Abstract
Macroautophagy (hereafter autophagy) is a ubiquitous process in eukaryotic cells that is integrally involved in various aspects of cellular and organismal physiology. The morphological hallmark of autophagy is the formation of double-membrane cytosolic vesicles, autophagosomes, which sequester cytoplasmic cargo and deliver it to the lysosome or vacuole. Thus, autophagy involves dynamic membrane mobilization, yet the source of the lipid that forms the autophagosomes and the mechanism of membrane delivery are poorly characterized. The TRAPP complexes are multimeric guanine nucleotide exchange factors (GEFs) that activate the Rab GTPase Ypt1, which is required for secretion. Here we describe another form of this complex (TRAPPIII) that acts as an autophagy-specific GEF for Ypt1. The Trs85 subunit of the TRAPPIII complex directs this Ypt1 GEF to the phagophore assembly site (PAS) that is involved in autophagosome formation. Consistent with the observation that a Ypt1 GEF is directed to the PAS, we find that Ypt1 is essential for autophagy. This is an example of a Rab GEF that is specifically targeted for canonical autophagosome formation.
Keywords: Rab, stress, trafficking, vacuole, yeast
Autophagy is a catabolic process in which damaged or superfluous cytoplasmic components are degraded in response to stress conditions; it is evolutionarily conserved in eukaryotes and is integrally involved in development and physiology (1, 2). The morphological hallmark of autophagy is the formation of double-membrane cytosolic vesicles, autophagosomes, which sequester cytoplasm. The autophagosomes then fuse with the lysosome, resulting in the degradation of the cargo. The mechanism of autophagosome formation is distinct from that used for vesicle formation in the secretory or endocytic pathways and is said to be de novo in that it does not occur by direct budding from a preexisting organelle. Instead, a nucleating structure, the phagophore, appears to expand by the addition of membrane possibly through vesicular fusion. One consequence of this mechanism is that it allows the sequestration of essentially any sized cargo, including intact organelles or invasive microbes, and this capability is critical to autophagic function. When autophagy is induced there is a substantial demand for membrane, and a major question in the field concerns the membrane origin; nearly every organelle has been implicated in this role (3). The early secretory pathway is likely one such membrane source for autophagy (4, 5).
Rab GTPases are key regulators of membrane traffic that mediate multiple events including vesicle tethering and membrane fusion. These molecular switches cycle between an inactive (GDP-bound) and active (GTP-bound) conformation. The yeast Rab Ypt1, which is essential for ER-Golgi and Golgi traffic (6), is activated by the multimeric guanine nucleotide exchange factor (GEF) called TRAPP (7, 8). Two forms of the TRAPP complexes have been identified (9). These two complexes share several subunits, including four (Bet3, Bet5, Trs23, and Trs31) that are essential to activate Ypt1. How each of these subunits contributes to nucleotide exchange activity has recently been described (10). The first and smaller form of the complex, TRAPPI, mediates ER-Golgi traffic (11). The second and larger complex, TRAPPII, mediates Golgi traffic (12, 13). Both complexes are tethering factors that are needed to tether vesicles to their acceptor compartment. The TRAPPI complex recognizes the coat (COPII) on ER-derived vesicles (11), whereas the TRAPPII complex recognizes the coat (COPI) on Golgi-derived vesicles (12, 13). Subunits specific to TRAPPII, bind to the COPI coat complex (13). We have proposed that TRAPPII-specific subunits mask the COPII binding site on TRAPPI to convert this GEF into a tethering factor that recognizes a new class of vesicles (10, 13).
The TRAPP complexes include three nonessential subunits, Trs33, Trs65, and Trs85 (9, 14). Previous studies demonstrate that Trs85 is required for the cytoplasm to vacuole targeting (Cvt) pathway, a specific type of autophagy, and macroautophagy, the nonspecific autophagy of cytoplasm (15, 16). These earlier studies did not resolve if a free pool of Trs85 or TRAPP was required for the Cvt pathway and autophagy. Here, we show that Trs85 is part of a third TRAPP complex, TRAPPIII, which specifically acts in autophagy. TRAPPIII is a Ypt1 GEF that is targeted to the PAS by the Trs85 subunit. Consistent with this proposal, we also show that Ypt1 is required for both specific and nonspecific types of autophagy.
Results
Trs85 Is a Component of a Third TRAPP Complex That Is a Ypt1 GEF.
