Abstract
Therapeutic strategies that augment insulin release from pancreatic β-cells are considered beneficial in the treatment of type 2 diabetes. We previously demonstrated that activation of β-cell M3 muscarinic receptors (M3Rs) greatly promotes glucose-stimulated insulin secretion (GSIS), suggesting that strategies aimed at enhancing signaling through β-cell M3Rs may become therapeutically useful. M3R activation leads to the stimulation of G proteins of the Gq family, which are under the inhibitory control of proteins known as regulators of G protein signaling (RGS proteins). At present, it remains unknown whether RGS proteins play a role in regulating insulin release. To address this issue, we initially demonstrated that MIN6 insulinoma cells express functional M3Rs and that RGS4 was by far the most abundant RGS protein expressed by these cells. Strikingly, siRNA-mediated knockdown of RGS4 expression in MIN6 cells greatly enhanced M3R-mediated augmentation of GSIS and calcium release. We obtained similar findings using pancreatic islets prepared from RGS4-deficient mice. Interestingly, RGS4 deficiency had little effect on insulin release caused by activation of other β-cell GPCRs. Finally, treatment of mutant mice selectively lacking RGS4 in pancreatic β-cells with a muscarinic agonist (bethanechol) led to significantly increased plasma insulin and reduced blood glucose levels, as compared to control littermates. Studies with β-cell-specific M3R knockout mice showed that these responses were mediated by β-cell M3Rs. These findings indicate that RGS4 is a potent negative regulator of M3R function in pancreatic β-cells, suggesting that RGS4 may represent a potential target to promote insulin release for therapeutic purposes.
Keywords: knockout mice, muscarinic receptor, RGS proteins, G protein-coupled receptor
Type 2 diabetes (T2D) has emerged as a major threat to human health worldwide. Besides peripheral insulin resistance, T2D is usually associated with β-cell dysfunction (1). Thus, the development of new drugs aimed at improving β-cell function, including stimulation of insulin release, is the focus of many laboratories (2).
β-cell function is regulated by many hormones and neurotransmitters most of which act on specific G protein-coupled receptors (GPCRs) that are expressed on the surface of pancreatic β-cells (3, 4). Following ligand-induced activation, a specific GPCR interacts with and activates one or more classes of heterotrimeric G proteins (consisting of α, β, and γ subunits), which in turn modulate various intracellular signal transduction pathways (5).
Stimulation of the muscarinic cholinergic (parasympathetic) nerves innervating the endocrine pancreas leads to a pronounced increase in glucose-stimulated insulin secretion (GSIS) (4, 6). Studies with pancreatic islets prepared from M3 muscarinic acetylcholine receptor (M3R) KO mice demonstrated that muscarinic enhancement of GSIS is mediated by the M3R subtype (7, 8). The M3R is a member of the muscarinic receptor family (M1–M5) that is selectively coupled to Gq-type G proteins (9).
Additional studies showed that mutant mice lacking M3Rs in pancreatic β-cells displayed impaired glucose tolerance and significantly reduced insulin release (10). On the other hand, transgenic mice overexpressing M3 receptors in pancreatic β-cells showed greatly improved glucose tolerance and GSIS (10). Moreover, these mutant mice were resistant to diet-induced glucose intolerance and hyperglycemia (10). These findings suggested that strategies aimed at enhancing signaling through β-cell M3Rs might become therapeutically useful for the treatment of T2D.
To gain insight into the cellular mechanisms that regulate M3R function in pancreatic β-cells and to identify potential therapeutic targets, we set out to identify proteins that act as negative regulators of β-cell M3Rs. As is the case with other GPCRs, agonist activation of the M3R triggers a number of cellular events, including M3R phosphorylation by various kinases that serve to inhibit or terminate M3R signaling (11, 12). In addition, distinct members of the superfamily of regulators of G protein signaling (RGS proteins) act as GTPase-activating proteins (GAPs) to greatly accelerate the rate of Gα-GTP hydrolysis, thus limiting the lifetime of active Gα-GTP G protein subunits (13, 14).
The role of RGS proteins in modulating GPCR function in β-cells has not been studied so far. Moreover, it remains unknown which specific RGS proteins are expressed by pancreatic β-cells. In the present study, we initially demonstrated that RGS4 is abundantly expressed in MIN6 cells, an insulinoma cell line that expresses functional M3Rs, as well as mouse pancreatic islets. To study the role of RGS4 in modulating M3R function in MIN6 cells, we carried out RGS4 siRNA knockdown experiments. Moreover, we performed in vitro and in vivo studies with whole body RGS4 KO mice and mutant mice that selectively lacked RGS4 in pancreatic β-cells only. These studies demonstrated that RGS4 acts as a potent negative regulator of M3R-mediated GSIS in MIN6 cells and mouse islets.
This study provides direct experimental evidence that RGS proteins play an important role in regulating β-cell function. Because M3R-mediated augmentation of GSIS plays a central role in maintaining normal glucose homeostasis (10), our results underscore the potential usefulness of RGS protein (RGS4) inhibitors as drugs for the treatment of T2D.
Results
MIN6 Cells Express Functional M3 Muscarinic Receptors (M3Rs).
