Abstract
Huntington’s disease (HD), an incurable neurodegenerative disorder, has a complex pathogenesis including protein aggregation and the dysregulation of neuronal transcription and metabolism. Here, we demonstrate that inhibition of sirtuin 2 (SIRT2) achieves neuroprotection in cellular and invertebrate models of HD. Genetic or pharmacologic inhibition of SIRT2 in a striatal neuron model of HD resulted in gene expression changes including significant down-regulation of RNAs responsible for sterol biosynthesis. Whereas mutant huntingtin fragments increased sterols in neuronal cells, SIRT2 inhibition reduced sterol levels via decreased nuclear trafficking of SREBP-2. Importantly, manipulation of sterol biosynthesis at the transcriptional level mimicked SIRT2 inhibition, demonstrating that the metabolic effects of SIRT2 inhibition are sufficient to diminish mutant huntingtin toxicity. These data identify SIRT2 inhibition as a promising avenue for HD therapy and elucidate a unique mechanism of SIRT2-inhibitor-mediated neuroprotection. Furthermore, the ascertainment of SIRT2’s role in regulating cellular metabolism demonstrates a central function shared with other sirtuin proteins.
Keywords: cholesterol, Huntington’s disease, metabolism, sirtuin, transcription factor SREBP-2
Polyglutamine (polyQ) expansions in the HD gene product, huntingtin (Htt), cause a slowly progressive and fatal neurological phenotype associated with neuronal loss in the cortex and striatum (1). The relative contributions of proposed etiologic mechanisms involving multiple biochemical and cellular pathways remain uncertain (2, 3). In the last decade, cell and animal models recapitulating distinct features of Huntington’s disease (HD) pathology have been generated and successfully employed in preclinical drug trials (4–7).
Sirtuins comprise a family of protein deacetylase enzymes that have been shown to impact longevity in a number of eukaryotic species (reviewed in ref. 8). Enhancement of organismal longevity and other health-promoting effects of sirtuins have frequently been attributed to the regulation of metabolism. The attractive properties of sirtuins in lower organisms have ignited intensive investigation of their biological and therapeutic roles in mammals, particularly for the purposes of combating metabolic and age-dependent human diseases. There are seven known mammalian sirtuins, SIRTs 1–7, the most studied of which is SIRT1, a close structural and functional homolog of Sir2 found in yeast and Drosophila. Another mammalian sirtuin, SIRT2, has been shown to be a tubulin deacetylase and an important regulator of cell division and myelinogenesis (9–11). However, roles for SIRT2 in neurons, a nondividing cell type, have remained largely unknown.
Previous work from our group has shown that chemical inhibitors of SIRT2 change protein inclusion body characteristics and increase neuronal survival in models of Parkinson's disease (12). Nonetheless, elucidation of the full spectrum of cellular and molecular mechanisms underlying SIRT2-inhibitor-mediated neuroprotection and whether SIRT2 inhibition would be beneficial in other neurodegenerative conditions remained to be determined. This study reveals a unique role for SIRT2 in the control of neuronal metabolism and shows the potential benefit of targeting this sirtuin pharmacologically to treat HD.
Results
Genetic or Pharmacologic Inhibition of SIRT2 Is Neuroprotective in Models of HD.
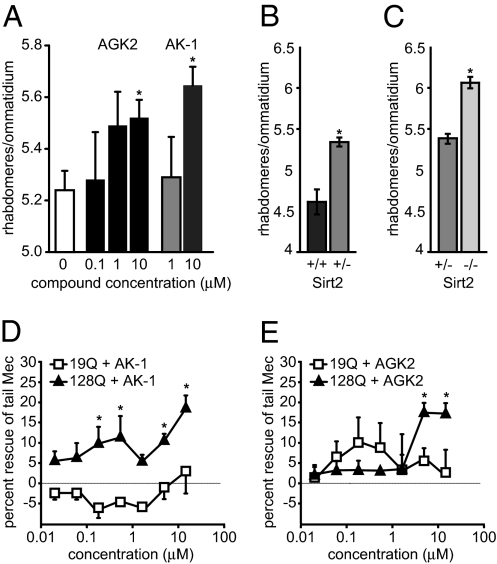
Given previous evidence that SIRT2 inhibitors ameliorate the neurodegenerative phenotypes of cell and animal models of Parkinson’s disease (12), we asked whether a similar effect could be observed in models of HD. Thus, we first evaluated the recently identified selective and structurally diverse SIRT2 inhibitors AGK2 and AK-1 (12) for their disease-rescuing effects in Drosophila melanogaster expressing N-terminal Htt fragments (N-ter Htt) from human HD exon1 (Httex1) (5, 13). Freshly eclosed flies expressing Httex1 Q93 in all neurons were fed medium supplemented with AK-1 or AGK2, and neuronal degeneration was assessed 7 days later by using the pseudopupil technique [which scores the number of surviving rhabdomeres (photoreceptor neurons) per ommatidium]. Both inhibitors achieved significant neuroprotection in HD flies at 10 μM (Fig. 1A), improving the number of rhabdomeres from 5.2 to 5.5 and 5.6, respectively. Genetic ablation of SIRT2 also rescued Httex1 Q93-induced photoreceptor neuron death in a dose-dependent manner (Fig. 1 B and C; ref. 13).