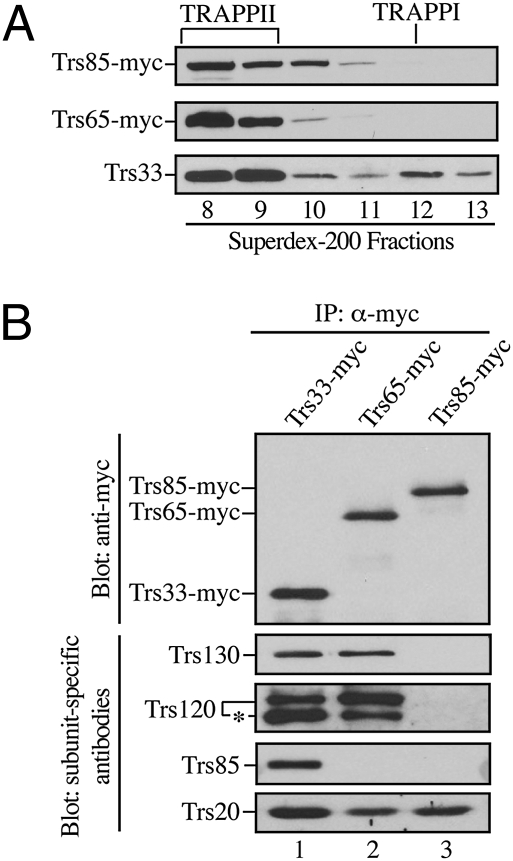
The finding that Trs85 is required for autophagy in yeast (15, 16) raised the possibility that there may be a separate pool of Trs85 that functions in autophagy-related processes. To begin to address this possibility, we fractionated cytosol on a Superdex-200 column and blotted the column fractions for Trs85, Trs65, and Trs33. Trs33 and Trs65 (TRAPPII-specific subunit) marked the location of the TRAPPI and TRAPPII complexes in these fractions (Fig. 1A, Middle and Bottom). There are 10 TRAPP subunits (Trs130, Trs120, Trs85, Trs65, Trs33, Trs31, Trs23, Trs20, Bet5, and Bet3). Three of these subunits (Trs130, Trs120, and Trs65) are unique to TRAPPII (9, 14). Trs33 peaked in fractions 8, 9 (TRAPPII), and 12 (TRAPPI) (Fig. 1A), whereas Trs65-myc was largely found in fractions 8 and 9 (TRAPPII). The location of Trs85 in these fractions was determined by monitoring an epitope tagged version of Trs85 (Trs85-myc) that was shown to be functional (Fig. S1 A and B). Trs85-myc trailed more than Trs65-myc on the Superdex-200 column and was primarily present in column fractions 8–10 (Fig. 1A, Top and Middle).
Fig. 1.
Trs85 does not coprecipitate with Trs130, Trs120, and Trs65. (A) Trs85 is found in Superdex-200 column fractions 8–10. Cleared lysates (from strains SFNY1295 and SFNY1302) were fractionated and analyzed by Western blot analysis with anti-myc antibody (Top and Middle) or anti-Trs33 antibody (Bottom). (B) Trs130, Trs120, and Trs65 do not coprecipitate with Trs85 from lysates. Cleared lysates were immunoprecipitated with anti-myc antibody and immunoblotted for the presence of the indicated TRAPP subunits. Top was blotted with anti-myc antibody and Lower with subunit-specific antibodies as indicated. The asterisk marks a degradation product of Trs120.
The fractionation of Trs85-myc suggested there may be a pool of Trs85 that is not present in either TRAPPI or TRAPPII. To begin to address this possibility, we immunoprecipitated Trs85-myc from lysates and compared the precipitated TRAPP subunits to precipitates of Trs33-myc and Trs65-myc. These data showed that Trs85 coprecipitated with Trs33-myc and Trs20 but not Trs65-myc, Trs130, or Trs120 (or a breakdown product of Trs120, see starred band in Fig. 1B and ref. 9) (Fig. 1B).
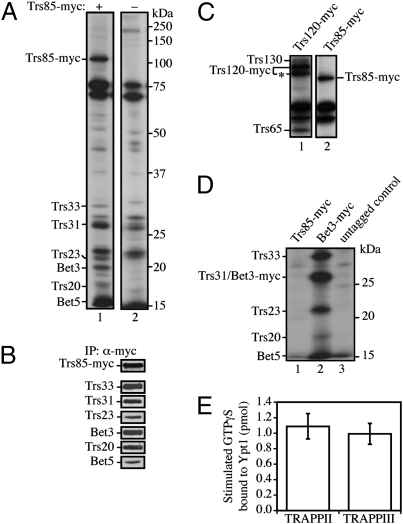
To identify the other TRAPP subunits that coprecipitate with Trs85, we fractionated a radiolabeled lysate prepared from the Trs85-myc strain and precipitated Trs85 from Superdex-200 column fraction 8; contaminating proteins in the precipitate were identified by fractionating an untagged lysate. This analysis revealed that Trs85-myc coprecipitates with Trs33, Trs31, Trs23, Bet3, Trs20, and Bet5 (Fig. 2A, compare lane 1 to untagged control in lane 2). The identity of these coprecipitating bands was confirmed by precipitating Trs85-myc from fraction 8 and blotting for Trs33, Trs31, Trs23, Bet3, Trs20, and Bet5 (Fig. 2B). The TRAPPII-specific subunits Trs65, Trs130, Trs120, and a breakdown product of Trs120 (see starred band in Fig. 2C) were not detected in the Trs85-myc precipitate from fraction 8 (compare lane 2 with the Trs120-myc precipitate in lane 1 in Fig. 2C). Together, these findings show that Trs85 is not a component of the TRAPPII complex. Additionally, none of the small TRAPP subunits could be precipitated from fraction 12 (TRAPPI) when a Trs85-myc-containing lysate was fractionated (Fig. 2D, compare the TRAPPI complex in lane 2 with lane 1 and the untagged control in lane 3), and Trs85 was not detected when Bet3-myc was precipitated from fraction 12 (Fig. S2). Together, these findings indicate that Trs85 is not a component of the TRAPPI or TRAPPII complexes, which are required for ER-Golgi and Golgi traffic (9, 12). Consistent with this observation, we observed no significant delay in the trafficking of the vacuolar protease Prc1 from the ER through the Golgi complex in trs85Δ cells (Fig. S3A) or an affect on cell growth (Fig. S3B). Although an earlier study suggested that Trs85 is a subunit of the TRAPPI and TRAPPII complexes (9), the data we report here imply this is not the case. Instead, Trs85 appears to be a specific component of a third TRAPP complex, called TRAPPIII, which is required for autophagy and the Cvt pathway (see Fig. S4 for a summary of the subunits in the different TRAPP complexes). Earlier characterization of the TRAPP complexes was done by precipitating Bet3-myc from Superdex-200 column fractions (9); because Bet3 is present in all three TRAPP complexes, we speculate that TRAPPIII was not resolved from TRAPPI and TRAPPII in the earlier study.