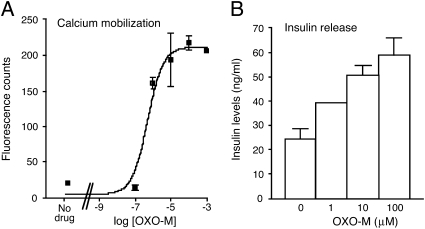
Our first goal was to establish a cell culture system that would allow us to identify proteins regulating M3R-mediated facilitation of insulin release. MIN6 cells are a well-established immortalized adherent murine β-cell line (15). Initially, we examined whether MIN6 cells were endowed with functional M3Rs. Specifically, we incubated MIN6 cells in the presence of 16.7 mM glucose with oxotremorine-M (OXO-M), a hydrolytically stable muscarinic agonist, and then measured the resulting increases in intracellular calcium levels [Ca2+]i and GSIS (Fig. 1). Fig. 1A shows that OXO-M treatment led to concentration-dependent increases in [Ca2+]i, as studied by FLIPR technology. Moreover, this response was accompanied by a significant increase (approximately 2.5-fold) in GSIS (Fig. 1B). OXO-M had no significant effect on basal insulin release measured in the absence of glucose.
Fig. 1.
OXO-M-mediated increases in [Ca2+]i and insulin release in MIN6 cells. (A) Calcium measurements. OXO-M-induced increases in [Ca2+]i were measured in MIN6 cells grown in 96-well plates by using FLIPR technology. (B) Insulin release measurements. OXO-M–mediated increases in insulin secretion were determined in the presence of 16.7 mM glucose. Representative concentration-response curves are shown. Data points represent means ± SEM of assays carried out in triplicate. Nine additional experiments gave similar results.
To test the hypothesis that the OXO-M-mediated calcium and insulin responses were mediated by M3Rs, we transfected MIN6 cells with two different M3R siRNAs (M3-1 and M3-2) and two different negative control siRNAs. The two control siRNAs were targeted against the GPR40 receptor (16), another Gq-coupled receptor known to be expressed in pancreatic β-cells, or represented scrambled negative control siRNA (Ambion). Real-time qRT-PCR studies showed that electroporation of MIN6 cells with M3-1 siRNA led to an approximately 75% reduction in M3R mRNA levels (Fig. S1)(for primer sequences, see SI Materials and Methods). Similar results were obtained with M3-2 siRNA. To examine actual muscarinic receptor densities (Bmax values), we carried out radioligand binding studies using a saturating concentration (5 nM) of the nonsubtype-selective muscarinic antagonist, [3H]-N-methylscopolamine ([3H]-NMS). MIN6 cells that had been electroporated with negative control siRNA displayed a Bmax value of 196 ± 31 fmoles/mg membrane protein (n = 5). This value was reduced to only 60 ± 17 fmoles/mg in MIN6 cells that had been treated with M3-1 siRNA (n = 5), indicating that most muscarinic receptors expressed by MIN6 cells represent M3Rs.
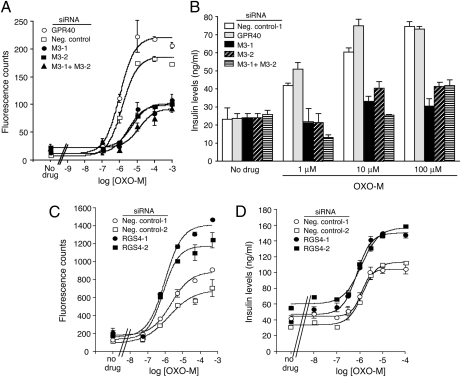
In agreement with the qRT-PCR experiments and the [3H]-NMS binding studies, treatment of MIN6 cells with either of the two M3R siRNAs (or a combination of the two M3R siRNAs) led to a pronounced reduction (>60%) in OXO-M-induced increases in [Ca2+]i and GSIS (Fig. 2 A and B). This observation indicates that muscarinic stimulation of [Ca2+]i and GSIS is mediated mainly by the M3R subtype in MIN6 cells. Real-time qRT-PCR studies with M1–M5 muscarinic receptor-specific primers indicated that MIN6 cells virtually exclusively express M3R mRNA (Fig. S2). This finding suggests that the residual OXO-M responses seen in M3R siRNA-treated MIN6 cells (Fig. 2 A and B) are due a small population of M3Rs that escaped siRNA-mediated knockdown. Taken together, our results indicate that MIN6 cells represent an excellent in vitro model system to study factors regulating M3R-mediated stimulation of insulin release.
Fig. 2.
Effect of siRNA-mediated M3R or RGS4 knockdown in MIN6 cells on OXO-M-induced calcium and insulin responses. Following treatment of MIN6 cells with the indicated siRNAs, OXO-M-induced increases in [Ca2+]i and insulin secretion were determined. (A–D) Effect of M3R knockdown (A and B) or RGS4 knockdown (C and D) on OXO-M-induced calcium and insulin responses. Assays were carried out with MIN6 cells that had been electroporated with the indicated siRNAs. Negative control siRNAs represent scrambled siRNAs. In the M3R knockdown assays, GPR40 siRNA was included as an additional negative control (see main text for details). OXO-M-induced increases in [Ca2+]i were measured by using FLIPR technology. OXO-M-stimulated enhancement of insulin secretion was assessed in the presence of 16.7 mM glucose. Representative concentration-response curves are shown. Data points represent means ± SEM of assays carried out in triplicate. At least three additional experiments gave similar results.