Fig. 1.
Neuroprotective effects of SIRT2 inhibitors in invertebrate models of HD. (A) AGK2 and AK-1 decrease the degeneration of light-sensing rhabdomeres in the Drosophila eye. *, P < 0.02 (for 10 μM AGK2 or AK-1, respectively). (B and C) Heterozygous and homozygous deletion of Sirt2 shows a dose-dependent reduction in the Drosophila model of HD. AK-1 (D) and AGK2 (E) rescued the defective touch response in C. elegans expressing polyQ N-ter Htt fused to CFP in touch receptor neurons. (*, P < 0.05).
We next tested whether SIRT2 inhibitors would modulate the neuronal dysfunction associated with expression of N-ter Htt in Caenorhabditis elegans touch receptor neurons (7). Both AGK2 and AK-1 showed significant rescue of mutant polyQ cytotoxicity as measured by improvement in the worms' defective response to a light touch at the tail (Fig. 1 D and E). The improvements of touch response by AGK2 and AK-1 were not attributable to decreased Htt transgene expression (96 ± 12 and 133 ± 85% of control for AGK2 and AK-1, respectively) and were specific to the mutant Htt fragment-expressing animals (128Q vs. 19Q). These results show significant disease-modulating effects of SIRT2 inhibitors in two in vivo models of polyQ disease pathogenesis, warranting further study of SIRT2 as a therapeutic target in HD.
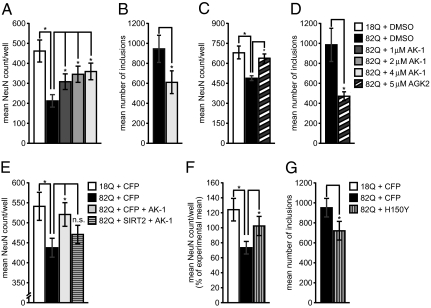
We next examined the effects of SIRT2 inhibition in a primary neuron model of HD. The first 171 amino acids of Htt containing a stretch of 18 or 82 polyglutamines (nonpathologic Htt171-18Q or mutant Htt171-82Q, respectively) was expressed by lentiviral transduction of primary striatal neurons, whose function and survival was monitored by NeuN-positive cell counts (14). A significant dose-dependent rescue of Htt171-82Q neurons was observed with AK-1 treatment (Fig. 2A). AGK2 treatment also significantly rescued striatal neurons from mutant Htt toxicity (Fig. 2C). Because we had previously observed that SIRT2 inhibitors dramatically modulate the formation of alpha-synuclein protein inclusions (12) and soluble and aggregated polyQ proteins showed an increased affinity to acetylated α-tubulin in a PC12 cell model of HD (15) (Fig. S1 A–D), we assessed whether SIRT2 inhibition might also alter mutant Htt inclusion characteristics. Treatment with AK-1 and AGK2 significantly reduced the presence of mutant Htt inclusions (Fig. 2 B and D). However, in contrast to effects of SIRT2 inhibition on alpha-synuclein inclusions, no effects on Htt inclusion size or morphology were observed (Fig. S1 E–O). The efficacy of two chemically distinct inhibitors against Htt171-82Q aggregation and toxicity in striatal neurons supports the argument that SIRT2 inhibition prevents HD-related neurodegeneration.
Fig. 2.
SIRT2 inhibition protects against Htt171-82Q toxicity in primary striatal neurons. (A) As shown previously, Htt171-82Q (black bar) exhibits toxicity toward striatal neurons as compared with Htt171-18Q (white bar). AK-1 rescued Htt171-82Q-expressing cells in a dose-dependent manner (at concentrations of 1, 2, and 4 μM) (A) and significantly reduced the number of mutant Htt positive inclusions (B). AGK2 also rescued striatal neurons from Htt171-82Q toxicity (C) and significantly reduced the number of inclusions (D). Overexpression of SIRT2WT abrogates neuroprotection by AK-1 (E). A lentiviral vector encoding CFP is used as a coinfection control. A dominant negative deacetylase mutant SIRT2H150Y significantly decreases Htt171-82Q toxicity in primary striatal neurons (F) and also significantly reduced the number of inclusions (G). A lentiviral vector encoding CFP is used as a coinfection control (* P < 0.05).