Fig. 2.
Identification of a third TRAPP complex that activates Ypt1. (A) Trs85-myc coprecipitates with Trs33, Trs31, Trs23, Bet3, Trs20, and Bet5. Radiolabeled lysates (from strains SFNY1295 and NY915, respectively) were fractionated, and fraction 8 was immunoprecipitated as described in Materials and Methods. (B) Confirmation of TRAPP subunits that coprecipitate with Trs85. A lysate prepared from strain SFNY1295 was fractionated on a Superdex-200 column. Fraction 8 was immunoprecipitated with anti-myc antibody and analyzed by Western blot analysis using antibody to the indicated TRAPP subunit. (C) Trs130, Trs120, and Trs65 do not coprecipitate with Trs85 from fraction 8. Radiolabeled lysates (from strains SFNY1301 and SFNY1295) were fractionated, and fraction 8 was immunoprecipitated. The two dark bands that appear above Trs65 are contaminants (see the untagged control in A lane 2). The asterisk marks a degradation product of Trs120. (D) Trs85 is not a component of the TRAPPI complex. Radiolabeled lysates (from strains SFNY1295, SFNY656, and NY915) were fractionated, and fraction 12 was immunoprecipitated. (E) TRAPPIII is a Ypt1 GEF. TRAPPII was purified from a strain containing TAP-tagged Trs65 (SFNY1075), and TRAPPIII was purified from a strain expressing TAP-tagged Trs85 (SFNY1080) by incubating lysate with IgG-Sepharose beads as described previously (8). The beads were then used to assay for the uptake of GTPγS onto Ypt1. The data shown are normalized to the amount of Trs33 present on IgG-Sepharose beads.
The TRAPPIII complex contains all of the subunits that are required for Ypt1 GEF activity (10). To determine if Trs85 is a component of a functional Ypt1 GEF, we immobilized TAP-tagged Trs85 on IgG-Sepharose beads and assessed its ability to stimulate the uptake of GTPγS onto Ypt1. As a control, we also assayed TRAPPII (TAP-tagged Trs65), which has comparable Ypt1 GEF activity to TRAPPI (10). Both TAP-tagged Trs85 and Trs65 stimulated the uptake of GTPγS onto Ypt1 to approximately the same level (Fig. 2E). These findings show that Trs85 is a component of a Ypt1 GEF that is distinct from TRAPPI and TRAPPII.
Ypt1 Is Required for Nonspecific Autophagy.