MIN6 Cells Predominantly Express RGS4.
To identify RGS proteins that regulate M3R signaling in MIN6 cells, we used qRT-PCR to examine the mRNA levels of 19 RGS proteins using primers that had been validated previously (17). The 19 tested RGS proteins included all members of the A/RZ, B/R4, C/R7, and D/R12 subfamilies (13, 14). In MIN6 cells, RGS4 mRNA was clearly the most abundant RGS transcript (Fig. S3A). Mouse pancreatic islets expressed several RGS transcripts at moderate to high levels including RGS4 mRNA (Fig. S3B). Pancreatic islets do not only contain insulin-producing β-cells but also several other cell types, providing a possible explanation for the different RGS mRNA expression patterns observed between MIN6 cells and mouse pancreatic islets.
RGS4 Knockdown Selectively Enhances M3R-Mediated Calcium and Insulin Responses in MIN6 Cells.
To examine the potential role of RGS4 in regulating M3R signaling in MIN6 cells, we electroporated MIN6 cells with two different RGS4 siRNAs (RGS4-1 and RGS4-2; Ambion). Western blotting studies showed that either of the two siRNAs was able to knock down RGS4 protein expression with high efficiency (by approximately 70–75%, as compared to control samples) (Fig. S4). Strikingly, following electroporation of MIN6 cells with either RGS4-1 or RGS4-2 siRNA, OXO-M treatment resulted in concentration-dependent increases in [Ca2+]i levels and GSIS, as compared to cells treated with either of the two negative control siRNAs (Ambion; Fig. 2 C and D). RGS4 knockdown also increased basal insulin release (measured in the absence of ligands and the presence of 16.7 mM glucose) by approximately 40–100% (P < 0.05). One possible explanation for this observation is that reduced RGS4 activity increases the lifetime of activated G protein α subunits which can form spontaneously under basal conditions.
Besides the M3R, pancreatic β-cells express several other GPCRs that are able to promote GSIS, including the GLP-1, different P2Y, and the V1b vasopressin receptor subtypes (4). We initially demonstrated that stimulation of these latter receptors in MIN6 cells with their corresponding ligands [GLP-1, ADP, and arginine vasopressin (AVP), respectively] led to concentration-dependent enhancements of GSIS (Fig. S5). However, the observed Emax values were smaller than the maximum response to OXO-M (Fig. S5). Interestingly, siRNA-mediated knockdown of RGS4 expression had no significant effect on the Emax values observed after simulation of MIN6 cells with GLP-1, ADP, or AVP (Fig. S6). Similarly, agonist EC50 values were not significantly affected following the reduction of RGS4 expression.
M3Rs Can Be Coimmunoprecipitated with RGS4 in Cotransfected COS-7 Cells.
To examine whether M3Rs exist in a complex with RGS4 in insulin-producing cells, we initially attempted to coimmunoprecipitate M3Rs and RGS4 from MIN6 cell lysates. However, because of the relatively low expression levels of M3Rs in this cell line (see above), we were unable to reproducibly detect endogenous M3Rs in Western blotting studies. To circumvent these difficulties, we cotransfected COS-7 cells with human RGS4 and an HA-epitope tagged version of the human M3R. In this case, coimmunoprecipitation studies revealed the existence of an M3R/RGS4 complex (Fig. 3).
Fig. 3.

Coimmunoprecipitation of RGS4 protein and the M3R in cotransfected COS-7 cells. COS-7 cells were transfected with plasmids coding for human RGS4 and an HA-epitope-tagged version of the human M3R, either in combination (lane 1) or individually (lanes 3 and 4). For control purposes, cell membranes prepared from COS-cells expressing either RGS4 or M3R (lane 2) were mixed and processed in the same fashion as all other samples. Forty-eight hours after transfection, membrane extracts were prepared (approximately 1 mg of total protein) and subjected to immunoprecipitation (IP) using an agarose-conjugated anti-HA monoclonal antibody. RGS4 was detected via Western blotting analysis using a rabbit polyclonal antibody raised against human RGS4 (size of the immunoreactive band: approximately 24 kDa). Please note that RGS4 was only detectable in samples derived from COS-7 cells that had been cotransfected with the RGS4 and M3R plasmids. Two additional experiments gave similar results. “Input” represents cell lysates used for IP studies.
M3R-Mediated Augmentation of GSIS Is Selectively Enhanced in RGS4-Deficient Pancreatic Islets.
