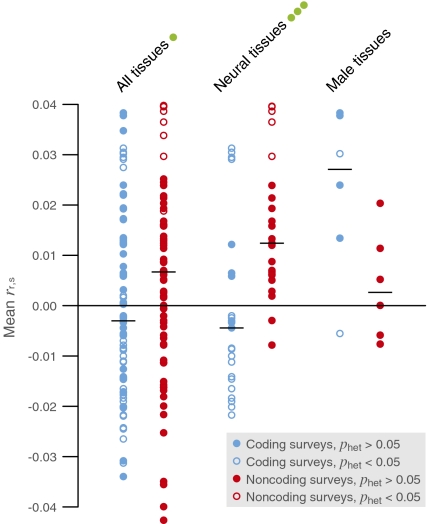
For each tissue, we computed the rank correlation between score for positive selection and specificity to the tissue (rr,s) and its SE within each survey. We then computed the weighted mean of this correlation, its SE, and the heterogeneity statistic across coding surveys and across noncoding surveys. Figure 3 plots the results for all tissues and highlights two groups of tissues exhibiting strong contrasts, and Table S8 presents the complete results. Consonant with the results presented above, specificity to neural tissues (mainly components of brain plus ganglia) is more correlated with adaptive noncoding changes than with adaptive coding changes (P < 10−6, Wilcoxon signed-rank test), whereas specificity to male reproductive tissues (components of testis plus prostate gland) is marginally the opposite (P = 0.063). These contrasts prevail almost without exception, because mean rr,s is greater across noncoding surveys than across coding surveys for all but 1 of the 23 neural tissues, whereas the opposite holds for all but 1 of the 6 male tissues.
Fig. 3.
Expression specificity and positive selection across coding and noncoding surveys. Neural tissues and male (reproductive) tissues are highlighted. Circles represent mean rank correlation (rr,s) between the score for positive selection and specificity to tissues in the Novartis Gene Expression Atlas. Horizontal lines indicate median of means. Green dots on group names indicate two-tailed P values for the Wilcoxon signed-rank test of equal median across coding versus noncoding surveys: one dot, 5 × 10−4 < P < 0.05; three dots, P < 5 × 10−6.
We similarly computed the rank correlation between score for positive selection and evenness across all tissues (rr,e) within each survey, the weighted mean of this correlation across coding and across noncoding surveys, and the associated SEs and heterogeneity statistics. We repeated this computation restricting attention to the 25%, 10%, or 5% most evenly and least evenly expressed genes per survey. Table S9 presents the results. As the evenness tail fraction decreases, the mean rr,e across coding surveys becomes more negative, becoming significantly so when the fraction is 10% or 5% (Pdep = 0.0057 and 0.0076, respectively). This indicates that very evenly expressed genes have experienced less positive selection on their coding sequences than very unevenly expressed genes, a phenomenon reported by Kosiol et al. (9). This pattern persists when only genes in the significantly enriched PANTHER and GO categories are included in the computations. No such pattern is discernible across the noncoding surveys.
Discussion
All of the patterns that we have mentioned persist when any one of the six surveys is excluded (Tables S3, S4, S5, S6, S7, S8, and S9). There are, of course, potential sources of noise or error both in the surveys and in our analyses of them. For example, some sequences exhibiting accelerated evolution in the human lineage may do so due to nonadaptive processes, and many genes have multiple functions or functions missing from current annotations. Moreover, human-specific gene duplications and deletions are beyond the scope of the surveys analyzed here but may have contributed much to human adaptation (20). But assuming that the strong contrasts that we have identified are genuinely characteristic of adaptive evolution, at least in humans, what might account for them?