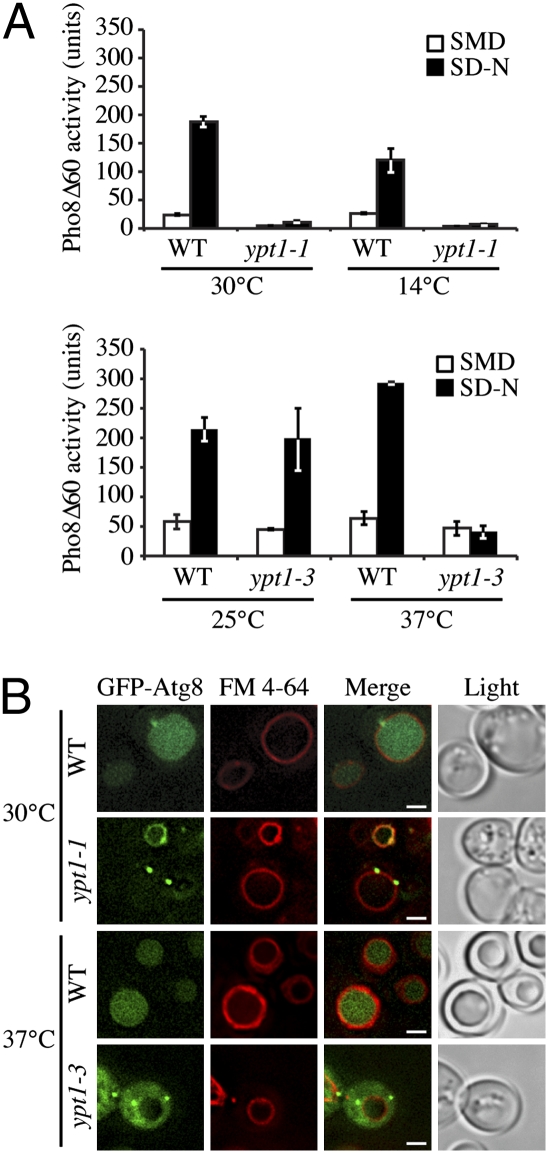
The observation that a component of a Ypt1 GEF is required for autophagy (15, 16) suggests that Ypt1 may also be required for this event. To address this possibility, the role of this GTPase in autophagy was examined. Ypt1 is an essential component of the ER-Golgi trafficking machinery, and its loss leads to cell death (6, 17). To circumvent this problem, we used conditional ypt1 mutants. The ypt1-1, ypt1-3 and ypt1A136D mutants are temperature-conditional partial loss-of-function alleles of YPT1 that block ER-Golgi traffic at the nonpermissive temperature (18). To begin our analysis, we used the Pho8Δ60 (vacuolar alkaline phosphatase lacking the N-terminal 60 amino acids) assay, which provides a quantitative method to measure autophagic activity (19). In wild-type yeast, nitrogen-starvation induces autophagy and delivery of Pho8Δ60 from the cytosol into the vacuole lumen, resulting in activation of Pho8. The ypt1-1 mutant, which grows poorly at all temperatures, was defective in Pho8Δ60 activation at the permissive (30 °C) and nonpermissive (14 °C) temperatures (Fig. 3A). The ypt1A136D mutant showed some reduction in activity at the permissive (25 °C) temperature relative to the isogenic parental strain and essentially a complete block in Pho8Δ60 activity at 37 °C (Fig. S5A), whereas ypt1-3 was essentially normal at the permissive (25 °C) temperature, but blocked at 37 °C (Fig. 3A), indicating impaired autophagy in both cases. These results also suggest that the defect was not allele-specific.
Fig. 3.
Ypt1 is involved in nonspecific autophagy in yeast. (A) Isogenic wild-type (WT) and the indicated ypt1 mutant strains were cultured in growth medium (SMD) at the permissive temperature (30 °C or 25 °C), to exponential phase and then shifted to a nonpermissive temperature (14 °C or 37 °C); the ypt1-1 mutant is cold sensitive for growth and was analyzed at 14 °C. In parallel, cells were switched to nitrogen starvation (SD-N) medium for 4 h at each temperature. Cell lysates from each condition were collected and assayed for Pho8Δ60 activity. Error bars indicate SE. (B) The isogenic wild-type and ypt1 mutant strains were transformed with GFP-Atg8, grown to exponential phase as in A and then shifted to the nonpermissive temperature for 30 min. Rapamycin (0.2 μM) was added for 4 h. Shown are epifluorescent images of GFP-Atg8 and the vacuolar limiting membrane (stained with FM 4–64). (Scale bars, 2.5 μm.)
As a second method to analyze autophagy, we examined translocation of the autophagy protein Atg8 (fused to GFP) to the vacuole. In wild-type cells, GFP-Atg8 is localized to the PAS and the cytosol. Rapamycin treatment induces autophagy and results in delivery of GFP-Atg8 into the vacuole. This process is evidenced by a GFP signal in the vacuole lumen. At the permissive or nonpermissive temperature, all of the wild-type strains showed GFP-Atg8 localization at the PAS and/or in the vacuole lumen (Fig. 3B, Fig. S5B). In contrast, the ypt1-3 and ypt1A136D mutants at the nonpermissive temperature, and the ypt1-1 strain at either temperature, displayed blocks in GFP-Atg8 transport, as shown by a lack of GFP signal in the vacuole lumen of the mutant cells after rapamycin treatment (Fig. 3B and Fig. S5B).
Membrane flow through the secretory pathway is required for autophagy (4, 5), leaving open the possibility that any defect in autophagy that we observe may be an indirect consequence of blocking secretion. For this reason, we also analyzed the ypt1-2 mutant, which is not temperature-sensitive for growth and blocks membrane traffic in vitro but not in vivo (17). The ypt1-2 mutant displayed a defect in Pho8Δ60 activity (Fig. S5A) and in the delivery of GFP-Atg8 to the vacuole at elevated temperatures (Fig. S5B). Although we observed occasional GFP fluorescence in the vacuole lumen with the ypt1-2 mutant, the percentage of cells exhibiting vacuolar GFP-Atg8 localization was 17% for the ypt1-2 strain compared to 90% for wild type at 37 °C.