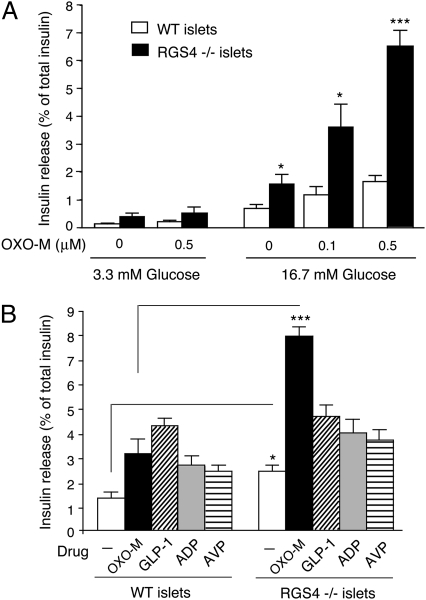
To examine whether RGS4 also acts as a negative regulator of M3R function in mouse islets (β-cells), we carried out a series of insulin release studies using isolated pancreatic islets from RGS4-deficient mice (RGS4 KO mice) and their WT littermates. In the presence of a low concentration of glucose (3.3 mM), OXO-M (0.5 μM) had no significant effect on basal insulin release in islets from either WT or RGS4 KO mice (RGS4−/− mice; Fig. 4A). Strikingly, in the presence of a high concentration of glucose (16.7 mM), OXO-M (0.1 and 0.5 μM) treatment led to a significantly greater enhancement of GSIS in RGS4-deficient than in WT islets (Fig. 4A). In addition, insulin secretion in the absence of OXO-M was also significantly increased (by approximately twofold) in the presence of 16.7 mM glucose. The total insulin content of islets prepared from RGS4 KO mice was not significantly different from that of WT control islets (25.2 ± 2.5 vs. 18.9 ± 2.0 ng/islet, respectively; 24 independent batches of islets from eight mice per strain were tested).
Fig. 4.
Selective augmentation of OXO-M-induced insulin release in pancreatic islets prepared from RGS4-deficient mice. (A) Concentration-dependent enhancement of OXO-M-induced insulin release in islets prepared from RGS4 KO mice (RGS4−/− mice). Isolated pancreatic islets prepared from WT and RGS4 KO mice were incubated for 1 h at 37 °C in Krebs solution containing the indicated glucose concentrations, either in the absence or the presence of OXO-M (0.1 and 0.5 μM). (B) Selective augmentation of insulin secretion from RGS4-deficient pancreatic islets by OXO-M. Isolated pancreatic islets prepared from WT and RGS4−/− mice were incubated for 1 h at 37 °C in Krebs solution containing 16.7 mM glucose. In addition, the incubation medium contained one of the following four ligands known to promote insulin secretion via activation of specific β-cell GPCRs: OXO-M (0.5 μM), GLP-1 (10 nM), ADP (100 μM), or AVP (100 nM). The amount of insulin secreted into the medium during the 1-h incubation period was normalized to the total insulin content of each well (islets plus medium). Data are expressed as means ± SEM of three independent experiments, each carried out in triplicate. *P < 0.05, ***P < 0.001, as compared to the corresponding WT value.
To examine whether RGS4 also regulated insulin release caused by activation of other GPCRs expressed by pancreatic β-cells, we incubated pancreatic islets from WT and RGS4 KO mice with GLP-1, ADP, or AVP (in presence of 16.7 mM glucose), using ligand concentrations known to significantly enhance GSIS in mouse pancreatic islets. In contrast to the results obtained with OXO-M-treated islets, no significant changes in GSIS were observed after stimulation of RGS4-deficient islets with any of these three ligands (Fig. 4B).
Muscarinic Agonist-Induced Insulin Release in Vivo Is Increased in Mice Lacking RGS4 in Pancreatic β-Cells Only.
RGS4 is not only expressed by pancreatic β-cells but is found in many tissues and cell types (18). To generate mice that lack RGS4 in pancreatic β-cells only (β-RGS4-KO mice), we crossed mutant mice carrying a “floxed” version of the RGS4 gene (19) with transgenic mice expressing Cre recombinase under the control of the rat insulin promoter II (RIP-Cre mice). Specifically, we mated RGS4 fl/+ mice with RGS4 fl/+ mice that were hemizygous for the RIP-Cre transgene. This mating strategy produced β-RGS4-KO mice (RGS4 fl/fl mice carrying the RIP-Cre transgene) and RGS4 fl/fl littermates which served as control animals. Real-time qRT-PCR studies showed that RGS4 mRNA levels were selectively reduced (by approximately 60%) in islets prepared from β-RGS4-KO mice (Fig. S7). The residual RGS4 expression observed with islets from β-RGS4-KO mice may be due to the presence of RGS4 in non-β islet cells and/or incomplete excision of the RGS4 gene in β-cells. β-RGS4-KO mice showed normal body weight (Fig. S8 A and C), were fertile and displayed no obvious developmental, behavioral, or morphological deficits.
Previous studies demonstrated that treatment of WT mice with bethanechol, a nonsubtype-selective muscarinic agonist, leads to a significant increase in insulin release in vivo, associated with reduced blood glucose levels (20). To examine whether these responses were due to activation of β-cell M3Rs, we injected freely fed WT mice and mice lacking M3Rs in pancreatic β-cells only (β-M3-KO mice) (ref. 10) with bethanechol (2 μg/g, s.c.). Whereas bethanechol-treated WT mice showed significantly increased plasma insulin and reduced blood glucose levels, these effects were abolished in β-M3-KO mice (Fig. S9). This observation indicates that systemic administration of the muscarinic agonist, bethanechol, triggers insulin release predominantly via activation of β-cell M3Rs.