To further rule out off-target effects of our compounds, we used genetic approaches to validate the neuroprotective effects of SIRT2 inhibition. First, we showed that overexpression of wild-type SIRT2 (SIRT2WT) counteracted the neuroprotection of AK-1 (Fig. 2E). Second, we demonstrated that expression of a dominant negative, deacetylase-deficient SIRT2 mutant that diminishes cellular SIRT2 activity (SIRT2H150Y) (Fig. S2B) also rescued primary striatal neurons from Htt171-82Q toxicity (Fig. 2F) and reduced the number of mutant Htt inclusions (Fig. 2G). These data strongly support the perspective that SIRT2 inhibition accounts for the observed AK-1- and AGK2-mediated neuroprotection.
SIRT2 Is Not Overexpressed in HD.
Because distinct changes in RNA and protein expression have been associated with mutant Htt toxicity (3), we next explored whether a pathological increase in SIRT2 might be the disease-related effect targeted by SIRT2 inhibition. Because previous descriptions of the expression of SIRT2 in the mammalian brain showed it to be localized primarily to oligodendrocytes (11, 16), we first verified SIRT2 expression in neurons, where mutant Htt’s effects are greatest. Mouse brain sections showed extensive colocalization of SIRT2 and CNPase (2′,3′-cyclic nucleotide 3′-phosphodiesterase) immunoreactivities, including in white matter bundles within the striatum, in concordance with the reported localization of SIRT2 in oligodendrocytes (Fig. S3A), but we also observed colocalization of SIRT2 immunoreactivity with the neuronal marker NeuN in cortical and striatal neurons (Fig. S3 B and C). Furthermore, Western blots also detected ≈43 and ≈37 kDa anti-SIRT2-immunoreactive bands in neuron-like cell lines, mouse cerebral cortex, and rat primary neurons (Fig. S3 D–F). However, equivalent expression and localization of SIRT2-immunoreactive species were observed in both normal and HD model conditions [wild-type and transgenic HD (R6/2) mice and Htt171-18Q- and Htt171-82Q-expressing striatal neurons] (Fig. 3 B–D). Thus, we concluded that SIRT2 is present in cortical and striatal neurons but is not up-regulated in HD.
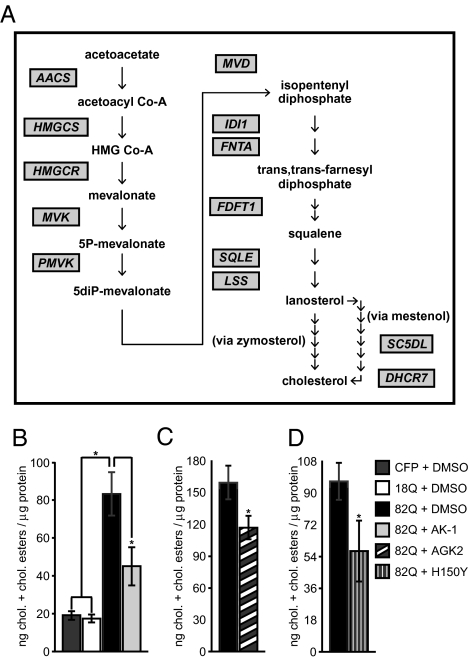
Fig. 3.
Gene expression and metabolic effects of SIRT2 inhibition in neurons. (A) Multiple genes controlling sterol biosynthesis are down-regulated by AK-1 treatment. Scheme depicts enzymes in sterol biosynthesis whose RNAs are down-regulated by AK-1 treatment by the criterion of FDR P < 0.05. (The segment of the pathway between acetoacetate and cholesterol is shown.) Full analyses of the data, including statistical measures, fold-changes, and Gene Ontology analyses, are presented in Dataset S1 and S2. (B–D) SIRT2 inhibition reverses Htt171-82Q-mediated increase in sterols. (B) DMSO-treated neurons expressing Htt171-82Q had significantly higher sterol levels (cholesterol and cholesteryl esters) than those expressing either CFP or Htt171-18Q and this effect was reversed by treatment with AK-1, AGK2 (5 μM, 48 hrs) (C), and SIRT2H150Y (D) (*P < 0.05).
Inhibition of SIRT2 Down-Regulates Genes Involved in Sterol Biosynthesis.
The known histone deacetylase activity (17), nucleo-cytoplasmic shuttling (9, 18, 19), and experimentally verified neuronal expression of SIRT2 (see above) led us to hypothesize that SIRT2 might regulate neuronal transcription; we therefore postulated that modulation of neuronal gene expression might be the cellular mechanism of SIRT2 inhibitor-mediated neuroprotection. Mutant Htt is known to wield major effects on steady-state mRNA levels through either direct or indirect transcriptional regulation (20). Primary neurons expressing mutant Htt fragments faithfully recapitulate HD-related changes in RNA expression (21) and, thus, provide a suitable model in which to evaluate modulation of gene expression by SIRT2 inhibition. As expected, Htt171-82Q expression in striatal neurons resulted in significant gene expression effects as compared with Htt171-18Q-expressing cells (116 decreases and 36 increases by criteria of FDR P < 0.05; Dataset S1). However, these effects remained globally uncorrected by SIRT2 inhibitor treatment (Fig. S4 and Dataset S1).