Previous studies indicate that the trs85Δ mutant is defective in autophagosome formation (16) and in GFP-Atg8 localization to the PAS (15). To place Ypt1 and Trs85 within the autophagy pathway we examined GFP-Atg8 localization in two different ypt1 alleles. We found an approximately 24%, 47%, and 26% reduction in GFP-Atg8 puncta at the PAS in the trs85Δ, ypt1-2, and ypt1-3 mutants, respectively (Fig. S6A). In addition, when compared to wild type, these mutants displayed a substantial increase in the number of cells with more than one GFP-Atg8 punctum per cell (Fig. S6B), suggesting a defect in both the recruitment and localization of Atg8 to the PAS. Combined with the previous data, these results imply that Ypt1 and Trs85 play a role in autophagosome biogenesis.
Ypt1 Is Required for Specific Autophagy.
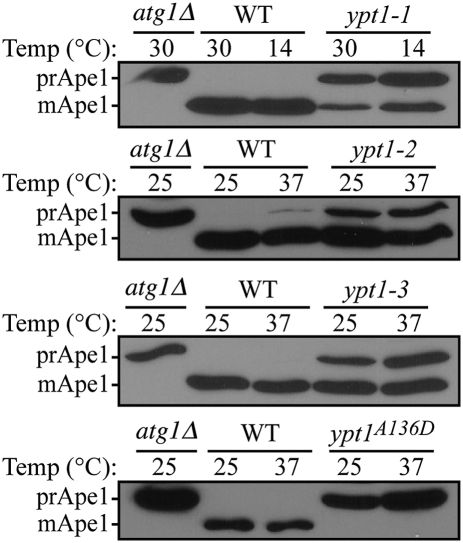
To extend our analysis of the role of Ypt1 in autophagy, we examined a selective type of autophagy, the Cvt pathway, which delivers the precursor form of the resident hydrolase aminopeptidase I (prApe1) to the vacuole, where it is proteolytically activated. As expected, atg1Δ mutant cells that are defective in the Cvt pathway accumulated prApe1 (Fig. 4). In contrast, wild-type cells contained primarily the mature form of Ape1 (Fig. 4). The ypt1 mutant strains displayed variable defects in the processing of prApe1, ranging from partial (ypt1-1, ypt1-2, and ypt1-3) to complete blocks (ypt1A136D). In general, equivalent defects were seen at the permissive and nonpermissive temperatures, suggesting that the Cvt pathway was particularly sensitive to the functional status of Ypt1. It is likely that the mutant protein does not retain complete function even at the permissive temperatures. These results indicate a role for Ypt1 in specific autophagy.
Fig. 4.
The ypt1 mutants are defective for the Cvt pathway. The isogenic wild-type and ypt1 mutant strains used in Fig. 3 and an atg1Δ control strain were cultured in rich medium (SMD) at the indicated permissive temperature to exponential phase and then shifted to the indicated nonpermissive temperature for 60 min. Protein extracts were resolved by SDS/PAGE and Western blot analysis was performed with anti-Ape1 antiserum. The positions of prApe1 and mature Ape1 are as indicated.
A Constitutively Active ypt1 Mutant Suppresses the trs85Δ Defect.
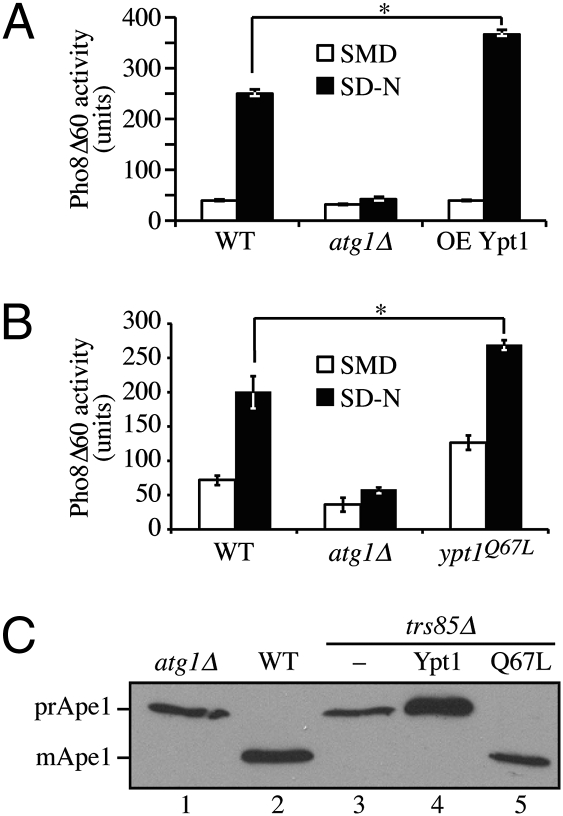
If Ypt1 is involved in directing membrane flow to the autophagy pathway, we hypothesized that elevated expression of Ypt1 might display enhanced autophagic activity. Therefore, we overexpressed Ypt1 and examined autophagy using the Pho8Δ60 assay. There was no change in autophagy activity in rich medium; however, following starvation we observed an approximately 30% increase in Pho8Δ60 activity (Fig. 5A), suggesting that Ypt1 is a limiting factor for the autophagy process.
Fig. 5.