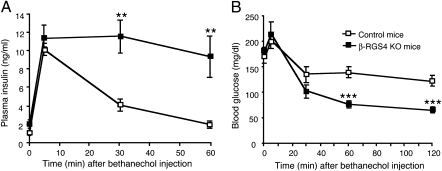
On the basis of these findings, we injected β-RGS4-KO mice and control mice with a single dose of bethanechol (2 μg/g, s.c.) and then measured the resulting changes in plasma insulin and blood glucose levels. Basal glucose levels were not significantly different between β-RGS4-KO and control mice (Fig. S8 B and D). However, there was a clear trend toward increased plasma insulin levels in RGS4-KO mice (control, 1.06 ± 0.25 ng/mL; β-RGS4-KO, 2.06 ± 0.42 ng/mL; n = 6 per group (males); P = 0.069). Interestingly, bethanechol-induced insulin release was greatly augmented in β-RGS4-KO mice, as compared to control littermates (Fig. 5A). As a result, bethanechol-mediated reductions in blood glucose levels were also more pronounced in β-RGS4-KO mice (Fig. 5B).
Fig. 5.
Enhanced insulin secretion and reduced blood glucose levels following bethanechol treatment of β-RGS4-KO mice in vivo. (A and B) β-RGS4-KO mice and control littermates received a single dose of bethanechol (2 μg/g, s.c.). Plasma insulin (A) and blood glucose (B) levels were measured at the indicated time points. Values are given as means ± SEM (n = 6 per group; freely fed 9-month-old males). **P < 0.01, ***P < 0.001, as compared to the corresponding values obtained with control mice.
Discussion
In the present study, we initially demonstrated that RGS4 is enriched in MIN6 insulinoma cells and mouse pancreatic islets. RGS4 is a member of the B/R4 subfamily of RGS proteins, acting as a GAP for Gq- and Gi-type G protein α-subunits (13, 14).
To explore the potential role of RGS4 in regulating β-cell function, we studied the effect of RGS4 deficiency on M3R-mediated augmentation of intracellular signaling and insulin release. First, we demonstrated that siRNA-mediated knockdown of RGS4 in MIN6 cells enhanced M3R-mediated increases in [Ca2+]i levels and GSIS. Second, M3R-mediated augmentation of insulin release was also greatly increased in pancreatic islets prepared from RGS4-deficient mice. Third, systemic administration of a muscarinic agonist, bethanechol, caused increased insulin release and more prominent blood glucose-lowering effects in mice selectively lacking RGS4 in pancreatic β-cells, as compared with control littermates. Taken together, these findings strongly support the concept that RGS4 acts as a negative regulator of M3R signaling in pancreatic β-cells.
For control purposes, we also tested other GPCR ligands known to promote GSIS by acting on specific β-cell GPCRs. Specifically, we examined whether GLP-1-, ADP-, and AVP-mediated increases in GSIS were affected by RGS4 deficiency in MIN6 cells and pancreatic islets. Whereas the GLP-1 receptor exerts its cellular actions via coupling to Gs, ADP, and AVP act on β-cell P2Y (e.g., P2Y1) and vasopressin (V1b) receptor subtypes, respectively, which are selectively coupled to G proteins of the Gq family, similar to the M3R (4). Interestingly, RGS4 deficiency had little or no effect on GLP-1-, ADP-, and AVP-mediated increases in GSIS in MIN6 cells or pancreatic islets, indicating that RGS4 selectively interferes with M3R function in insulin-containing cells.
Previous studies using other experimental systems also demonstrated that RGS proteins can exert selective inhibitory actions on specific receptor/G protein combinations (13, 14, 18, 21, 22). For example, Xu et al. (23) studied the potency of RGS4 in modulating calcium signaling mediated by activation of different Gq-coupled receptors expressed by rodent pancreatic acinar cells. The authors found that RGS4 was considerably more effective in inhibiting muscarinic receptor-mediated calcium signaling, as compared to the corresponding cholecystokinin and bombesin receptor-mediated responses (23). Several studies suggest that RGS proteins can directly interact with specific GPCRs (13, 14, 18, 21, 22), providing a molecular basis for the observed receptor selectivity of certain RGS proteins. In agreement with this concept, we were able to immunoprecipitate M3Rs in a complex with RGS4 in cotransfected mammalian cells. However, questions that remain to be addressed in future studies are whether there are other proteins that are part of the M3R/RGS4 signaling complex, whether the composition of this complex changes upon M3R activation, and whether RGS4 is recruited to the plasma membrane in an M3R-dependent manner.
In fact, recent studies predict the existence of GPCR/RGS signaling complexes containing additional signaling or scaffolding proteins, including spinophilin, 14-3-3 proteins, or Ca2+/calmodulin (18, 21). Cells may therefore use a complex web of protein–protein interactions to regulate RGS protein activity and receptor/RGS selectivity. Thus, the precise molecular mechanisms underlying the ability of RGS4 to selectively interfere with M3R signaling in pancreatic β-cells remain to be elucidated.