We next assessed whether other gene regulatory effects might account for SIRT2-mediated neuroprotection. Interestingly, short-term treatment with SIRT2 inhibitor AK-1 produced large statistically significant changes in RNA expression in untransduced, Htt171-18Q- and Htt171-82Q-expressing neurons. To define the functional effects of SIRT2-related gene regulation, we assessed which biological pathways were represented by genes responding to SIRT2 inhibition. Strikingly, molecular pathway analysis of SIRT2-regulated genes according to Gene Ontology classification showed highly significant overrepresentation of metabolic cascades, the most prominent of these being decreased expression of genes associated with sterol biosynthesis (Fig. 3A and Dataset S2). Although these findings are consistent with previous data implicating other sirtuins in cellular metabolism (8), they are surprising and unique with respect to the previously known roles of SIRT2. These data raised the intriguing possibility that the cellular neuroprotective mechanism of SIRT2 inhibition might be the negative regulation of sterol production. We confirmed the microarray results in new samples of striatal neurons treated with AK-1 and AGK2; these assays reproduced the down-regulation of cholesterol biosynthetic genes (Fig. S5 A and C) and also showed a significant decrease in the levels of sterols (cholesterol and cholesteryl esters; see Materials and Methods) (Fig. S5 B and D). These data demonstrate that AK-1- and AGK2-mediated neuroprotection is correlated with the negative regulation of sterol biosynthesis.
We next asked whether the metabolic regulation mediated by AK-1 and AGK2 was specifically attributable to their inhibition of SIRT2. We therefore assessed whether inhibition of SIRT2 via the overexpression of the deacetylase-deficient SIRT2H150Y mutant would result in the same down-regulation of sterol biosynthesis genes and sterol levels. Although overexpression of wild-type SIRT2 had no effect on sterol biosynthesis, SIRT2H150Y significantly decreased sterol levels and RNAs encoding cholesterol biosynthetic enzymes (Fig. S5 E and F). The fact that genetic manipulation of SIRT2 activity reproduces the gene regulatory effects of SIRT2-targeting small molecules substantiates the on-target effects of these inhibitors. Moreover, these data convincingly demonstrate a role of SIRT2 in regulating cellular metabolism.
SIRT2 Inhibition Reduces polyQ-Induced Cholesterol Dyshomeostasis.
The correlation of decreased sterol biosynthesis and neuroprotection in our experiments raised the possibility that the negative regulation of sterol production by SIRT2 inhibitors directly opposed a toxic dysregulation of sterol homeostasis by Htt171-82Q. Moreover, previous literature has also described effects of mutant Htt on sterol-related pathways (22, 23). Therefore, we examined the effects of mutant Htt fragments on sterol content in our primary striatal neuron model of HD. Indeed, Htt171-82Q expression increased cellular sterol levels (cholesterol and cholesteryl esters; see Materials and Methods) compared to CFP or Htt171-18Q controls (Fig. 3B). Consistent with the previous results, AK-1, AGK2, and SIRT2H150Y were able to diminish sterol species in Htt171-82Q-expressing neurons (Fig. 3 B–D). These results support the model that polyQ-induced sterol dyshomeostasis contributes to HD-related neurotoxicity, whereas inhibition of SIRT2 down-regulates sterol biosynthesis and abates this toxicity.
SIRT2-Mediated Effects Are Driven by Extranuclear Events.
Our immnocytochemical studies (Figs. S2A and (S3 A–C) had shown that neuronal SIRT2 was localized to both nuclear and extranuclear compartments. Based on the gene regulatory effects of SIRT2 inhibition, we postulated that the observed neuroprotective and gene regulatory effects involved direct nuclear actions. We therefore overexpressed SIRT2 variants with nuclear import and export mutations to explore this question. Counter to our prediction, however, nuclear targeting of SIRT2 negated its ability to block neuroprotection by AK-1, whereas extranuclear targeting efficiently antagonized AK-1's effect (Fig. S6 A and B). Likewise, cellular sterol levels were increased by extranuclear compared with nucleus-targeted SIRT2 (Fig. S6C). These results show that both the neuroprotective and metabolic effects of SIRT2 inhibition are mediated by cellular events that occur in extranuclear compartment(s).
SIRT2 Regulates the Nuclear Trafficking of SREBP-2.