Ypt1 overexpression enhances autophagy and constitutively active Ypt1 bypasses the requirement for its GEF. (A) Wild-type (WT; TN124), atg1Δ, and Ypt1 overexpressing (OE Ypt1) cells were grown at 30 °C and shifted to SD-N medium for 4 h. Protein extracts were analyzed by the Pho8Δ60 assay. Error bars indicate SE. The asterisk indicates a significant difference from the WT level, P < 0.05. (B) WT, atg1Δ, and ypt1Q67L cells were grown and analyzed as in A. (C) WT, atg1Δ, and trs85Δ cells harboring an empty vector or a plasmid encoding Ypt1 or Ypt1Q67L were grown in rich medium to exponential phase. Protein extracts were resolved by SDS/PAGE and Western blot analysis was performed with anti-Ape1 antiserum.
We extended our analysis by determining whether a constitutively active (i.e., GTP-bound) form of Ypt1 affected autophagy. First, we expressed the ypt1Q67L mutant under the control of the GAL1 promoter and examined the effect on nonspecific autophagy. Even under basal conditions (nutrient-rich medium) there was an elevation in Pho8Δ60 activity in the presence of ypt1Q67L (Fig. 5B). A similar increase in activity was seen when autophagy was induced by starvation (Fig. 5B). Second, to further examine the role of the Trs85-containing TRAPPIII complex as an autophagy-specific GEF for Ypt1, we expressed the constitutively active ypt1Q67L mutant in a strain deleted for TRS85 and monitored the effect on the Cvt pathway. As seen previously, the trs85Δ mutant accumulated only the precursor form of Ape1 similar to the atg1Δ control strain (Fig. 5C, lanes 1 and 3). Overexpression of wild-type Ypt1 in the trs85Δ mutant had no effect on prApe1 processing (Fig. 5C, lane 4). In contrast, overexpression of the ypt1Q67L mutant resulted in complete maturation of prApe1, indicating efficient delivery to the vacuole (Fig. 5C, lane 5). Thus, the constitutively active form of Ypt1 suppressed the defect in the Cvt pathway that resulted from the loss of Trs85.
Trs85 Directs Ypt1 to the PAS.
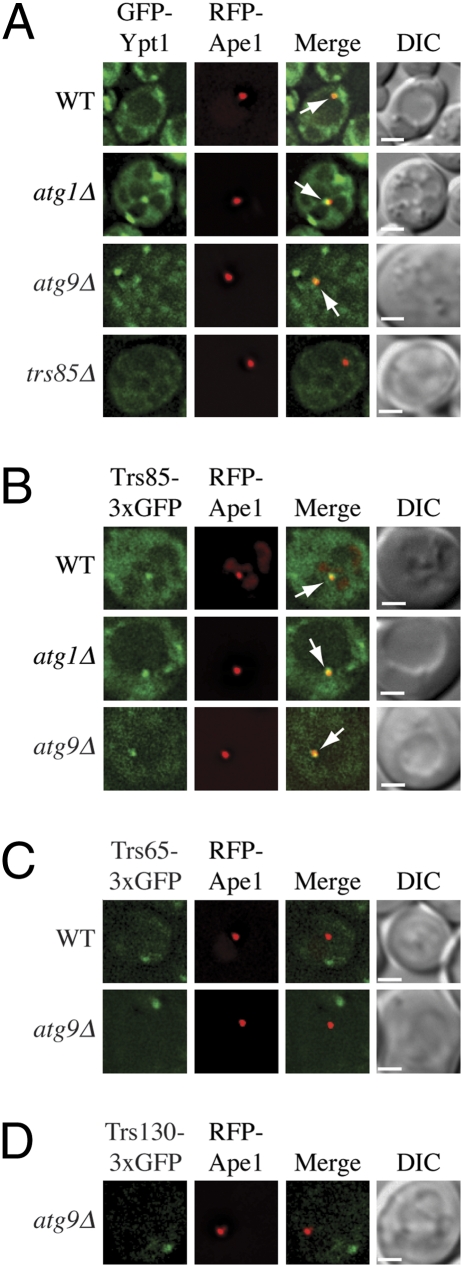
The observation that Trs85 is required for autophagy (15, 16) and is part of an autophagy-specific GEF for Ypt1 raised the possibility that TRAPPIII is targeted to the PAS. To address this point, we examined the localization of Trs85 and Ypt1. In yeast, autophagosomes are thought to form at the PAS. Thus, Ypt1 and Trs85 should localize to the PAS if they play a direct role in autophagy. Because of this, we compared the localization of Ypt1 and Trs85 to Trs65 and Trs130. Trs65 and Trs130 are only present in the TRAPPII complex, and Trs65 is not required for the Cvt pathway or autophagy (15, 16). All proteins were tagged with GFP and expressed in cells with RFP-Ape1, a marker for the PAS. The cells were grown to midlog phase and then starved for 45 min before determining the extent of colocalization between RFP-Ape1 and the GFP tagged proteins. Both Ypt1 and Trs85 colocalized to the PAS at a much higher rate than Trs65 or Trs130 (approximately 44%, 34%, 4%, and 10%, respectively) (Fig. 6 and Table S1). Cells lacking atg1Δ are defective in autophagosome formation, which leads to the accumulation of autophagic proteins at the PAS (20). Therefore, we examined the localization of Ypt1 and the TRAPP subunits in an atg1Δ strain. We found an increase in colocalization in the absence of Atg1, which was most apparent for Trs85 (Fig. 6 and Table S1); however, even in the atg1Δ background, Trs65 and Trs130 did not display significant colocalization with the PAS marker. To verify that the punctate appearance of Trs85 was not due to multimerization of the triple-GFP tag, we expressed Trs85-3xGFP in the multiple-knockout (MKO) strain. The MKO strain lacks the 24 ATG genes that are known to be required for autophagosome formation in Saccharomyces cerevisiae (21). When expressed in the MKO strain, Trs85-3xGFP displayed occasional puncta along with diffuse cytosolic staining (Fig. S7). The few Trs85-3xGFP puncta observed did not colocalize with RFP-Ape1 in either rich medium or starvation conditions, indicating that this chimera did not aggregate and did not colocalize with prApe1 in the absence of the Atg proteins. These findings indicate that Trs85, but not Trs65 or Trs130, localizes to the PAS and that this localization is dependent on Atg proteins. Consistent with the notion that Trs85 and Trs65 are in separate TRAPP complexes, we found a larger cytoplasmic pool of Trs85 (Fig. 6 and Fig. S8A).