Previous studies have shown that RGS4 is enriched and widely expressed in the brain (24–26), and various reports have linked changes in RGS4 expression or activity to several major CNS disorders, including Parkinson's disease (27), schizophrenia (28), and drug addiction (29). More recent studies demonstrated that RGS4 also modulates various peripheral functions including heart rate (30), renal blood flow (31), secretion of catecholamines from adrenal glands (32), and breast cancer migration (33).
Interestingly, a recent study showed that RGS7 can inhibit Ca2+ mobilization mediated by M3R stimulation but not by activation of other Gq-coupled receptors (34). The authors demonstrated that this inhibition was mediated by a complex between RGS7 and Gβ5 and involved binding of the DEP domain of RGS7 to the third intracellular loop of the M3R (34). Given the selectivity of this interaction, it should be of considerable interest to examine whether the RGS7/Gβ5 complex also regulates signaling through β-cell M3Rs.
In summary, we demonstrated that RGS4 plays a critical role in modulating β-cell function. Specifically, we showed that RGS4 acts as a selective inhibitor of M3R signaling in insulinoma cells and pancreatic islets and that RGS4 deficiency in β-cells leads to enhanced M3R-mediated insulin release in vivo. Given the large number of GPCRs expressed by pancreatic β-cells (3, 4), it is likely that other GPCR/RGS combinations will be identified that affect various aspects of β-cell function including insulin release and β-cell replication and survival. It has been proposed that RGS proteins represent potential targets for drug development (35). Our findings may lead to the design of drugs aimed at improving β-cell function for therapeutic purposes.
Materials and Methods
Drugs and Expression Plasmids Used.
OXO-M, carbamyl-β-methylcholine chloride (bethanechol), ADP, and AVP were obtained from Sigma-Aldrich, and glucagon-like-peptide 1 (GLP-1, 7–36) amide was from Bachem. [3H]-N-Methylscopolamine ([3H]NMS: 79–83 Ci/mmol) was purchased from PerkinElmer Life Sciences. Mammalian expression plasmids in the pcDNA3.1(+) vector coding for human RGS4 and the human M3R containing a string of three N-terminal HA-epitope tags were obtained from the UMR cDNA Resource Center.
Generation of Whole Body and β-Cell–Specific RGS4 KO Mice.
The generation of whole body RGS4 KO mice and floxed RGS4 mutant mice (RGS4 fl/fl mice) has been described very recently (19). These mice were backcrossed for more than 10 generations onto the C57BL/6J background (The Jackson Laboratory). The β-RGS4-KO mice were generated by crossing RGS4 fl/fl mice with transgenic mice expressing Cre recombinase under the control of the β-cell-specific rat insulin promoter II (RIP-Cre mice; genetic background: C57BL/6J; supplier: The Jackson Laboratories) (36). The resulting RGS4 fl/+ Cre and RGS4 fl/+ mice were intermated to generate β-RGS4-KO mice (RGS4 fl/fl Cre mice) and the corresponding control littermates (RGS4 fl/fl mice). RGS4 mutant mice were genotyped via PCR analysis of mouse tail DNA (for details, see SI Materials and Methods).
MIN6 Cell Culture and Electroporation with siRNAs.
MIN6 cells (a kind gift from Dr. Abner Notkins, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD) were cultured as described previously (15). For gene silencing studies, approximately 1 × 106 MIN6 cells were electroporated with 60 nmols of siRNA according to the manufacturer's instructions (Amaxa) and tested 48 h later. Duplex siRNAs for the mouse M3R, mouse GPR40, mouse RGS4, and negative control siRNAs were obtained from Ambion.
Calcium Assay (FLIPR).
MIN6 cells were electroporated with different siRNAs, and OXO-M-dependent increases in intracellular calcium levels were determined by the use of FLIPR technology (Molecular Devices) about 48 h later. Assays were carried out in duplicate in 96-well plates, as described previously (37). Emax and EC50 values were obtained from OXO-M concentration-response curves analyzed by using GraphPad Prism 4.0 software (GraphPad Software).
Insulin Secretion Studied in MIN6 Cells.
Electroporated MIN6 cells grown in 96-well plates were incubated with different concentrations of OXO-M, GLP-1, ADP, or AVP at 37 °C for 60 min in 16.7 mM glucose KRBH (Krebs–Ringer bicarbonate/hepes) buffer. The amount of insulin released into the medium was measured by using an ELISA kit (Crystal Chem Inc.). Emax and EC50 values were obtained as described in the previous paragraph.
Western Blot Analysis.
After electroporation with different siRNAs, MIN6 cell lysates were centrifuged at 4 °C at 45,000 rpm in a SWT55i rotor for 30 min. The supernatants were electrophoresed on 12% NuPAGE Bis-Tris gels, transferred to nitrocellulose membranes, and inmunoblotted using a polyclonal anti-RGS4 primary antibody (1:10,000 dilution) (38) and an anti-rabbit HRP-labeled secondary antibody(1:10,000 dilution; Amersham Biosciences). The secondary antibody was visualized via chemiluminescence. The blots were stripped and reprobed using an anti-β-actin polyclonal antibody (1:1,000 dilution; Cell Signaling).