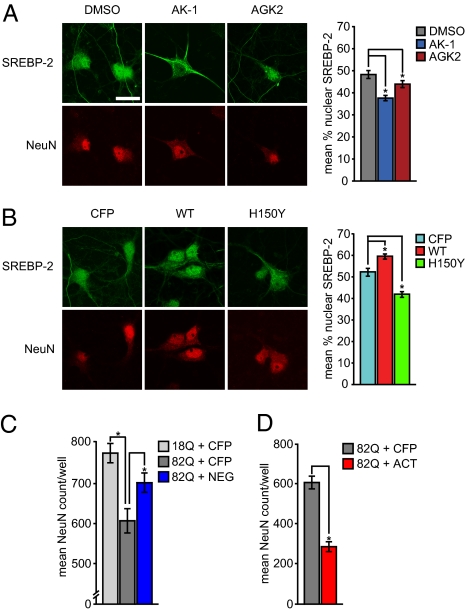
We next investigated the mechanism by which SIRT2 might regulate sterol biosynthesis. Visualization of gene networks with Ingenuity Pathway Analysis showed that many of the genes down-regulated by AK-1 were all controlled by a common transcription factor: the sterol response element binding protein 2 (SREBP-2); we therefore hypothesized that SREBP-2 might be an important mediator of SIRT2-related neuroprotection. SREBP-2 activity is known to be controlled by its necessary translocation from the endoplasmic reticulum to the nucleus, where it can bind to SRE enhancer elements in DNA and increase cholesterol biosynthesis. Thus, we tested whether SIRT2 activity regulated the nuclear trafficking of SREBP-2. Indeed, chemical inhibition of SIRT2 with AK-1 or AGK2 led to a reduction in the nuclear compartmentalization of SREBP-2 (Fig. 4A). Furthermore, overexpression of wild-type SIRT2 (or extranuclear SIRT2) increased the percentage of nuclear SREBP-2 labeling in primary neurons, whereas SIRT2H150Y (or nuclear SIRT2) had the opposite effect (Fig. 4B and Fig. S7). Combined with the observed down-regulation of a large set of known SREBP-2 target genes and decreased cellular sterol levels, these results identified SREBP-2 as a crucial mediator of the effects of SIRT2 on metabolism.
Fig. 4.
SIRT2 inhibition reduces nuclear trafficking of SREBP-2. (A) Both AK-1 and AGK2 decrease the trafficking of endogenous SREBP-2 to the nucleus. (B) Expression of wild-type SIRT2 increases endogenous nuclear SREBP-2 whereas deacetylase-deficient SIRT2H150Y reduces this effect. (Scale bar: 10 μM.) (C) Dominant negative mutant SREBP-2NEG protects neurons against Htt171-82Q toxicity. (D) Conversely, the constitutively active mutant SREBP-2ACT enhances Htt171-82Q toxicity (*, P < 0.05).
Metabolic Regulation via SREBP-2 Inhibition Conveys Neuroprotection.
Although the above results show that down-regulation of sterol biosynthesis correlates with SIRT2-inhibitor-mediated neuroprotection, they do not establish causality. Therefore, the remaining question to be addressed was whether the negative regulation of SREBP-2 by SIRT2 inhibition was sufficient to achieve neuroprotection. We tested this hypothesis by expressing dominant-negative (SREBP-2NEG) versus constitutively active SREBP-2 (SREBP-2ACT) mutants in cultured neurons (Fig. S8 A–D) and assessed their effects on polyQ toxicity. Consistent with our hypothesis, the dominant-negative SREBP-2 conveyed significant neuroprotection to Htt171-82Q cells (Fig. 4C) but had no effect on the number of mutant Htt-positive inclusions (Fig. S8E). Conversely, the constitutively active SREBP-2 enhanced polyQ toxicity (Fig. 4D). Moreover, the neuroprotective effect of AK-1, AGK2, and SIRT2H150Y was circumvented by addition of constitutively active SREBP-2, which is not targeted to the endoplasmic reticulum (Fig. S8 F–H). These data demonstrate that the regulation of metabolism is an important mechanism of the neuroprotective activity conveyed by SIRT2 inhibitors.
Discussion
SIRT2 Demonstrates Neuroprotection in Models of HD.
Previously we demonstrated that SIRT2 inhibition prevented neurodegeneration in models of Parkinson’s disease (12). Here, we further validate that our chemical SIRT2 inhibitors mediate neuroprotection through on-target activities and extend the potential applications for such inhibitors to Huntington’s disease, another fatal neurodegenerative disorder. We also report a breakthrough discovery regarding the metabolism-related mechanism(s) through which SIRT2 inhibition acts by elucidation of its gene regulatory effects.