A simple possibile explanation is that genes with functions enriched with positive selection across coding or noncoding surveys are associated with longer coding or noncoding sequences, respectively. All else being equal, longer sequences should undergo more adaptive changes. Consistent with this idea, genes in noncoding-enriched PANTHER and GO categories are associated with an average of 185 ± 5 kb of noncoding sequence (introns, UTRs, and half of flanking regions), compared with 157 ± 5 kb for genes in coding-enriched categories. Much of this sequence may be nonfunctional, but the surveys by Pollard et al. (10) and Prabhakar et al. (11) treated noncoding sequences that are conserved across nonhuman species and hence presumptively functional. Genes in noncoding-enriched categories were associated with an average of 1,058 ± 50 bp of Pollard et al.’s sequences and 4,367 ± 145 bp of Prabhakar et al.’s sequences, versus 937 ± 67 bp and 3,732 ± 180 bp, respectively, for genes in coding-enriched categories. These differences suggest that some functions might have adapted more through noncoding changes than through coding changes, in part because they are affected by more noncoding sequences per gene, constituting a larger “target” for mutations. Length variation is clearly not the only relevant factor, however. Some categories associated with longer noncoding sequences are not enriched with positive selection across noncoding surveys, for example, the GO biological processes “positive regulation of developmental process” and “cyclic-nucleotide-mediated signaling” (21). Moreover, genes in noncoding-enriched categories also have longer coding sequences than genes in coding-enriched categories (average, 1,974 ± 22 bp vs 1,909 ± 31 bp).
As mentioned earlier, it has been argued that for genes active in many contexts, noncoding changes are more likely than coding changes to be adaptive, because a noncoding mutation is more likely to enhance a gene’s function in one context without degrading it in other contexts, given that gene expression in different contexts is often governed by distinct noncoding sequences (3–5, 19). It follows that if most genes with certain functions are active in many contexts, then adaptation of these functions should occur mainly via noncoding changes. Although expression in many tissues need not entail conflicting demands in different contexts, our results regarding evenness offer modest support for these ideas. Very even genes, many of which presumably play important roles in many contexts, have tended to undergo fewer adaptive coding changes than very uneven genes, most of which presumably play important roles in fewer contexts. This pattern does not appear to hold for adaptive noncoding changes. (Conceivably, we may have lacked statistical power to detect the pattern across noncoding surveys; however, the pattern can be detected across coding surveys even when only the smaller set of genes in significantly enriched PANTHER and GO categories is analyzed.) Moreover, genes in noncoding-enriched PANTHER and GO categories have higher evenness scores on average than genes in coding-enriched categories, although the difference is small (average, 0.62 ± 0.0026 vs 0.59 ± 0.0037).
It also has been argued that selection is typically more efficient on noncoding mutations than on coding mutations, because coding mutations are typically recessive, whereas noncoding mutations are typically codominant with respect to gene expression, although whether the latter are also typically codominant with respect to organismal traits is unclear (2). This differential should be diminished for mutations on the X chromosome, which is hemizygous in males, suggesting that the X might be enriched with positive selection on coding changes but not on noncoding changes. The evidence regarding these ideas from the surveys analyzed here is mixed (Table S10). Across both coding and noncoding surveys, the X is significantly heterogeneous (Phet = 4.9 × 10−4 and 5.0 × 10−4), indicating appreciable discord among the surveys. The X is enriched in two of the three coding surveys (7, 8) but also in one of the three noncoding surveys (11).
A full understanding of which traits adapt mainly via coding changes versus noncoding changes and why must await the maturity of functional genomics. In particular, many more functional annotations of noncoding sequences are needed. Nonetheless, our current findings are important. At a minimum, they strongly suggest that both coding and noncoding changes have played important and distinctive roles in human adaptation. One implication is that studying adaptive coding changes alone, which has long been a major focus of research in evolutionary genetics, can yield an incomplete and unbalanced picture of adaptive evolution, which can be significantly extended and enriched by studies of adaptive noncoding changes. More such studies in nonhuman species are needed to reveal which of the patterns that we have found in humans exist in other species as well.
The finding that neural adaptation has occurred mainly via noncoding changes is particularly important in view of the remarkable cognitive innovations in the human lineage. It is consistent with the hypothesis advanced 35 years ago that the major phenotypic differences between humans and chimpanzees reflect changes in gene regulation rather than in protein structure (22). Noncoding sequences flagged by the surveys analyzed here and associated with neural development and function are excellent candidates for research into the genetics and evolution of human cognition.