Radioligand Binding Studies.
Membrane homogenates (approximately 250 μg/tube) prepared from MIN6 cells were incubated with a saturating concentration (5 nM) of [3H]-NMS, a nonsubtype-selective muscarinic antagonist. Binding assays were carried out and analyzed as described (37).
Real-Time qRT-PCR Analysis of RGS Protein Expression.
Total RNA was prepared from MIN6 cells as well as mouse pancreatic islets and other mouse tissues. Subsequently, RGS gene expression levels were determined via real-time qRT-PCR (for details, see SI Materials and Methods).
Transient Expression of RGS4 and the M3R in COS-7 Cells and Coimmunoprecipitation Studies.
COS-7 cells were grown as monolayers as described (37). Cells were transfected with plasmid DNA in 10-cm dishes using the Lipofectamine plus kit (Invitrogen). The interaction of human RGS4 with an HA-epitope tagged version of the human M3R was studied in cotransfected COS-7 cells using a coimmunoprecipitation strategy (for details, see SI Materials and Methods).
Static Islet Insulin Secretion Assay.
Pancreatic islets were isolated from whole body RGS4 KO mice (RGS4−/− mice) and WT littermates. Static islet insulin secretion assays were carried out as described in detail previously (10). Insulin concentrations were determined by using an ELISA kit (Crystal Chem Inc.).
Bethanechol Injection Experiments.
A single dose of bethanechol chloride (2 μg/g; s.c.) was administered to freely fed β-RGS4-KO mice and their corresponding control littermates (floxed RGS4 mice). Blood was collected from the tail vein at different time points (0, 5, 30, 60, and 120 min) for the measurement of blood glucose and plasma insulin levels. Blood glucose levels were determined using an automated blood glucose reader (Glucometer Elite Sensor; Bayer). Plasma insulin concentrations were determined via ELISA (Crystal Chem Inc.).
Statistics.
Data are expressed as means ± SEM for the indicated number of experiments. For comparisons between two groups, the unpaired Student's t test (two-tailed) was used. For multiple comparisons, the one-way analysis of variance (ANOVA) was used. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Department of Health and Human Services. We thank Ms. Yinghong Cui (National Institute of Diabetes and Digestive and Kidney Diseases) for excellent technical assistance and Dr. Susanne M. Mumby (University of Texas Southwestern Medical Center) for providing the anti-RGS4 antibody. I.R.A. was the recipient of a Basque Government Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1003655107/DCSupplemental.
References
- 1.Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003;46:3–19. doi: 10.1007/s00125-002-1009-0. [DOI] [PubMed] [Google Scholar]
- 2.Moller DE. New drug targets for type 2 diabetes and the metabolic syndrome. Nature. 2001;414:821–827. doi: 10.1038/414821a. [DOI] [PubMed] [Google Scholar]
- 3.Regard JB, et al. Probing cell type-specific functions of Gi in vivo identifies GPCR regulators of insulin secretion. J Clin Invest. 2007;117:4034–4043. doi: 10.1172/JCI32994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahrén B. Islet G protein-coupled receptors as potential targets for treatment of type 2 diabetes. Nat Rev Drug Discov. 2009;8:369–385. doi: 10.1038/nrd2782. [DOI] [PubMed] [Google Scholar]
- 5.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 6.Gilon P, Henquin JC. Mechanisms and physiological significance of the cholinergic control of pancreatic β-cell function. Endocr Rev. 2001;22:565–604. doi: 10.1210/edrv.22.5.0440. [DOI] [PubMed] [Google Scholar]
- 7.Zawalich WS, et al. Effects of muscarinic receptor type 3 knockout on mouse islet secretory responses. Biochem Biophys Res Commun. 2004;315:872–876. doi: 10.1016/j.bbrc.2004.01.139. [DOI] [PubMed] [Google Scholar]
- 8.Duttaroy A, et al. Muscarinic stimulation of pancreatic insulin and glucagon release is abolished in m3 muscarinic acetylcholine receptor-deficient mice. Diabetes. 2004;53:1714–1720. doi: 10.2337/diabetes.53.7.1714. [DOI] [PubMed] [Google Scholar]
- 9.Wess J. Molecular biology of muscarinic acetylcholine receptors. Crit Rev Neurobiol. 1996;10:69–99. doi: 10.1615/critrevneurobiol.v10.i1.40. [DOI] [PubMed] [Google Scholar]
- 10.Gautam D, et al. A critical role for beta cell M3 muscarinic acetylcholine receptors in regulating insulin release and blood glucose homeostasis in vivo. Cell Metab. 2006;3:449–461. doi: 10.1016/j.cmet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Tobin AB, Butcher AJ, Kong KC. Location, location, location…site-specific GPCR phosphorylation offers a mechanism for cell-type-specific signalling. Trends Pharmacol Sci. 2008;29:413–420. doi: 10.1016/j.tips.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo J, Busillo JM, Benovic JL. M3 muscarinic acetylcholine receptor-mediated signaling is regulated by distinct mechanisms. Mol Pharmacol. 2008;74:338–347. doi: 10.1124/mol.107.044750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 14.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 15.Ishihara H, et al. Pancreatic beta cell line MIN6 exhibits characteristics of glucose metabolism and glucose-stimulated insulin secretion similar to those of normal islets. Diabetologia. 1993;36:1139–1145. doi: 10.1007/BF00401058. [DOI] [PubMed] [Google Scholar]