Fig. 6.
Ypt1 and Trs85 localize to the PAS. (A) Wild-type (WT, SEY6210), atg1Δ (WHY1), atg9Δ (JKY007), and trs85Δ (YJH3) cells transformed with plasmids expressing RFP-Ape1 and GFP-Ypt1; (B) WT (YJH9), atg1Δ (YCB151), and atg9Δ (MDY15) cells expressing integrated RFP-Ape1 and Trs85-3xGFP; (C) WT (SFNY1573) and atg9Δ strains expressing Trs65-3xGFP (MDY12); or (D) the atg9Δ strain expressing Trs130-3xGFP (MDY10) were cultured in SMD medium to exponential phase before being transferred to SD-N medium for 45 min. The cells were then analyzed by fluorescence microscopy. Arrows mark overlapping GFP-Ypt1 and RFP-Ape1 puncta, and overlapping Trs85-3xGFP and RFP-Ape1 puncta. (Scale bar, 2.5 μm.)
We also examined localization of these subunits in the atg9Δ and atg11Δ strains. The absence of either Atg9 or Atg11 caused a decrease in the level of Trs85 or Ypt1 that colocalized with RFP-Ape1 relative to the atg1Δ strain, but neither deletion had a significant effect on the low level of colocalization seen with Trs65 or Trs130 (Table S1). Atg11 is required for the movement of Atg9, a putative membrane carrier for autophagy-related pathways (22, 23), to the PAS. Decreased colocalization of Trs85 and Ypt1 in the atg9Δ and atg11Δ mutants relative to the atg1Δ strain is consistent with the hypothesis that Ypt1 and its GEF tethers Atg9-containing membranes needed for the biogenesis of autophagic sequestering vesicles. In agreement with this proposal, we found that the peripheral pools of Atg9, which are thought to mark the donor membranes involved in autophagosome biogenesis, colocalize with Ypt1 (85% ± 6 of the cells have at least one overlapping punctum, n = 211 cells; Fig. S8B). Finally, we monitored the localization of Ypt1 in the absence of Trs85. GFP-Ypt1 colocalization with RFP-Ape1 dropped from approximately 44% in the wild-type strain to 12% in the trs85Δ background (Fig. 6 and Table S1). Together, these findings imply that Trs85 plays a role in directing Ypt1 to the PAS.
Discussion
Here we describe a third form of the TRAPP complex, TRAPPIII, which contains Trs85 and is a GEF for Ypt1. Additionally, we show that TRAPPIII and Ypt1 are required for the Cvt pathway and nonselective autophagy. The defect in autophagy-related processes in the ypt1 mutants we analyzed is not an indirect consequence of blocking ER-to-Golgi traffic because these phenotypes are seen under conditions where secretory traffic is normal. Similarly, the loss of Trs85 has no effect on the secretory pathway but disrupts specific and nonspecific autophagy. We also found that Trs85 and Ypt1, and not the TRAPPII-specific subunits Trs65 and Trs130, localize to the PAS. Although previous studies implicated Trs85 in these processes (15, 16), the relationship between Trs85, TRAPP and autophagy has remained unclear until now. The findings we report here indicate that TRAPPIII and Ypt1 play a direct role in the Cvt pathway and autophagy. Additionally, they imply that Trs85 directs the Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy.