Methods
Surveys and Scores.
To analyze the noncoding surveys (10–12), we associated each noncoding sequence treated in those surveys with the nearest coding sequence in the University of California Santa Cruz Known Genes collection (23). The survey of Pollard et al. (10) includes sequences overlapping known coding sequences, which we excluded from our analyses. For the surveys of Pollard et al. (10) and Prabhakar et al. (11), some genes were associated with multiple sequences, in which case we combined the P values of Pollard et al. or Prabhakar et al. for the sequences into a P value for the gene using the method of Simes (24); the P value of the gene is the minimum Benjamini-Hochberg–adjusted P value of the sequences. This is important, because although these surveys have no a priori bias regarding function, genes with certain functions tend to be larger or more isolated and hence associated with more sequences treated in these surveys (21). If this phenomenon were not accounted for when converting sequence scores into gene scores, such functions would tend to be enriched with high-scoring genes purely by chance, in the absence of positive selection or any other evolution-accelerating process. Simes's method avoids this potential bias, because the more sequences are associated with a gene, the more their P values are discounted.
No sequence-based test for positive selection is perfectly reliable. Along with false-positives occurring purely by chance, nonadaptive processes can mimic signatures of positive selection (10, 13–15). An important motivation for our analyses is that to a substantial extent, different tests are sensitive to different confounding factors (13–15). For example, the survey of Pollard et al. (10) detected accelerated evolution in the human lineage relative to several other lineages, assessing relative acceleration in a region of interest with respect to a set of reference regions. Such acceleration might result from relaxation of negative selection or biased gene conversion (BGC), as Pollard et al. discuss (10). In contrast, the surveys of Prabhakar et al. (11) and Haygood et al. (12) assessed acceleration with respect to estimates of the local rate of neutral evolution, which is unlikely to reflect relaxation of negative selection or BGC (11, 12, 14, 15). Indeed, Haygood et al.’s null model explicitly accommodates relaxation of negative selection, and these authors found no evidence that BGC influenced their results (12).
Four of the six surveys are sensitive to positive selection in the human lineage alone, but the surveys of Bustamante et al. (7) and Nielsen et al. (8) are sensitive to positive selection in the chimpanzee lineage as well. However, of the patterns discussed here, only enrichment of the X chromosome is supported by these two surveys, but not by the coding survey of Kosiol et al. (9).
Table S1 provides more information about the surveys.
Functional Categories and Expression Domains.
In each survey, we mapped as many genes as we could first to UniProt (25) identifiers and then to PANTHER (16) and GO (17) categories (excluding placeholder categories, e.g., “biological process unclassified”). For each category, we computed the rank-biserial correlation, rrb, between the score for positive selection and membership in the category. These computations involved only genes that were treated in the given survey and that we were able to map to at least one category of the relevant kind. rrb measures the association between an ordinal variable and a dichotomous variable (26); it is proportional to the standard (Pearson) correlation coefficient between the ranks of the ordinal variable and any two values, say 0 and 1, for the dichotomous variable. We treated the score for positive selection as an ordinal variable rather than as a continuous variable, to avoid issues that might arise from the diversity of scoring functions among the surveys. For each category, we estimated the SE of rrb as the SD of rrb over 1,000 bootstrap replicates per survey.
Similarly, in each survey we mapped as many genes as we could to Novartis (18) probes. We took means over multiple arrays per tissue and maxima over multiple probes per gene to obtain the expression of a gene in a tissue. For each gene, we computed specificity and evenness scores, as illustrated in Fig. 2, but in the context of all 73 noncancerous tissues in the atlas. For each tissue, we computed the rank (Spearman) correlation, rr,s, between the score for positive selection and specificity to the tissue. We also computed the rank correlation, rr,e, between the score for positive selection and evenness across all tissues. We estimated the SEs of rr,s and rr,e from 1,000 bootstrap replicates per survey.