- 16.Itoh Y, et al. Free fatty acids regulate insulin secretion from pancreatic β cells through GPR40. Nature. 2003;422:173–176. doi: 10.1038/nature01478. [DOI] [PubMed] [Google Scholar]
- 17.Kurrasch DM, Huang J, Wilkie TM, Repa JJ. Quantitative real-time polymerase chain reaction measurement of regulators of G-protein signaling mRNA levels in mouse tissues. Methods Enzymol. 2004;389:3–15. doi: 10.1016/S0076-6879(04)89001-3. [DOI] [PubMed] [Google Scholar]
- 18.Bansal G, Druey KM, Xie Z. R4 RGS proteins: Regulation of G-protein signaling and beyond. Pharmacol Ther. 2007;116:473–495. doi: 10.1016/j.pharmthera.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winden KD, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukudo S, et al. Muscarinic stimulation and antagonism and glucoregulation in nondiabetic and obese hyperglycemic mice. Diabetes. 1989;38:1433–1438. doi: 10.2337/diab.38.11.1433. [DOI] [PubMed] [Google Scholar]
- 21.Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. RGS proteins have a signalling complex: Interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell Signal. 2006;18:579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Neitzel KL, Hepler JR. Cellular mechanisms that determine selective RGS protein regulation of G protein-coupled receptor signaling. Semin Cell Dev Biol. 2006;17:383–389. doi: 10.1016/j.semcdb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Xu X, et al. RGS proteins determine signaling specificity of Gq-coupled receptors. J Biol Chem. 1999;274:3549–3556. doi: 10.1074/jbc.274.6.3549. [DOI] [PubMed] [Google Scholar]
- 24.Erdely HA, et al. Regional expression of RGS4 mRNA in human brain. Eur J Neurosci. 2004;19:3125–3128. doi: 10.1111/j.0953-816X.2004.03364.x. [DOI] [PubMed] [Google Scholar]
- 25.Ebert PJ, Campbell DB, Levitt P. Bacterial artificial chromosome transgenic analysis of dynamic expression patterns of regulator of G-protein signaling 4 during development. I. Cerebral cortex. Neuroscience. 2006;142:1145–1161. doi: 10.1016/j.neuroscience.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert PJ, Campbell DB, Levitt P. Bacterial artificial chromosome transgenic analysis of dynamic expression patterns of regulator of G-protein signaling 4 during development. II. Subcortical regions. Neuroscience. 2006;142:1163–1181. doi: 10.1016/j.neuroscience.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Ding J, et al. RGS4-dependent attenuation of M4 autoreceptor function in striatal cholinergic interneurons following dopamine depletion. Nat Neurosci. 2006;9:832–842. doi: 10.1038/nn1700. [DOI] [PubMed] [Google Scholar]
- 28.Levitt P, Ebert P, Mirnics K, Nimgaonkar VL, Lewis DA. Making the case for a candidate vulnerability gene in schizophrenia: Convergent evidence for regulator of G-protein signaling 4 (RGS4) Biol Psychiatry. 2006;60:534–537. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 29.Hooks SB, Martemyanov K, Zachariou V. A role of RGS proteins in drug addiction. Biochem Pharmacol. 2008;75:76–84. doi: 10.1016/j.bcp.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 30.Cifelli C, et al. RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node. Circ Res. 2008;103:527–535. doi: 10.1161/CIRCRESAHA.108.180984. [DOI] [PubMed] [Google Scholar]
- 31.Siedlecki A, et al. RGS4 controls renal blood flow and inhibits cyclosporine-mediated nephrotoxicity. Am J Transplant. 2010;10:231–241. doi: 10.1111/j.1600-6143.2009.02930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iankova I, et al. Regulator of G protein signaling-4 controls fatty acid and glucose homeostasis. Endocrinology. 2008;149:5706–5712. doi: 10.1210/en.2008-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Y, et al. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 2009;69:5743–5751. doi: 10.1158/0008-5472.CAN-08-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandiford SL, Slepak VZ. The Gbeta5-RGS7 complex selectively inhibits muscarinic M3 receptor signaling via the interaction between the third intracellular loop of the receptor and the DEP domain of RGS7. Biochemistry. 2009;48:2282–2289. doi: 10.1021/bi801989c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neubig RR, Siderovski DP. Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov. 2002;1:187–197. doi: 10.1038/nrd747. [DOI] [PubMed] [Google Scholar]
- 36.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 37.Scarselli M, Li B, Kim SK, Wess J. Multiple residues in the second extracellular loop are critical for M3 muscarinic acetylcholine receptor activation. J Biol Chem. 2007;282:7385–7396. doi: 10.1074/jbc.M610394200. [DOI] [PubMed] [Google Scholar]
- 38.Krumins AM, et al. Differentially regulated expression of endogenous RGS4 and RGS7. J Biol Chem. 2004;279:2593–2599. doi: 10.1074/jbc.M311600200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.