Our experiments show that Ypt1 is essential for both specific and nonspecific autophagy (Fig. 3–5). Based on the results we report here and previous findings, we propose that TRAPPIII and Ypt1 are required for a membrane tethering event that is needed for Cvt vesicle and autophagosome formation. It was recently reported that the Ypt1 effector COG is also needed for the Cvt pathway and autophagy (24). The COG complex contains two lobes, A and B (25). The A lobe, but not the B lobe, is required for autophagosome formation (24). Consistent with the proposal that TRAPPIII and Ypt1 are required for Cvt vesicle formation, prApe1 is sensitive to exogenously added protease in trs85Δ cells (15, 16) and Atg8 is mislocalized in trs85Δ and ypt1 mutants (15, 16) (Fig. S6). No defect in the localization of Atg9 was previously reported in trs85Δ cells (15). Together, these results imply that TRAPPIII and Ypt1 are needed after Atg9 is recruited to the PAS but before or at the stage of Atg8 recruitment.
Our findings show that yeast cells contain three GEFs for Ypt1: TRAPPI, TRAPPII and TRAPPIII. The TRAPP complexes act in ER-Golgi traffic (TRAPPI) (9, 11), Golgi traffic (TRAPPII) (9, 12), and autophagy (TRAPPIII). All three complexes share several subunits (Bet3, Trs23, Bet5, and Trs31) that are essential for Ypt1 GEF activity, as well as Trs20 and Trs33. Trs65 (not present in higher eukaryotes), Trs120, and Trs130 are specific to TRAPPII, whereas Trs85 is only in TRAPPIII (Fig. S4). We previously postulated that the TRAPPII-specific subunits Trs120 and Trs130 target Ypt1/Rab1 GEF activity to COPI coated vesicles (10, 13). Here we show that the TRAPP subunit Trs85 targets Ypt1 GEF activity to the PAS. Thus, certain TRAPP subunits act as adaptors to bring core GEF components to different parts of the cell (10, 13). The cellular components that interact with Trs85 at the PAS are the focus of current studies.
Materials and Methods
Strains and Media.
Yeast strains used in this study are listed in Table S2. Strains were grown in media (SMD, YPD, YPL, and YTO) as described previously (20, 26). For autophagy induction, cells were shifted to SD-N or treated with rapamycin (20).
Immunoblotting and Quantitative Analysis.
Protein samples for Western blot analysis were analyzed as described previously (20).
Fluorescence Microscopy.
Cells were cultured in SMD selective medium to midlog phase. For starvation experiments, cells were shifted to SD-N for 45 min. Fluorescence signals were visualized on a DeltaVision system using an Olympus IX71 fluorescence microscope (Olympus). The images were captured by a Photometrics CoolSNAP HQ camera (Roper Scientific, Inc.) and deconvolved using softWoRx software (Applied Precision).
Nucleotide Exchange Assay.
TRAPP was purified from strains SFNY1080 and SFNY1075 and the uptake of GTPγS was measured as described previously (8).
Gel Filtration Analysis and Immunoprecipitation.
Yeast cells were radiolabeled as described previously (9). An aliquot (200 × 106 cpm) of radiolabeled lysate or 10 mg of cleared lysate was applied to a Superdex 200 gel filtration column and fractions of 1 mL were collected. Fractions (8, 12), or 2 mg of cleared lysate, were immunoprecipitated with anti-myc antibody and analyzed by SDS-polyacrylamide gel electrophoresis.
Other Assays.
The Pho8Δ60 assay was done as previously described (16).
Supplementary Material
Acknowledgments
This work was supported by Public Health Service Grant GM53396 (D.J.K.) and the Howard Hughes Medical Institute (S.F.-N.). S.F.-N. is an investigator, and D.B. and S.M. are postdoctoral associates, of the Howard Hughes Medical Institute. J.H.B. holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. J.H. holds a postdoctoral fellowship from the Canadian Association of Gastroenterology Canadian Institutes of Health Research/Crohn's and Colitis Foundation of Canada.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000063107/DCSupplemental.
References
- 1.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reggiori F. 1. Membrane origin for autophagy. Curr Top Dev Biol. 2006;74:1–30. doi: 10.1016/S0070-2153(06)74001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamasaki M, Noda T, Ohsumi Y. The early secretory pathway contributes to autophagy in yeast. Cell Struct Funct. 2003;28:49–54. doi: 10.1247/csf.28.49. [DOI] [PubMed] [Google Scholar]
- 5.Reggiori F, et al. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- 7.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sacher M, et al. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 10.Cai Y, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai H, et al. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 12.Cai H, Zhang Y, Pypaert M, Walker L, Ferro-Novick S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J Cell Biol. 2005;171:823–833. doi: 10.1083/jcb.200505145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamasaki A, et al. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell. 2009;20:4205–4215. doi: 10.1091/mbc.E09-05-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sacher M, Barrowman J, Schieltz D, Yates JR, III, Ferro-Novick S. Identification and characterization of five new subunits of TRAPP. Eur J Cell Biol. 2000;79:71–80. doi: 10.1078/S0171-9335(04)70009-6. [DOI] [PubMed] [Google Scholar]
- 15.Meiling-Wesse K, et al. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- 16.Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy. 2005;1:37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacon RA, Salminen A, Ruohola H, Novick P, Ferro-Novick S. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J Cell Biol. 1989;109:1015–1022. doi: 10.1083/jcb.109.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 20.Cheong H, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008;182:703–713. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He C, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yen W-L, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungar D, et al. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.