Fixed-Effects Meta-Analyses.
Our analyses of patterns across surveys are essentially standard fixed-effects meta-analyses (27). Given a category and a set of surveys, the weighted mean of rrb is ∑i wi (rrb)i, where wi = [1/SE(rrb)i]2/∑k [1/SE(rrb)k]2. It follows that the SEM is 1/{∑i[1/SE(rrb)i]2}1/2. We tested mean rrb = 0 by comparing (mean rrb)/(SEM rrb) to the standard normal distribution; we denote the upper and lower one-tailed P values by Penr and Pdep, respectively. Under the null hypothesis, (rrb)i = (rrb)k for every i and k, the heterogeneity statistic ∑i wi [(rrb)i − mean rrb]2 has approximately a χ2 distribution with 2 degrees of freedom, assuming three surveys; we denote the χ2 P value by Phet. Analogous formulas and procedures apply to mean rr,s and mean rr,e.
Random-Effects Meta-Analyses.
Fixed-effects meta-analyses are sometimes criticized on the grounds that homogeneity across studies is unrealistic, and Phet is an unsatisfactory indicator (28). Tables S3, S4, S5, S6, S7, S8, and S9 also present results from a random-effects approach (28). Here mean rrb is as above but with wi = ni/∑k nk, where ni is the number of categorized genes in the ith survey. More importantly, rrb is considered a random variable over surveys, the SEM is {∑i ni [(rrb)i − mean rrb]2/3∑k nk}1/2, and the test of mean rrb = 0 is a t test with 2 degrees of freedom, assuming three surveys. Again, analogous formulas and procedures apply to mean rr,s and mean rr,e.
Although the fixed- and random-effects results differ in detail, they display the same major patterns. For example, according to both sets of results, the PANTHER biological processes “neurogenesis,” “other neuronal activity,” and “muscle development” are enriched with positive selection across noncoding surveys; “immunity and defense,” “chemosensory perception,” and “spermatogenesis and motility” are enriched across coding surveys; and “T cell–mediated immunity” is enriched across both noncoding and coding surveys.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911249107/DCSupplemental.
References
- 1.Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 2.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 3.Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 4.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:1159–1166. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 6.Kelley JL, Swanson WJ. Positive selection in the human genome: From genome scans to biological significance. Annu Rev Genomics Hum Genet. 2008;9:143–160. doi: 10.1146/annurev.genom.9.081307.164411. [DOI] [PubMed] [Google Scholar]
- 7.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:976–985. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosiol C, et al. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4:1–17. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard KS, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:1599–1611. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 12.Haygood R, Fedrigo O, Hanson B, Yokoyama K-D, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- 13.Li YF, Costello JC, Holloway AK, Hahn MW. “Reverse ecology” and the power of population genomics. Evolution. 2008;62:2984–2994. doi: 10.1111/j.1558-5646.2008.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabhakar S, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prabhakar S, et al. Response to comment on “Human-specific gain of function in a developmental enhancer”. Science. 2009;323:714. doi: 10.1126/science.1165848. [DOI] [PubMed] [Google Scholar]
- 16.Mi H, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gene Ontology Consortium. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 20.Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taher L, Ovcharenko I. Variable locus length in the human genome leads to ascertainment bias in functional inference for noncoding elements. Bioinformatics. 2009;25:578–584. doi: 10.1093/bioinformatics/btp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King M-C, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 23.Karolchik D, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. [Google Scholar]
- 25.UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraemer HC. In: EncycloPedia of Statistical Sciences. Kotz S, Johnson NL, editors. New York: Wiley; 1982. pp. 276–280. [Google Scholar]
- 27.Hedges LV. In: The Handbook of Research Synthesis. Cooper H, Hedges LV, editors. New York: Russell Sage Foundation; 1994. pp. 285–299. [Google Scholar]
- 28.Hunter JE, Schmidt FL. Methods of Meta-Analysis. 2nd Ed. Thousand Oaks, CA: Sage; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.