SIRT2 inhibition by either genetic or chemical means resulted in a decrease in Htt inclusion accumulation and increased neuronal viability. The recapitulation of SIRT2 inhibitor-mediated neuroprotection through negative regulation of sterol biosynthesis and the blockade of neuroprotection by circumventing this regulation demonstrate that this metabolic effect contributes significantly to Htt pathology. Previous studies have also assessed mutant Htt’s effects on sterol homeostasis. These analyses reported the accumulation of cellular cholesterol attributed to the binding of mutant Htt to caveolae (23), altered levels of cholesterol biosynthesis-related RNAs and sterols, including decreased 24-OH-cholesterol (22, 24, 26, 27), and a direct (albeit weak) interaction with SREBP-2 (28). The evidence available to date does not yet provide a complete understanding of mutant Htt's effects on sterol homeostasis, however. Although differences in assay measures may account for some heterogeneity in findings, we believe that a more comprehensive explanation resides in the fact that the mechanisms of brain sterol regulation, distinct contributions of different cell types, and varying effects depending on nutrient status still remain to be elucidated. Our working hypothesis is that a plurality of sterol-pathway-related mitigatory effects exists and that the balance of these activities and specific nutrient conditions determines the net outcome. This perspective is consistent with recent findings by Valenza and Cattaneo demonstrating complex changes in sterol biosynthesis and compensatory homeostasis in the brains of R6/2 mice (24). Given the generally accepted importance of sterol regulation to brain function, it seems likely that a more detailed understanding of the mechanisms of sterol-related neuroprotection will allow important insights for HD therapy.
Unique Role of SIRT2 in the Regulation of Neuronal Cholesterol.
Here we provide strong evidence of a unique and important regulatory function of the SIRT2 deacetylase in neuronal cells. Biological pathway analysis indicated that prominent effects of SIRT2 comprise the regulation of genes controlling metabolism, including sterol and fatty acid biosynthesis, carbohydrate metabolism, and purine metabolism (Dataset S2). An apparent role for SIRT2 in regulating lipid and sterol biogenesis is consistent with its previously reported localization in oligodendrocytic myelin sheaths (16) and role in adipocyte differentiation (29). Conversely, however, no previous studies implicate SIRT2 in the regulation of cholesterol biosynthesis and, thus, the findings of this study motivate further examination of SIRT2 in this role.
Studies of late have shown detrimental effects of cholesterol accumulation in neurons and substantiate the potential benefit of decreasing neuronal cholesterol (or other sterol species) as a neuroprotective strategy. Although there has been ambivalent evidence in the more distant past, a flurry of recent reports have clarified the detrimental effects of cholesterol accumulation in neurons (30–32). These studies are paralleled by evidence that increased neuronal cholesterol presents a risk factor for Alzheimer’s and Parkinson’s diseases (33–35). Although debated, it has also been proposed that neuronal cholesterol may be due to instrinsic biosynthesis (36), rather than transport from extracellular sources. Moreover, other aspects of the regulation of neuronal cholesterol homeostasis are also unique, such as the important role of 24-OH-cholesterol and its biosynthesis by CYP46A1. In fact, CYP46A1 has been deemed a master regulator of neuronal cholesterol biosynthesis based on the evidence that 24-OH-cholesterol has a more significant role than the SREBP cleavage-activating proteins (SCAPs 1 and 2) in regulating the nuclear translocation of SREBP-2 in this cell type (37, 38).
Molecular Mechanisms of SIRT2-Related Metabolic and Neuroprotective Activities.
Although we have established that SIRT2 modulation controls the nuclear trafficking and transcriptional activity of SREBP-2, the specific molecular determinants of this regulation remain to be elucidated. A priori, the assumption would be that the deacetylation of a SIRT2 substrate (or substrates) is responsible for this effect. It is well established that α-tubulin is a substrate of both SIRT2 and its cytoplasmic interacting protein HDAC6 and, thus, inhibition of SIRT2–HDAC6 complexes might affect microtubule-dependent trafficking. Therefore, it is plausible to propose α-tubulin as the effector whose increased acetylation inhibits SREBP-2 translocation. Alternatively, other proteins, although not previously named as SIRT2 substrates, would also comprise obvious candidate effectors because of their known roles in the regulation of neuronal cholesterol homeostasis. These candidates include SREBP-2 itself or the proteins regulating the sequestration of SREBP-2 in the endoplasmic reticulum, namely the SCAPs, the site 1 and 2 proteases, and CYP46A1. Intriguingly, we note that both SREBP-2 and CYP46A1 contain lysines with high context prediction scores for acetylation (obtained using Prediction of Acetyation on Internal Lysines [PAIL] [http://bdmpail.biocuckoo.org/prediction.php]).
Although we have provided evidence for the contribution of decreased SREBP-2 activity in SIRT2 inhibitor-mediated neuroprotection, we have not ruled out parallel contributions of other SIRT2-dependent effects. For example, increasing α-tubulin acetylation could ameliorate HD pathogenesis by independent mechanisms such as regulating mutant Htt aggregation or degradation (39) or the transport of membranous vesicles, including those containing BDNF (40). In fact, our observation that both genetic and pharmacologic inhibition of SIRT2 leads to reduced numbers of mutant Htt inclusions, whereas inhibition of SREBP-2, although protective, does not alter inclusion numbers, further supports the perspective that other SIRT2-dependent processes may have independent mitigating effects.
Role of SIRT2 in the Regulation of Metabolism Draws Parallels to Other Sirtuin Activities.
The newly identified role of SIRT2 as a regulator of metabolism in neurons establishes a previously undisclosed function shared with other members of the sir2/SIRT family (8). Roles for sir2 and SIRTs 1, 3, 4, 5, and 6 in the control of cellular metabolism have been well established in lower organisms and in nonneural mammalian tissues, respectively (8). Previous data as well as those of the present study suggest that sirtuin proteins comprise a complex network of metabolic regulators whose activities are present in a number of molecular pathways in a variety of tissues. Although we have provided substantial evidence for SIRT2’s role in cholesterol biosynthesis, the other potential metabolic activities suggested by the pharmacogenomic profile of AK-1 remain to be determined.
Implications for Other Age-Related Diseases.
Whereas increased activity of cholesterol biosynthetic enzymes and cholesterol accumulation generally occurs during aging, lower cholesterol levels have been associated with peripheral and central benefits, including increased lifespan and decreased amyloid accumulation (41, 42). Because the brain is previously known to be less sensitive to dietary modulation than peripheral tissues (36), it is particularly interesting to consider unique ways to regulate brain cholesterol. As shown in the present study, this regulation might be achieved through SIRT2 inhibition, barring the remaining challenges of developing brain-permeable small-molecule SIRT2 inhibitor compounds and assessing potential complications of SIRT2 inhibition in nonneuronal cells (most notably, oligodendrocytes). However, if SIRT2 also regulates cholesterol synthesis beyond the nervous system, then even non-brain-permeable SIRT2 inhibitors may be considered for the management of peripheral disorders involving hypercholesterolemia or vascular disease. Thus, the unique facets of SIRT2 activity identified in this report warrant further study in a broader health-related context.
Materials and Methods
Detailed methods appear in the SI Materials and Methods. These include Drosophila feeding and ommatidial analysis, drug evaluation in C. elegans, antibodies and reagents, plasmids, lentiviral vectors, primary cultures, immunocytochemistry, inclusion analysis, Western blot, mouse extract preparation, immunohistochemistry, gene expression profiling, cholesterol assay, evaluation of SREBP-2 compartmentalization and statistical analysis.
Supplementary Material
Acknowledgments
We thank A. Pailluson, O. Hagenbuchle, and the Lausanne DNA Array facility; C. Wellington and M. Hayden (University of British Columbia, Vancouver) for Htt constructs; M. Rey, M. Dixon, M. Forrest, and L. Feletti for cloning and primary cultures; and the National Drosophila Stock Center in Bloomington, IN, for clones and support. We also thank J. Auwerx, H. Yamamoto, and L. Quinti for helpful discussions and critical reading of the manuscript. This work was supported by a generous gift for Discovery Novel Neurodegenerative Disease Therapeutics to MassGeneral Institute for Neurodegenerative Disease and grants from the CHDI Foundation (to A.A., A.G.K., G.P.B., L.M.T., and J.L.M.), the Swiss National Science Foundation (to R.L.-C. and H.R.), the Ecole Polytechnique Fédérale de Lausanne (to R.L.-C. and A.K.), Institut National de la Santé et de la Recherche Médicale (to J.P. and C.N.), the Wellcome Trust (to G.P.B.), Medical Research Council (to G.P.B.), the Hereditary Disease Foundation (to L.M.T. and J.L.M.), the Huntington’s Disease Society of America Coalition for the Cure (to L.M.T.), and National Institutes of Health Awards NS52789 (to L.M.T. and J.L.M.), HD36081 (to J.L.M.), and NS045283 (to J.L.M. and L.M.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002924107/DCSupplemental.
References
- 1.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Kazantsev AG, Hersch SM. Drug targeting of dysregulated transcription in Huntington’s disease. Prog Neurobiol. 2007;83:249–259. doi: 10.1016/j.pneurobio.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luthi-Carter R. Huntington’s and other polyglutamine diseases: Many effects of single gene mutations. Drug Discov Today Dis Mech. 2007;4:111–119. [Google Scholar]
- 4.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 5.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 6.Ferrante RJ, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker JA, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 8.Taylor DM, Maxwell MM, Luthi-Carter R, Kazantsev AG. Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol Life Sci. 2008;65:4000–4018. doi: 10.1007/s00018-008-8357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner HB, et al. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci. 2007;27:7717–7730. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. 2007;27:2606–2616. doi: 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Outeiro TF, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 13.Pallos J, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington’s disease. Hum Mol Genet. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudinskiy N, et al. Diminished hippocalcin expression in Huntington’s disease brain does not account for increased striatal neuron vulnerability as assessed in primary neurons. J Neurochem. 2009;111:460–472. doi: 10.1111/j.1471-4159.2009.06344.x. [DOI] [PubMed] [Google Scholar]
- 15.Apostol BL, et al. A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc Natl Acad Sci USA. 2003;100:5950–5955. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Southwood CM, Peppi M, Dryden S, Tainsky MA, Gow A. Microtubule deacetylases, SirT2 and HDAC6, in the nervous system. Neurochem Res. 2007;32:187–195. doi: 10.1007/s11064-006-9127-6. [DOI] [PubMed] [Google Scholar]
- 17.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Nguyen M, Qin FX-F, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 19.Vaquero A, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benn CL, et al. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J Neurosci. 2008;28:10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Runne H, et al. Dysregulation of gene expression in primary neuron models of Huntington’s disease shows that polyglutamine-related effects on the striatal transcriptome may not be dependent on brain circuitry. J Neurosci. 2008;28:9723–9731. doi: 10.1523/JNEUROSCI.3044-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valenza M, et al. Dysfunction of the cholesterol biosynthetic pathway in Huntington’s disease. J Neurosci. 2005;25:9932–9939. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trushina E, et al. Mutant huntingtin inhibits clathrin-independent endocytosis and causes accumulation of cholesterol in vitro and in vivo. Hum Mol Genet. 2006;15:3578–3591. doi: 10.1093/hmg/ddl434. [DOI] [PubMed] [Google Scholar]
- 24.Valenza M, et al. Progressive dysfunction of the cholesterol biosynthesis pathway in the R6/2 mouse model of Huntington’s disease. Neurobiol Dis. 2007;28:133–142. doi: 10.1016/j.nbd.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 26.Valenza M, et al. Cholesterol biosynthesis pathway is disturbed in YAC128 mice and is modulated by huntingtin mutation. Hum Mol Genet. 2007;16:2187–2198. doi: 10.1093/hmg/ddm170. [DOI] [PubMed] [Google Scholar]
- 27.Leoni V, et al. Plasma 24S-hydroxycholesterol and caudate MRI in pre-manifest and early Huntington’s disease. Brain. 2008;131:2851–2859. doi: 10.1093/brain/awn212. [DOI] [PubMed] [Google Scholar]
- 28.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karasinska JM, et al. Specific loss of brain ABCA1 increases brain cholesterol uptake and influences neuronal structure and function. J Neurosci. 2009;29:3579–3589. doi: 10.1523/JNEUROSCI.4741-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M, et al. Cyp46-mediated cholesterol loss promotes survival in stressed hippocampal neurons. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.04.022. 10.1016/j.neurobiolaging.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 32.Fernández A, Llacuna L, Fernández-Checa JC, Colell A. Mitochondrial cholesterol loading exacerbates amyloid beta peptide-induced inflammation and neurotoxicity. J Neurosci. 2009;29:6394–6405. doi: 10.1523/JNEUROSCI.4909-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-On P, et al. Statins reduce neuronal alpha-synuclein aggregation in in vitro models of Parkinson’s disease. J Neurochem. 2008;105:1656–1667. doi: 10.1111/j.1471-4159.2008.05254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barceló-Coblijn G, Golovko MY, Weinhofer I, Berger J, Murphy EJ. Brain neutral lipids mass is increased in alpha-synuclein gene-ablated mice. J Neurochem. 2007;101:132–141. doi: 10.1111/j.1471-4159.2006.04348.x. [DOI] [PubMed] [Google Scholar]
- 35.Stefani M, Liguri G. Cholesterol in Alzheimer’s disease: Unresolved questions. Curr Alzheimer Res. 2009;6:15–29. doi: 10.2174/156720509787313899. [DOI] [PubMed] [Google Scholar]
- 36.Korade Z, Kenworthy AK, Mirnics K. Molecular consequences of altered neuronal cholesterol biosynthesis. J Neurosci Res. 2009;87:866–875. doi: 10.1002/jnr.21917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lund EG, et al. Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J Biol Chem. 2003;278:22980–22988. doi: 10.1074/jbc.M303415200. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Muneton S, Sjövall J, Jovanovic JN, Griffiths WJ. The effect of 24S-hydroxycholesterol on cholesterol homeostasis in neurons: Quantitative changes to the cortical neuron proteome. J Proteome Res. 2008;7:1606–1614. doi: 10.1021/pr7006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 40.Dompierre JP, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halford RW, Russell DW. Reduction of cholesterol synthesis in the mouse brain does not affect amyloid formation in Alzheimer’s disease, but does extend lifespan. Proc Natl Acad Sci USA. 2009;106:3502–3506. doi: 10.1073/pnas.0813349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fassbender K, et al. Simvastatin strongly reduces levels of Alzheimer’s disease beta-amyloid peptides Abeta 42 and Abeta 40 in vitro and in vivo. Proc Natl Acad Sci USA. 2001;98:5856–5861. doi: 10.1073/pnas.081620098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.